ABSTRACT
Sequence diversity and immunodominance are major obstacles in the design of an effective vaccine against HIV. HIV Env is a highly-glycosylated protein composed of ‘conserved’ and ‘variable’ regions. The latter contains immunodominant epitopes that are frequently targeted by the immune system resulting in the generation of immune escape variants. This work describes 12 regions in HIV Env that are highly conserved throughout the known HIV M Group sequences (Env CE), and are poorly immunogenic in macaques vaccinated with full-length Env expressing DNA vaccines. Two versions of plasmids encoding the 12 Env CE were generated, differing by 0–5 AA per CE to maximize the inclusion of commonly detected variants. In contrast to the full-length env DNA vaccine, vaccination of macaques with a combination of these 2 Env CE DNA induced robust, durable cellular immune responses with a significant fraction of CD8+ T cells with cytotoxic phenotype (Granzyme B+ and CD107a+). Although inefficient in generating primary responses to the CE, boosting of the Env CE DNA primed macaques with the intact env DNA vaccine potently augmented pre-existing immunity, increasing magnitude, breadth and cytotoxicity of the cellular responses. Fine mapping showed that 7 of the 12 CE elicited T cell responses. Env CE DNA also induced humoral responses able to recognize the full-length Env. Env CE plasmids are therefore capable of inducing durable responses to highly conserved regions of Env that are frequently absent after Env vaccination or immunologically subdominant. These modified antigens are candidates for use as prophylactic and therapeutic HIV vaccines.
Introduction
The variability of HIV is a major stumbling block for the design of a universal vaccine. The plasticity of HIV allows for an enormous number of viable mutant strains, which in turn allows the virus to escape from immune responses while preserving viral function. Presentation of variable epitopes results in immune response easily overcome by virus mutation and escape. Immunodominance (i.e., hierarchical preference for recognition of one epitope of the vaccine over another) and generation of immune responses targeting non-protective epitopes can interfere or even suppress responses toward conserved, and ideally protective, segments in the viral proteome. Non-protective epitopes are those whose recognition is associated with a lack of virologic containment in vivo (e.g., high viral load) or those that can readily escape without impairing viral fitness. Immunodominant epitopes with a high degree of conservation do not necessarily confer immune control of viral replication, as they may represent viral adaptation to human HLA types at the population level, and at minimal fitness cost.Citation1-3 It seems likely that if HIV segments were capable of mutating without limiting virus functionality, they would not contribute substantially to a vaccine's protective response. Variable epitopes could serve as immunodominant “decoys” that can absorb immune reactivity and potentially preclude responses against protective epitopes. The mechanism responsible for the immunodominance of the variable regions is poorly understood but, clearly, provides an advantage for the persistence of the virus. For example, the most significant difference between HIV strains infecting vaccine vs. placebo recipients in the Step (HVTN 502) vaccine trial occurred within a region including the SLYNTVATL epitope in Gag,Citation4 an immunodominant epitope, and immune responses to which are not associated with diminution of viral load.Citation5 The SLYNTVATL reactivity may therefore represent an immunodominant decoy response that is detrimental to vaccine efficacy.Citation6
HIV vaccine immunogens have generally been derived from individual viral isolates. It may not be possible in a vaccine to cover all the viral antigenic diversity required to block viable or transition escape forms of the virus—as rapid, sequential epitope evolution, including uncommon intermediates, is commonplace early in infection.Citation7,8 The immunogen sequence diversity that may be required to block viable or transition escape mutants may be too large to accommodate for a single or a manageably sized variation inclusive vaccine. To maximize immunologic breadth, different strategies are being explored to block common escape forms of viruses and thus effectively block immunological escape pathways that (i) use consensus and deduced ancestral sequences,Citation9-11 and (ii) create antigenic combinations of common variants that evolve as a result of immune pressure (e.g., mosaicsCitation12-14,15 and similar structures.Citation16,17 WeCitation18-21 and othersCitation22-25 have designed vaccine approaches aiming to focus the immune responses to conserved regions of the virus.
