ABSTRACT
The use of antibiotics to target bacteria is a well-validated approach for controlling infections in animals and humans. Peptidoglycan biosynthesis is a crucial process in bacteria, and the conserved peptidoglycan synthase MraY is an attractive target for drug design. However, due to the lack of detailed MraY structural information, antibiotics targeting MraY have not yet been developed. In the present study, 2 hydrophilic regions of MraY from Escherichia coli were expressed as a fusion protein and used to raise a monoclonal antibody in mice. We confirmed that the MraY amino acid sequence PESHFSKRGTPT forms the core epitope recognized by the monoclonal antibody M-H11. Furthermore, our results show that M-H11 effectively controls Escherichia coli BL21 (DE3) plysS infection, both in vitro and in vivo. Our results may be of great value in the search for novel approaches used to control bacterial infections.
Introduction
Escherichia coli (E. coli) is one of the most important human pathogens. E. coli infections, especially extraintestinal infections, represent a major public health problem.Citation1 Antimicrobial resistance in E. coli is increasingly common and causes significant economic burdens on families and society.Citation2 Monoclonal antibody (mAb) based strategies for treating infections is an approach with considerable potential. MAbs have been demonstrated to enhance bacterial clearance, prevent colonization and invasion, and prevent damage caused by cytotoxic factors.Citation3-5
Bacterial cell walls have a critical role in maintaining cell shape and protecting bacteria from environmental challenges. The cell wall is composed of the polymer peptidoglycan, and most bacterial pathogens require continuous cell wall synthesis for survival.Citation6,7 Peptidoglycan biosynthesis is a complex process that takes place in the cytoplasm. MraY, or phosphor-MurNAc-pentapeptide translocase, is an integral membrane protein involved in this process, and it catalyzes the transfer of the peptidoglycan precursor phosphor-MurNAc-pentapeptide to the lipid carrier undecaprenyl phosphate. In 2013, the crystal structure of MraY from Aquifex aeolicus was determined at 3.3 Å resolution.Citation8,9 The biosynthesis of the peptidoglycan layer of bacterial cell walls is a well-proven target for antibiotic development. MraY catalyzes an essential step of peptidoglycan biosynthesis. MraY has long been considered a very promising target for the development of antibiotics, as many naturally occurring nucleoside inhibitors with antibacterial activity target this enzyme. But so far there have been no antibiotics targeting MraY developed for clinical uses. MraY-targeted natural products have gained more attention because of their in vivo efficacy against pathogenic bacteria including vancomycin-resistant Enterococcus (VRE), Mycobacterium tuberculosis, and methicillin-resistant Staphylococcus aureus (MRSA).Citation8,10 In this study, we characterized a novel anti-MraY mAb, M-H11, which showed efficacy in controlling bacterial infections both in vitro and in vivo.
Results
Generation of mAb
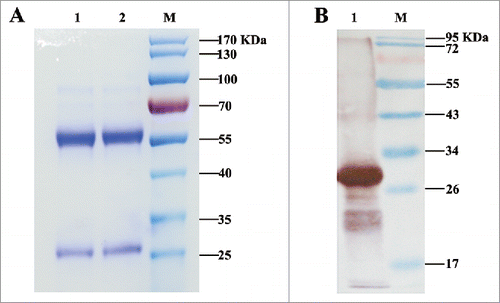
One stable hybridoma cell line secreting M-H11 mAbs was obtained. SDS-PAGE of purified M-H11 showed to be of high purity (). M-H11 was characterized using the SBA Clonotyping System-HRP kit, which showed that the M-H11 isotype was IgG2b and that the light chain was the κ chain. M-H11 could bind the MraY-AB recombinant protein, producing a band at 32 KDa (). The relative titer of the mAb was measured by indirect ELISA using serial dilutions and was found to be 1:128,000.
Figure 1. Analysis of purified IgG by SDS-PAGE and immunoblot analysis of mAb secreted by hybridomas. (A) Analysis of purified antibody by SDS-PAGE. Lanes 1, 2: purified mAb M-H11; Lane M: molecular mass markers (B) Immunoblot analysis of mAb secreted by hybridomas. Lane 1: purified fusion protein reacted with mAb M-H11; Lane M: molecular mass markers.