We previously reported the generation of novel immunogens that focus the immune responses to conserved regions in HIV gag.Citation19,20 These Conserved Elements (CE) were defined based on stringent conservation, functional importance and independence of ‘protective’ haplotypes associated with HIV control.Citation26-29 In this report, we expanded this vaccine strategy to include conserved regions of Env. We continue to use DNA as vaccine platform since it has several advantages such as simplicity, scalability and possibility for repeated applications due to the lack of immunity against the vector. The combination of DNA intramuscular delivery following by in vivo electroporation has induced promising immune responses in macaques and in humans (reviewed inCitation30, 31).
Results
Identification of Env CE
Sequence data set and amino acid database frequencies. Full-length HIV-1 group M Env coding sequences were downloaded from the HIV database (HIVDB, http://www.hiv.lanl.gov/). Any sequences with hypermutationsCitation32 early stop codons, frame-shift mutations or ambiguous amino acids were excluded. A multiple sequence alignment was prepared using MUSCLECitation33 and then manually edited. The database frequency of each amino acid at each site in the final alignment was then calculated using a perl script (http://indra.mullins.microbiol.washington.edu/perlscript/docs/CountAAFreq.html). The degree of conservation required for inclusion as an Env CE was at least 90% across the entire HIV-1 M group, and usually at least 98%. Regions for inclusion/exclusion from the vaccine were also selected based on whether immune responses to such regions were associated with virologic control or lack of control, and whether mutations at a given site had been shown to result in a loss of viral fitness ex vivo. Other features that resulted in a relaxation of the 90% requirement was an association with known function or CTL escape resulting in a loss in viral fitness, or to substantially extend the length of a CE. For example, the CD4 binding loop region of the HIV Env protein corresponds to a region that binds to broadly neutralizing antibodies and this region is included as CE10. To include this region, 7 toggle sites were used and one residue was included in CE10 without a toggle that had a conservation level of only 79%. In another example, a very long CE (CE14; 43 AA) was included by allowing 5 toggle sites and one residue was conserved only at a level of 84% across the HIV-1 M group. However, this site was conserved at 97% and 99% in HIV-1 M group subtypes B and C and no obvious toggle site was required. A set of 12 conserved elements (CE) was identified in the HIV Env protein () spanning 11, 14, 21, 15, 23, 21, 13, 12, 14, 43, 20, and 13 AA in length. The localization of the CE within HXB2 Env as reference is shown (). Toggle sites were used to create 2 versions of all the CE, except CE6 (gray shaded in ). A toggle site represents an amino acid site at which conservation may be low but at which 2 amino acids combined account for most of sequences of all HIV-1 M group sequences known, usually 98–100%. HIV-1 M group subtypes B, primarily found in the Americas and Western Europe, and subtype C primarily found in South Africa and India, represent most of the available sequence data, and together they represent > 60% of all HIV-1 infections. Toggle sites often represent AA that are highly conserved in either subtype B or C, and the Env CE1 and CE2 sequences correspond to the AA most associated with subtypes B and C, respectively, if the consensus or second most common variant residues differed.
Table 1. Sequence and localization of the Env CE.
Env CE are poorly immunogenic in env DNA vaccinated macaques
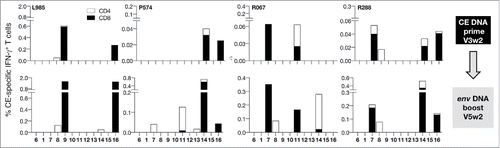
The immunogenicity of the 12 CE identified within HIV env was evaluated in samples from 16 macaques vaccinated with DNA expressing intact Env as part of other studies (). The animals received a mixture of clade B (including BaL) and C env DNA administered via the IM route followed by in vivo electroporation. PBMC collected 2 weeks after the 2nd or 3rd vaccination were stimulated with (i) Env-specific peptide pools (15-mer peptides overlapping by 11 AA derived from BaL) spanning the complete Env and (ii) a CE-specific peptide pool consisting of 10-mer peptides overlapping by 9 AA and 15-mer peptides overlapping by 11 AA designed to maximize the detection of CD4+ and CD8+ T cell responses (). displays the CE- and Env-specific T cell responses in these animals ranked according to the magnitude of Env-specific responses, and the presence or lack of CE-specific responses. Despite mounting robust Env-specific T cell reponses, ∼50% of the macaques failed to develop Env CE-specific responses (). The absence of CE-specific T cell responses was not related to the number of vaccinations (2 or 3) or the form of Env used in the vaccine (gp160, soluble trimeric gp140 or membrane-associated gp145 (). Comparison of the magnitude of the T cell responses to gp120 Env and CE further showed that, expect one animal (T145), there is a skewing of the responses toward gp120, indicating that the majority of the responses target epitopes in the more variable regions. Mapping of individual Env CE responses in 5 of the animals (using 12 individual peptides pools covering both Env CE variants) showed that only 1 or 2 CE were recognized (). These data indicate that the 12 Env CE are poorly immunogenic when present within the complete Env protein, perhaps due to immunological interference with other epitopes. This finding is reminiscent of our reported data on the HIV Gag CE and SIV Gag CE.Citation18, 20
Table 2. env DNA vaccine and T cell responses in vaccinated macaques.