Phage panning yield and phage clone detection by indirect ELISA
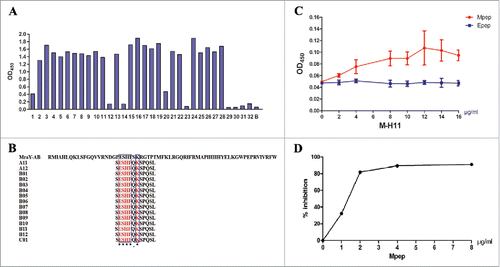
The phage percent yield for M-H11 increased during 3 rounds of panning, rising from 10−6 to 10−2, showing successful enrichment of positive phage. Subsequently, random phages were amplified, and their reactivities with the mAb were assessed by indirect ELISA analysis. The results showed that 23 single phage displayed peptides bound to mAb M-H11 (OD450 > 1.2) among which 15 clones (OD450 > 1.5) were selected and sequenced. Generally, greater binding force means higher affinity. We chose 15 phage clones out of the 23 clones, because the OD450 value of them were greater than the others. Besides, the sample capacity is sufficient and it can reduce the cost of the experiment. These interactions were specific since no interaction was detected in the wells coating with bovine serum albumin (BSA) (OD450 < 0.08) ().
Figure 2. Analysis and characterization of Mpep. (A) Detection of the binding activity of the phage clones by ELISA. 1–32: phage clones from the third round of biopanning; B: BSA control. (B) The sequences of 12 peptides displayed by 15 phage clones. Multiple sequence alignment was performed using Clustal Omega. The symbol * means the site has the same amino acid, the symbol. means the amino acids belong to one group. Glutarnine (Q) and Serine (S) are both of uncharged polar side chains group. The blue box represents the same amino acid. (C) Affinity reaction of Mpep and M-H11 (Epep as control). Data were expressed as the mean ± standard deviation (S.D). The experiment was repeated 3 times. (D) Competitive inhibition of Mpep against MraY-AB and M-H11.
Sequencing of positive phage clones and homology comparison
The DNA sequences of 15 positive clones were determined, and a sequence alignment showed that 15 phage clones shared the same sequence (SESHFQKSPQSL). A comparison of these sequences with the MraY-AB protein sequence revealed that the amino acid sequence PESHFSKRGTPT (designated as Mpep) was conserved in MraY-AB (). This result suggested that Mpep is the core sequence of MraY-AB that forms the epitope recognized by M-H11.
Peptide synthesis and ELISA analysis
To confirm that Mpep could be recognized by M-H11, the interaction between the 2 proteins was assessed using an indirect ELISA. The ELISA plate was coated with 10 µg/ml Mpep and incubated with increasing concentrations of M-H11 (0, 2, 4, 6, 8, 10, 12, 14, and 16 µg/ml). The results showed that as the mAb concentration increased, the OD values became larger, and the affinity was very strong (). These interactions were concluded to be specific because no interaction was detected using Epep (VRLKPLNCSR, a unrelated synthetic peptide from E protein of bacteriophage phiX174) (OD450 < 0.08).
Competitive inhibition experiment using increasing peptide concentrations
The result of the competitive inhibition experiment using increasing concentrations of Mpep is presented in . With the MraY-AB protein coated at 5 µg/ml and a M-H11 concentration of 0.2 µg/ml, the binding of M-H11 to the MraY-AB protein was significantly inhibited by Mpep. The strength of the inhibition was dependent on the concentration; the strength of the inhibition increased as the concentration of the synthetic peptide increased. We conclude that the synthesized peptide was the epitope of the MraY-AB protein, which was specifically recognized by the mAb M-H11.
M-H11 could inhibit E. coli BL21 (DE3) plysS in vitro
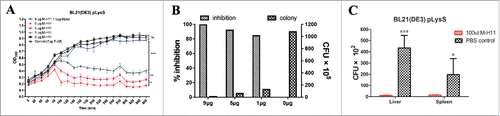
After obtaining M-H11 mAb, we were interested in determining whether M-H11 could inhibit the growth of E. coli in vitro; therefore an in vitro bacteriostatic test was performed. E-A5 was a specific mAb against E protein of bacteriophage phiX174. We used this irrelevant mAb as a control to ensure that there is no non-specific antibody effect. In addition, Mpep as a competitive inhibitor to confirm the specificity of inhibition of M-H11 mAb activity. The bacterial growth was inhibited after 100 min when incubated with M-H11. However, the bacteria incubated with PBS or the E-A5 mAb grew well (). Notably, the bacteria incubated with Mpep also grew well, indicating the specificity of the activity of M-H11. These results suggest that mAb M-H11 inhibits the growth of E. coli BL21 (DE3) plysS. The colony forming units (CFUs) and inhibitory rates were determined when the experiments went on for 6 h. Bacterial growth inhibition rates were 99.3%, 91.9% and 84.3% when incubated with 9 µg, 5 µg and 1 µg of M-H11, respectively (), suggesting that M-H11 inhibition occurs in a concentration dependent manner.
Figure 3. M-H11 has the ability to inhibit the E. coli BL21 (DE3) plysS growth in vitro and in vivo. (A) Line chart showing M-H11 inhibition of E. coli BL21 (DE3) plysS growth in vitro. (B) Histogram showing the inhibition rate of M-H11 on E. coli BL21 (DE3) plysS growth in vitro. (C) Histogram showing the inhibition ability of M-H11 on E. coli BL21 (DE3) plysS growth in vivo. Six-week-old female BALB/c mice were randomly divided into 2 groups (5 in each group) in vivo experiment.
M-H11 could inhibit E. coli BL21 (DE3) plysS in vivo
The in vitro inhibition of E. coli BL21 (DE3) plysS growth by M-H11 led us to determine whether mAb M-H11 could inhibit the growth of E. coli BL21 (DE3) plysS in vivo. We first determined the infection dynamics of E. coli BL21 (DE3) plysS alone in mice without mAb M-H11 treatment. The bacterial load in the liver and spleen of mice were higher than those in lung and kidney, and the amount of bacteria was significant higher after 3–5 h post-infection (). We therefore dissected mice at 3.5 h post-infection in subsequent experiments. The bacterial loads present in both in the liver and spleen of mice in the experimental group were significantly decreased compared with control group (). These results demonstrate that mAb M-H11 is able to inhibit the growth of E. coli BL21 (DE3) plysS in vivo.
Table 1. The colony count of organs in mice infected with E. coli BL21 (DE3) plysS alone.
Discussion
Antimicrobial resistance worldwide is increasingly prevalent and threatens the health of both humans and animals. The continued emergence of antibiotic-resistant bacterial pathogens, combined with limited available therapies, underscores the need to identify novel antibacterial targets, with bacterial peptidoglycan biosynthesis enzymes remaining excellent targets for antibacterial action.Citation12 The structural determination of A. aeolicus MraY has revealed new possibilities for structure-based drug design against MraY and related enzymes.Citation9 Monoclonal antibodies are increasingly being developed as therapeutics to complement drugs and vaccines or to fill gaps where no options exist. These therapeutic antibodies (ThAb) may be especially important for infections in which increasing antibiotic resistance, toxin-mediated pathogenesis, or emerging pathogens are of concern.Citation13 Characterization of a panel of novel anti-PcrV mAbs was recently generated and characterized. The researchers determined that some anti-PcrV mAbs exhibited potent inhibition of P. aeruginosa in vitro, while others provided strong prophylactic protection in several murine models of infection and therapeutic post-infection models.Citation14