Figure 1. CE are poorly immunogenic in env DNA vaccinated macaques. Analysis of Env CE T cell responses in 16 macaques immunized with a mixture of DNA expressing full-length Env including HIV-1 BaL. Env-specific IFN-γ T cell responses were measured using a matching peptide pool of HIV Env clade B strain BaL spanning gp120. CE-specific IFN-γ+ T responses were measured using a peptide pool (mixture of 15-mer overlapping by 11 AA and 10-mer overlapping by 9 AA) covering the 12 CE.
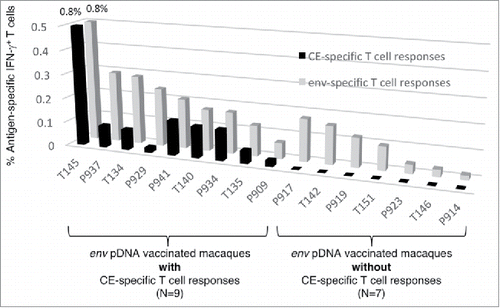
Table 3. Mapping of CE responses in selected macaques after vaccination with full-length HIV env DNA.
Generation of Env CE DNA vaccine
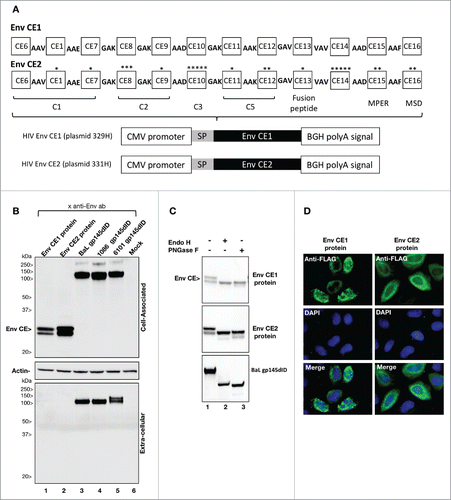
To promote the induction of Env CE-specific cellular immune responses, we designed and synthesized 2 versions of synthetic proteins (Env CE1 and Env CE2), each of which is composed of 12 conserved elements (). Env CE1 and Env CE2 proteins differ by 0–5 amino acid per CE to maximize the coverage of common HIV variants. The CE were collinearly arranged and separated by short amino acid linkers (e.g.,3 amino acids) designed to facilitate processing of the protein and avoidance of neo-antigens.Citation34,35 The coding sequences for Env CE1 and Env CE2 proteins were RNA/codon-optimized to maximize expression in mammalian cells and placed into a eukaryotic DNA plasmid vector, pCMVkan,Citation36 between the human CMV promoter and bovine growth hormone poly A signal. The pCMVkan expression vector is optimized for optimal growth in bacteria (kanR) and expression of the insert in mammalian cells. The Env CE1 and Env CE2 proteins were expressed upon transient transfection of HEK293 cells (). Env CE2 accumulated to higher level than Env CE1, possibly due to the 24 AA difference in these proteins, which could affect protein stability. In comparison, transfection of plasmids expressing gp145 Env proteins of BaL, 6101 and 1086 produced proteins that are found in both the cell-associated fraction (gp145) and the supernatant (processed gp120). Using FLAG-tagged version of the expression vectors to visualize the proteins efficiently, Env CE1 and Env CE2 proteins were found in the cytoplasm in a punctuate pattern in the perinuclear area of transfected HeLa cells (). Western immunoblot analysis further showed that Env CE1 and Env CE2 proteins migrate as 2 bands found exclusively in the cell-associated fraction (). To understand the nature of the 2 bands, cell extracts from HEK293 transfected cells were subjected to in vitro digestion with Endoglycosidase H (EndoH) and N-Glycosidase F (PNGase F), commonly used to interrogate protein glycosylation (). Endo H removes mannose-rich oligosaccharides, but cannot cleave complex oligosaccharide structures, whereas PNGase F removes all N-linked carbohydrates. Both Env CE1 and Env CE2 proteins were sensitive to both endoglycosidases indicating that these proteins underwent posttranslational modifications. Thus, the 2 bands represent different glycosylation forms of the Env CE proteins. As positive control, an extract containing the intact gp145 Env protein (BaL) was used (, bottom panel). We noted that the anti-gp120 serum used to visualize Env on the Western blot recognized the deglycosylated gp145 to a lesser extent as expected, since many antibodies are known to recognize the glycosylated forms of HIV Env.