E. coli is commonly found in the gut of humans and warm-blooded animals. While most E. coli strains are harmless, some, such as enterohemorrhagic E. coli, can cause severe foodborne disease.Citation15 In this work, a stable hybridoma cell line expressing the mAb M-H11, which targets the MraY-AB protein, was obtained. Phage display is a laboratory technique allows expression of exogenous (poly) peptides on the surface of phage particles. The Ph.D.-12 library of phage particles expressing a wide diversity of peptides, is commonly used to select those that bind the desired target. Phage display technology used in epitope analysis suggested that PESHFSKRGTPT was the core amino acid sequence epitope of MraY-AB recognized by M-H11. The reason of only one single peptide was identified during the library screen could be explained in 2 aspects. On the one hand, the procedure of biopanning was optimized. The time of panning washing was increased to 10 min and the rounds of panning washing were increased to 15 times, respectively, to remove clones with insufficient binding force. In the previous study, we found that sometimes we could not get clear consensus binding sequence after 3 rounds of panning washing. Therefore, we have been optimized the procedure of biopanning in this experiment. On the other hand, OD450 values of the 15 sequenced phage clones were more than 1.5, higher than the others. The sample capacity is sufficient and it can reduce the cost of the experiment. Notably, our results demonstrate that the mAb M-H11 significantly inhibits the growth of E. coli BL21 (DE3) plysS both in vitro and vivo. The main reason of using E. coli strain BL21(DE3) pLysS for the animal challenge studies could be explained from the following aspects. We found that MraY was highly conserved in all E. coli strains by bioinformatics analysis. E. coli strain BL21 (DE3) pLysS is a derivative of the wild strain for protein expression. In vitro experiments, E. coli strain BL21 (DE3) pLysS was used and we found that mAb M-H11 had antibacterial activity. Therefore, we continued to use this strain in vivo experiments. We found that even this modified strain can be transported to the liver and spleen after challenge. The mice were morbid after challenge indicating that this strain was pathogenic. For this reason, we did not choose more virulent strains. We think there may be several possibilities that mAb can inhibit the growing bacteria. Firstly, we speculate that mAb may be exposed to MraY protein through a pathway that is not yet known at a particular point-in-time in the process of peptidoglycan biosynthesis. In vitro experiments have shown that antibacterial activity was dose-dependent, suggesting that there must be enough mAb to inhibit the growing bacteria. Of course, we do not rule out another possibility that the epitope recognized by the mAb M-H11 is also present in some outer membrane protein of E. coli. MAb has a neutralizing activity, and the antigen-antibody reaction results in inhibition of bacterial growth. However, the current data we got could not elaborate on the exact antimicrobial mechanism of mAb. Future investigations will be required to determine the exact nature of this interaction.
Obviously, this is a primary assessment of the potential therapeutic of the mAb in mice model. We consider the following aspects maybe the potential weaknesses on limitations of the mAb therapy. In our experiment, only BALB/c mice were used as animal models of infection, no other types of mice were used, such as C57BL/6 mice. Besides, mAb was used in single dosage. The anti-bacterial experiments were performed only in accordance with the dose according to the reference,Citation16 and no more therapeutic dose gradient was designed. All of these are needed to supplement in later experiments.
The mAb generated in this work could potentially be evaluated for efficacy as therapeutic antibodies in the future. We are planning to extend these studies using more appropriate strains of E. coli, such as O157 which is very important to public health security. In addition, considering the relative conservation of MraY between gram negative and gram positive bacteria,Citation9 we try to explore the antibacterial potential of the mAb against infection of Salmonella, Shigella, Mycobacterium tuberculosis, Streptococcus, Staphylococcus or other bacteria. For the further experiments, we chose to focus on the molecular mechanism of anti-bacterial activity and evaluation of the effect of mAb combined with antibiotics. Furthermore, the dose and route of injection should be optimized, to increase the likelihood of anti-bacterial agents development. We have reason to believe that this work contributes to a growing body of targeted mAb design research and opens new avenues of drug development.
Methods
Ethics in animal experimentation
The specific pathogen-free (SPF) BALB/c mice used in this study were purchased from Vital River, Beijing, China. All experiments with live animals were approved by the Review Board of Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences. All efforts were made to minimize suffering.