Figure 2. Env CE1 and Env CE2 expression vectors. (A) Env-CE1 and Env CE2 proteins span 220 amino acids distributed among 12 different CE. They are highly related, differing by 24 amino acids symbolized by asterisks, indicating number and location of the toggle AA. The Env CE1 and Env CE2 complete sequences are each 282 amino acids in length, including linkers (3 amino acids in length each) and a 29-amino acid BaL Env signal peptide. The linker sequence includes AAV, AAE, GAK, AAD, AAK, GAV, VAV, or AAF. The Env CE1 and Env CE2 sequences were inserted in the CMVkan expression vector between the CMV promoter and the bovine growth hormone polyadenylation signal. (B) HEK293T cells were transfected with HIV pDNAs expressing Env CE1 (lane 1), Env CE2 (lane 2), Env gp145dID (lane 3–5). Lane 6 contains a sample from mock-transfected cells. Proteins from the cell-associated (top panel: 1/100 of the sample) and extra-cellular (bottom panel: 1/200 of the sample) were analyzed. Western immunoblots were probed using the rabbit anti-gp120 sera. Equal loading of the blot with the cell-associated fraction was controlled by probing the membrane with an anti-actin antibody (middle panel). (C) Cell lysates from HEK293 cells transfected with plasmids expressing the HIV CE1, HIV CE2 and BaL gp145dID were treated with Endo H, PNGase F or left untreated. The samples were analyzed by Western immunoblot assay using a rabbit anti-HIV gp120 antibody. (D) Subcellular localization of HIV Env CE proteins is shown in HeLa-derived HLtat cells transfected with Env CE-FLAG plasmids. The HIV Env CE-FLAG proteins were visualized with anti-FLAG primary antibody followed by Alexa-Fluor 488 conjugated secondary antibody, and the nuclei were stained with DAPI (separate and merged images are shown).
Immunogenicity of Env CE DNA vaccine in macaques
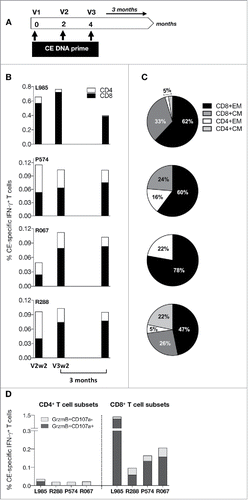
To test immunogenicity of the Env CE DNA vaccine, a combination of the 2 Env CE plasmids was used to immunize 4 macaques () at 0, 2 and 4 months. The animals were monitored for the development of CE-specific T cell responses 2 weeks after the 2nd and the 3rd vaccination (V2, V3), and the durability of the responses was monitored for 3 months after the 3rd vaccination (). This analysis demonstrated the presence of robust IFN-γ+ CE-specific cellular immune responses (0.1–0.8% of T cells), mediated primarily by CD8+ T cells, in all 4 animals. We interrogated both IFN-γ as well as TNF-α T cell responses. Since the TNF-α positive cells were also positive for IFN-γ, we focused our analysis on IFN-γ responses which covers all the T cell responses. In agreement with our previously reported data, maximal T cell responses were obtained with 2 to 3 DNA vaccinations using EP as delivery method. The Env CE vaccine induced both CD8+ and CD4+ memory T cell responses of the central and effector phenotype (). The Env CE DNA vaccine also induced CE-specific responses with a significant fraction of CD8+ T cells with cytotoxic phenotype (Granzyme B+ and CD107a+) (), while the CD4+ T cell responses with a cyotoxic phenotype were very low. We further found that CE-specific responses measured both as total as well as the subset of cytotoxic T cells showed robust durability with ∼2-fold contraction over the 2 months following the prime (V3wk2 to V4). Thus, the Env-CE DNA vaccine induced cellular immune responses with the desired features for an effective T cell vaccine.