Preparation of mAb
DNA encoding 2 hydrophilic MraY fragments, designated A (Arg43-Met74 amino acid (aa)) and B (Phe311-Trp343 aa), were amplified (primers shown in ) from E. coli DH5α genomic DNA. The A and B fragments were fused together by overlapping PCR. The PCR product was then cloned into the BamHІ and SalІ sites of the vector pGEX-6p-1 (Novagen, Germany). MraY-AB was expressed in E. coli BL21 cells (Novagen, Germany). The insert was verified by sequencing. MraY-AB protein expression and purification, as well as hybridoma generation, were performed as described previously.Citation11 Each of the 5 BALB/c female mice (N = 5) approximately 4–6 weeks old was immunized intraperitoneally with 100 µg purified MraY-AB protein emulsified with an equal volume of Complete Freund's Adjuvant (CFA; Sigma, St. Louis, USA). Then 2 further injections were administered using incomplete adjuvant at 2-week intervals. Three days after the last injection, the mice were injected with 100 µg of purified MraY-AB protein without adjuvant. Immunoglobulin G (IgG) was purified from mouse ascitic fluid using protein G-agarose affinity chromatography (GE Healthcare, United States). The mAb isotypes were determined using an SBA Clonotyping System-HRP Kit (Southern Biotechnology Associates, Inc., Birmingham, AL, USA). Immunoblot analysis was used to assess the reactivity and specificity of the mAb.
Table 2. Primers used in this study.
Peptide library screening, biopanning and indirect ELISA
The Ph.D.-12 library (Ph.D.-12 Phage Display Peptide Library Kit, New England Biolabs Inc., USA) displays 12-mer peptides at the N-terminus of the PШ M13 phage protein, which was amplified from E. coli 2738. Three rounds of biopanning were performed according to the manufacturer's instructions, as described previously.Citation11 The yields from the 3 rounds of panning were compared with assess the phage-enrichment efficiency. Individual phage clones were selected and assayed for target binding by ELISA according manufacturer's instructions.
Sequence analysis of phage single-stranded DNA
The positive phages selected from the ELISA were cloned and sequenced with the −96 gШ sequencing primer (5′-CCCTCATAGTTAGCGTAACG-3′) according to the manufacturer's instructions. Peptide sequences were deduced from the phage displayed sequences, which were compared with the native MraY protein sequence (GenBank, CP014225) using Lasergene software. Peptide synthesis was conducted using GenScript.
ELISA analysis of synthetic peptides
The binding affinity between the synthetic peptide PESHFSKRGTPT (Mpep) and M-H11 was assessed using an indirect ELISA. The synthetic peptide was dissolved in sterile deionized water and diluted in carbonate buffer (pH 9.6) to 10 µg/ml. The ELISA plate was coated with the synthetic peptide solution and then incubated with increasing concentrations of M-H11 (0, 2, 4, 6, 8, 10, 12, 14, and 16 µg/ml). A unrelated synthetic peptide (VRLKPLNCSR) (Epep, from E protein of bacteriophage phiX174) was used as a negative control.
Competitive inhibition assay using increasing concentrations of the Mpep peptide
Briefly, 96-well plates were coated with the MraY-AB protein (5 µg/ml) and then incubated with 0.2 µg/ml of mAb plus increasing concentrations of Mpep (5, 10, 15, 25, 50, 75, and 100 µg/ml). Subsequent procedures were performed according to standard ELISA protocols.