Figure 3. Env CE DNA vaccine induces T cell responses in macaques. (A) Cartoon shows the vaccination regimen used to prime of macaques. The animals were vaccinated 3x with a mixture of Env CE1 and Env CE2 DNA by intramuscular injection followed by in vivo electroporation. T cell responses were analyzed after the 2nd and 3rd vaccination and 3 months later. (B) CE-specific T cell responses (both CD4 and CD8) measured 2 weeks after the second and third vaccination, as well as 3 months later, in PBMC samples stimulated with a peptide pool covering all 12 CE. The percent of CE-specific IFN-γ+ T cells upon peptide stimulation is shown. (C) The % CE-specific IFN-γ+ producing CD4 and CD8 central memory (CM; CD28+CD95+) and effector memory (EM; CD28−CD95+) T cells was determined following peptide stimulation with the CE peptide pool. (D) CE-specific responses are cytotoxic. The percentage of IFN-γ+ producing CE-specific CD4 and CD8 T cells harboring Granzyme B and expressing CD107a was determined following stimulation by the CE peptide pool.
Env CE prime/complete env DNA booster vaccination
We previously reported that CE within HIV and SIV Gag were poorly immunogenic when present within the complete Gag protein, likely due to either suboptimal processing and presentation or immunological interference with other epitopes and thus, unable to induce de novo responses.Citation18,20 We further found that the Gag CE-specific responses could be augmented upon booster vaccination with DNA expressing full-length Gag. This opens the possibility that Env CE-specific responses could be subject to the same mechanism. To address this experimentally, Env-CE DNA primed macaques received 2 env DNA booster vaccinations (). To optimally cover the 12 CE sequences in Env CE1 and CE2, a mixture of 3 Env DNA, including BaL, 6101 and 1086, was used. shows the alignment of the Env CE1 and CE2 sequences and the corresponding sequences in the selected Env proteins. Three of the 12 CE (CE6, CE7, CE9) show 100% identity with the Env sequences used in the booster vaccination, while the other CE sequences match except for 1 or 2 toggle AA. We generated gp145 versions of these Env proteins which (i) span the sequence covered by the CE and (ii) lack the immunodominant (ID) region in the extracellular gp41. Expression of these Env proteins is shown in .
Figure 4. Env CE prime-boost vaccination. (A) Cartoon shows the vaccine regimen used in the prime-boost vaccination of the macaques. The boost was performed with a mixture of env DNA composed of clade B (BaL and 6101) and clade C (1086) env DNA covering the 23 CE sequences (including toggle AA) present in HIV Env CE1 and CE2 proteins. DNA was delivered by intramuscular injection followed by in vivo electroporation. (B) CE-specific T cell responses (both CD4 and CD8) measured at the time of the booster vaccination (V4, V5) and 2 weeks later. The longevity of the memory responses was monitored 8 months later. (C) The proportion of the memory responses was determined after the last booster vaccination. The % CE-specific IFN-γ+ producing CD4 and CD8 central memory (CM; CD28+CD95+) and effector memory (EM; CD28−CD95+) T cells was determined following stimulation with the peptide pool containing the 12 CE.
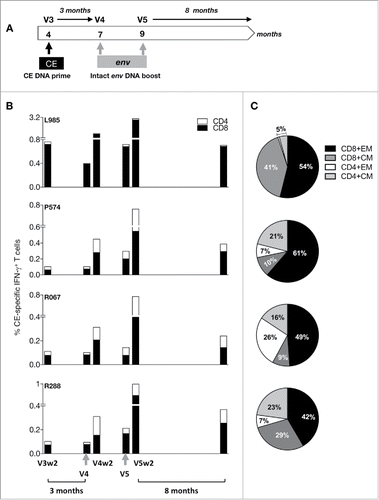
Table 4. Alignment of the Env CE and the HIV Env Sequences used in the booster vaccination.
Table 5. Mapping of CE responses after Env CE DNA prime and after env DNA boost.