Bacteriostatic test of mAb M-H11 anti-E. coli BL21 (DE3) plysS in vitro
E. coli BL21 (DE3) plysS (Novagen, Germany) cells were grown overnight in LB media (BD Biosciences) containing 34 µg/ml chloramphenicol (Amresco) at 37°C. A single clone was selected and grown at 37°C overnight, and then, the bacteria were inoculated into fresh LB medium with chloramphenicol until the OD600 reached 0.6. Bacteriostatic tests of the mAb M-H11 were performed in vitro in 200 µl reaction system containing 100 µl of E. coli BL21 (DE3) plysS and the purified mAb M-H11 at increasing concentrations (0, 1, 5 and 9 µg) in a 96-well cell culture plate. E-A5 was a specific mAb against E protein of bacteriophage phiX174. We used this irrelevant mAb as a control to ensure that there is no non-specific antibody effect. The mAb (E-A5) and PBS served as controls. Additionally, Mpep as a competitive inhibitor to confirm the specificity of the activity. The experiment was performed in triplicate at 37°C with shaking (220 rpm). Bacterial growth curves were generated by measuring the absorbances at 590 nm every 20 min, and this monitoring process lasted for 6 h. The reaction product was obtained after 6 h. The CFUs were identified by increasing dilution ratios (103, 104, 105, 106, 108) grown overnight in LB media containing 34 µg/ml chloramphenicol at 37°C. Inhibitory rates were calculated as follows: (1 - bacteria count of experiment group/control group) × 100%.
The bacteriostatic test of mAb M-H11 anti-E. coli BL21 (DE3) plysS in vivo
Firstly, 5 female BALB/c mice (6-week-old) in each group housed under specific pathogen-free conditions were used to determine the infectivity of E. coli BL21 (DE3) plysS in mice. Mice were injected with either 100 µl E. coli BL21 (DE3) plysS (OD600 reached 0.6) or 100 µl sterile PBS through the caudal vein. The challenge dose of 100 µl E. coli BL21 (DE3) plysS is about 1.0 × 108 CFU. The mice were dissected at 3 h, 5 h, 7 h and 9 h post-injection. Then, 1 ml of sterile PBS was added to 0.1 g of liver, lung, spleen or kidney tissue. After being ground on a sterile copper network, 100 µl of grinding fluid (a 103 dilution) was plated and grown overnight, and colony counting was conducted the following day. The comparison was conducted between the earlier time point and the later time point. Two-tailed Student's t test was used for the analysis of statistical significance (P value) in this study, and Prism software (version 5.0; GraphPad, San Diego, CA, USA) was used for these analyses. A P value of less than 0.05 was considered significant (* P < 0.05; ** P < 0.01; *** P < 0.001; ns, not significant). Subsequently, the bacteriostatic ability of mAb M-H11 was tested in vivo. Six-week-old female BALB/c mice were randomly divided into 2 groups (5 in each group) and maintained separately under specific pathogen-free conditions. Firstly, mice were injected with either 100 µl the purified mAb M-H11 or sterile PBS through the caudal vein. After 24 h, mice were injected with 100 µl E. coli BL21 (DE3) plysS (OD600 reached 0.6). The mice were dissected 3.5 h post-injection and bacterial loads in the liver and spleen tissue were determined as described above.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0501608), National Natural Science Foundation of China (31470893), Special Fund for Agro-scientific Research in the Public Interest (201403054).
References
- Bajaj P, Singh NS, Virdi JS. Escherichia coli beta-Lactamases: What really matters. Front Microbiol 2016; 7:417; PMID:27065978; https://doi.org/10.3389/fmicb.2016.00417
- Vila J, Saez-Lopez E, Johnson JR, Romling U, Dobrindt U, Canton R, et al. Escherichia coli: An old friend with new tidings. FEMS Microbiology Reviews 2016; 40:437-463
- Goure J, Pastor A, Faudry E, Chabert J, Dessen A, Attree I. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect Immun 2004; 72:4741-50; PMID:15271936; https://doi.org/10.1128/IAI.72.8.4741-4750.2004