Implementation of the prime-boost regimen using the Env-CE DNA prime followed by the intact Env DNA boost (vaccination 4 and 5, ), demonstrated a significant 4–10-fold increase of CE-specific cellular responses in the 4 animals (). In fact, 2 env DNA booster vaccinations lead to very robust levels of CE-specific immune responses (0.7–3% of total T cells). Thus, although inefficient in inducing primary responses to the CE, boosting with the intact Env DNA potently augmented pre-existing immunity demonstrating that the peptides within the CEs are properly processed and presented. This vaccine regimen achieved an effective shift in the immunological hierarchy, similar to our previously reported finding with HIV and SIV Gag vaccinations in macaques.Citation18,20 Furthermore, the Env CE responses were monitored 8 months after the 2nd booster vaccination (); these measurements showed 2- to 4-fold contraction compared with peak responses, supporting robust longevity of the CE-specific T cell memory responses.
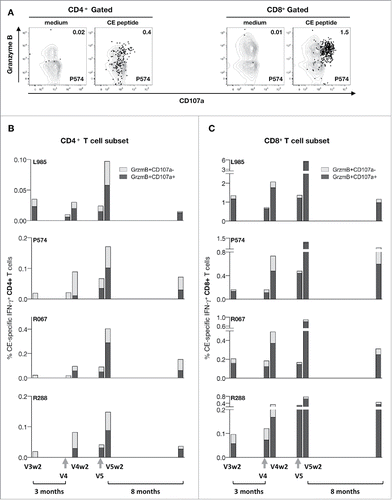
In addition, boosting with intact env DNA led to a proportional increase in CD4+ and CD8+ effector memory responses (). Boosting with intact env DNA further augmented the levels of cytotoxic (Granzyme B+ CD107a+) CD4+ and CD8+ T cell responses (; note that different scales were used). Flow analysis () showed the Granzyme B content and ability to degranulate (CD107a+) by the CE-specific CD4+ and CD8+ T cells. The levels of both cytotoxic CD4+ () and CD8+ () T cells increased. We found that the robust levels of the CD8+ T cell subset showed remarkable durability over the 8 months of follow-up, with a median 2-fold contraction.
Figure 5. CE prime-env DNA boost elicits potent and durable cytotoxic T cell responses. (A) Dot plots shows the Granzyme B content and degranulation activity (CD107a+) from unstimulated and peptide stimulated T lymphocytes from one vaccinated animals (P574) measured at 2 weeks after the 5th vaccination. The CD4+ (left panel) and CD8+ (right panel) CE-specific IFN-γ+ T cells are shown in black. (B) Frequency of CE-specific cytotoxic CD4+ T cell responses, and (C) CD8+ T cell responses after each booster vaccination are shown at 2 weeks after the priming (V3w2), after the 3 mo of rest at the day of the 1st booster vaccination (V4), 2 weeks later (V4w2), after a 2 mo rest period at the day of the 1st booster vaccination (V5), 2 weeks later (V5w2) and 8 months later.
Mapping of the Env CE responses
To evaluate the immunogenicity of individual CE, PBMC were stimulated with 12 CE-specific pools. IFN-γ+ T cell response is plotted () after the 3rd CE DNA prime and after the 2nd Env DNA boost and the responses are summarized in . Fine mapping showed that 6 of the 12 CE were immunogenic after the 3rd CE DNA prime (; prime) including CE7, CE8, CE9, CE11, CE14 and CE16, with 2–4 positive CE/animal. A comparison of responses to individual CE before and after boosting with DNA plasmids expressing intact Env showed that 7 of the 12 segments (60%) of the HIV Env CE are immunogenic. Responses to each individual CE were augmented further upon boost vaccination. Some of the CE (CE6, CE1, CE10, CE12 and CE15) did not show cellular responses in these animals. Interestingly, CE14, located in HR1 of gp41, appears to be immunogenic both within intact Env () and the Env CE DNA vaccine (). We noted that upon booster vaccination the appearance of additional positive CE in 3 of the animals (L985, P574 and R067). We speculate that ‘new’ responses detectable only after the boost may reflect responses below threshold of detection after the priming vaccination and maybe the result of an overall increase in CE-specific response upon intact env DNA booster vaccination. Of note, the mapping of 12 individual CE is likely affected by the overall magnitude of the total CE response. For this reason, we believe that assessment of median values of positive CE/animal best reflects the data. In comparison to env DNA vaccinated animals () which showed a median of 1 positive CE/animal among macaques mounting CE responses, the Env CE prime/env DNA boost vaccine induces significantly (p < 0.001) more positive CE/animal (median 4 CE) (). These data demonstrate that the CE prime/env DNA booster vaccination shifted the hierarchy toward broader Env CE-specific T cell responses.
Env CE DNA vaccine induced humoral immune responses
The Env CE DNA vaccine was also interrogated for its ability to induce humoral immune responses. We tested whether antibodies induced upon Env CE vaccination could recognize the immunogen by Western immunoblot assay (). The data from 2 of the 4 animals (L985, P574) showed that the priming vaccine induce antibodies that can detect the immobilized Env CE protein on the membrane. The responses developed by the other 2 macaques were below the threshold of detection. Upon boosting with complete env DNA both animals (L985, P574) showed stronger Env CE bands, indicating higher Env CE antibody titers. We next tested whether these antibodies could be detected by standard HIV gp120 Env ELISA. The antibodies induced by Env CE priming vaccinations in one of the 4 animals (P574) could be detected by ELISA () and, as expected, these antibodies further increased upon boost with intact Env. We further tested whether the Env CE DNA vaccine induced antibodies able to detect cross-clade Env (clade B and clade C) by Western blot assay (). Both animals (L985, P574) showed that priming with Env CE induces antibodies able to recognize not only the immunogen () but also clade B and C Env, supporting the notion the antibodies recognize shared conserved sequences, and these responses increased upon env DNA booster vaccinations. Env CE DNA priming alone did not induce neutralizing antibodies (Nab), likely reflected by the low ELISA titers. Upon 2 env DNA booster vaccinations, low Nab responses to tier 1A Env (MW965.26 in all 4 animals and SHIV SF162.P4 in 3 animals) were found. Overall low titers of Nab are expected in the absence of a protein in the vaccine.Citation31,42,43 We also analyzed whether the Env CE priming vaccination-induced antibodies could recognize linear peptides by ELISA screening of a library of 20-mer peptides with 14 AA overlap. As summarized in , all 4 animals showed responses to various linear peptides, although to different extents, and taken together peptides within CE6, CE9, CE10, CE12, CE14 and CE16 were recognized. Together, the analysis of the cellular and humoral immune responses induced by the Env CE DNA vaccine showed the development of immune responses able to recognize 10 of the 12 CE (located in C1, C2, C3, C5 and gp41 regions).
Figure 7. Humoral immune responses upon prime/boost. (A) Western immunoblot analysis was used to probe the Env CE proteins using plasma from vaccinated macaques. Proteins from cells transfected with a combination of HIV Env CE1 and Env CE2 plasmids were separated on denaturing gels and transferred onto membranes. Individual strips of membranes were incubated with plasma (1:100 dilution) from the vaccinated macaques and visualized using standard western blot methodology. (B) The Env CE DNA vaccine prime induced antibodies that can recognize gp120 Env by ELISA (HIV-1 IIIB) in animal P574. The responses were boosted by each of the full-length env DNA vaccination (4th and 5th vaccination V4w2, V5w2). (C) Antibodies induced by the HIV Env CE DNA vaccinated macaques recognize intact HIV Env proteins from both clade B (BaL gp145dID) and clade C (1086 gp145dID). HIV Env gp145 protein from transfected cells was separated on denaturing gels and transferred onto membranes. The membranes were incubated with plasma (dilution 1:100) from macaque L985 and P574 primed with Env CE DNA (3rd vaccination, V3wk2) and boosted with intact env DNA (5th vaccination, V5wk2). Proteins from mock-transfected cells served as a negative control.
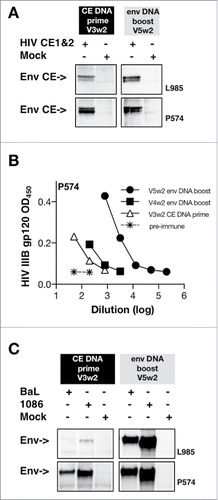
Table 6. Summary of CE immunity in the 4 env CE DNA vaccinated macaques.