- Horn MP, Zuercher AW, Imboden MA, Rudolf MP, Lazar H, Wu H, Hoiby N, Fas SC, Lang AB. Preclinical in vitro and in vivo characterization of the fully human monoclonal IgM antibody KBPA101 specific for Pseudomonas aeruginosa serotype IATS-O11. Antimicrob Agents Chemother 2010; 54:2338-44; PMID:20308370; https://doi.org/10.1128/AAC.01142-09
- Tkaczyk C, Hua L, Varkey R, Shi Y, Dettinger L, Woods R, Barnes A, MacGill RS, Wilson S, Chowdhury P, et al. Identification of anti-alpha toxin monoclonal antibodies that reduce the severity of Staphylococcus aureus dermonecrosis and exhibit a correlation between affinity and potency. Clin Vaccine Immunol 2012; 19:377-85; PMID:22237895; https://doi.org/10.1128/CVI.05589-11
- Bugg TD, Rodolis MT, Mihalyi A, Jamshidi S. Inhibition of phospho-MurNAc-pentapeptide translocase (MraY) by nucleoside natural product antibiotics, bacteriophage varphiX174 lysis protein E, and cationic antibacterial peptides. Bioorg Med Chem 2016; 24:6340-47; PMID:27021004; https://doi.org/10.1016/j.bmc.2016.03.018
- Shapiro AB, Jahic H, Gao N, Hajec L, Rivin O. A high-throughput, homogeneous, fluorescence resonance energy transfer-based assay for phospho-N-acetylmuramoyl-pentapeptide translocase (MraY). J Biomol Screen 2012; 17:662-72; PMID:22337656; https://doi.org/10.1177/1087057112436885
- Chung BC, Mashalidis EH, Tanino T, Kim M, Matsuda A, Hong J, Ichikawa S, Lee SY. Structural insights into inhibition of lipid I production in bacterial cell wall synthesis. Nature 2016; 533:557-60; PMID:27088606; https://doi.org/10.1038/nature17636
- Chung BC, Zhao J, Gillespie RA, Kwon DY, Guan Z, Hong J, Zhou P, Lee SY. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science 2013; 341:1012-6; PMID:23990562; https://doi.org/10.1126/science.1236501
- Koppermann S, Ducho C. Natural products at work: Structural insights into inhibition of the bacterial membrane protein MraY. Angewandte Chemie 2016; 55:11722-4; PMID:27511599; https://doi.org/10.1002/anie.201606396
- Wang H, Zhu T, Yu S, Liu H, Wang X, Chen L, Si W, Pang H, Liu S. A novel B cell epitope in cold-shock DEAD-box protein A from mycobacterium tuberculosis. Res Vet Sci 2013; 94:406-12; PMID:23261158; https://doi.org/10.1016/j.rvsc.2012.11.013
- Franz E, Veenman C, van Hoek AH, de Roda Husman A, Blaak H. Pathogenic escherichia coli producing extended-spectrum beta-lactamases isolated from surface water and wastewater. Sci Rep 2015; 5:14372; PMID:26399418; https://doi.org/10.1038/srep14372
- Irani V, Guy AJ, Andrew D, Beeson JG, Ramsland PA, Richards JS. Molecular properties of human IgG subclasses and their implications for designing therapeutic monoclonal antibodies against infectious diseases. Mol Immunol 2015; 67:171-82; PMID:25900877; https://doi.org/10.1016/j.molimm.2015.03.255
- Warrener P, Varkey R, Bonnell JC, DiGiandomenico A, Camara M, Cook K, Peng L, Zha J, Chowdury P, Sellman B, et al. A novel anti-pcrv antibody providing enhanced protection against pseudomonas aeruginosa in multiple animal infection models. Antimicrob Agents Chemother 2014; 58:4384-91; PMID:24841258; https://doi.org/10.1128/AAC.02643-14
- Poolman JT, Wacker M. Extraintestinal pathogenic escherichia coli, a common human pathogen: Challenges for vaccine development and progress in the field. J Infect Dis 2016; 213:6-13; PMID:26333944; https://doi.org/10.1093/infdis/jiv429
- Szijártó V, Guachalla LM, Visram ZC, Hartl K, Varga C, Mirkina I, Zmajkovic J, Badarau A, Zauner G, Pleban C, et al. Bactericidal monoclonal antibodies specific to the lipopolysaccharide O antigen from multidrug-resistant Escherichia coli clone ST131-O25b:H4 elicit protection in mice. Antimicrob Agents Chemother 2015; 59:3109-16; PMID:25779571; https://doi.org/10.1128/AAC.04494-14