Discussion
In this study, we expanded our HIV Conserved Element DNA vaccine concept and developed a DNA vaccine that focuses the immune responses to highly conserved sequences in Env. This DNA induces robust immune responses to epitopes that are only poorly immunogenic when present within intact Env. Addition of a booster vaccination with DNA expressing complete Env resulted in greatly increased Env immune responses focusing on the otherwise suboptimal Env CE regions. Thus, similarly to our reports with HIV and SIV Gag,Citation18, 20, 21 priming with a DNA vaccine expressing only CE alters the immune hierarchy resulting in recognition of these subdominant epitopes. Interestingly, both for Env and Gag, the magnitude, breadth and cytotoxic phenotype of the CE-specific responses are significantly increased upon boosting with DNA expressing the full-length molecules. We hypothesize that the details of the processing and presentation of epitopes from full-length Env may be superior in their ability to augment pre-existing CE immunity, however, they are unable to induce de novo CE-specific responses in ∼50% of the animals. The impairment of inducing de novo responses is thought to be due to the presence of dominant epitopes which suppress the development of the subdominant CE-specific T cell responses. Thus, priming with CE DNA and boosting with a plasmid expressing the intact protein may be generally applicable and may solve a major obstacle in HIV vaccine development, which is the focusing of responses to rapidly mutating immunodominant epitopes. This work demonstrates alteration of the hierarchy of epitope recognition and development of immune responses to potentially protective subdominant highly conserved epitopes.
The poor immunogenicity of the Env Conserved Elements reported in this study was found in macaques immunized with plasmid DNA encoding the complete Env protein. Several trials reported Env epitope mapping using samples from humans vaccinated with different forms of HIV-1 Env. Volunteers vaccinated with ADVAXCitation37 or DNA/NYVAC38 developed cellular immune responses targeting few epitopes with a total coverage of around 20 different peptides (mean of 3 epitopes per subject in the ADVAX study; 4.2 epitopes in the DNA/NYVAC) with some of the reported peptides partially overlapping or containing CE637-Citation38 and CE1 and CE13.Citation38 These data suggest that the CE identified in our work are poorly immunogenic also in persons vaccinated with the full-length Env. Interestingly, many of the reported epitopes identified in humans are MHC class II restricted and, therefore the majority of the volunteers responding developed T cell responses predominantly mediated by CD4+ T cells. Similar results were also obtained in the RV144 vaccine trial combining ALVAC and bivalent HIV-1 gp120 as protein.Citation39 In this study, which is the only trial that has identified low level of protection against infection, the vaccinated persons developed T cell responses targeting an average of 2 epitopes in Env, with a vast majority of these responses being mediated by CD4+ T cells, in many cases recognizing peptides in the V2 region containing the α4β7 integrin binding site.Citation40 Predominant CD4+ T cells responses in volunteers immunized with different forms of HIV-1 Env were already identified in the early 1990s by Orentas et al, using samples from humans immunized with recombinant HIV-1 gp160.Citation41 In contrast, the vaccine regimen described in this work (priming with Env CE DNA and boosting with the complete molecule) induces a balanced immune response mediated by both CD4+ and CD8+ T cells with cytotoxic phenotype and broad epitope recognition. It remains to be determined whether the macaque data reported here showing strong CD8+ T cell responses could be recapitulated after human immunization.
The data presented in this report provide a vaccine regimen able to induce potent T cell responses to subdominant epitopes. The question then arises whether the induced humoral immune responses are of better functional quality. A DNA only vaccine regimen is hampered by its inability to induce maximal antibody responses which can only achieve upon protein boost or co-immunization with protein.Citation42,43 Such studies will be necessary to evaluate the full potential of the CE DNA vaccine regimen and we have already found that the CE DNA vaccine can induce responses to regions in Env that are only poorly immunogenic in macaques vaccinated with full-length env DNA. Thus, the Env CE DNA vaccine regimen has the potential to further positively affect humoral immune response development.
Materials and methods
Plasmids
Plasmids HIV Env CE1 (plasmid 329H) and Env CE2 (plasmid 331H) contain the RNA/codon-optimized Env CE genes inserted into pCMVkan vectorCitation36 between the human CMV promoter and BGH polyadenylation signal. Both proteins contain the HIV BaL Env signal peptide at the N-terminus. Insertion of a FLAG-tag at the C–terminus of Env CE1 and Env CE2 generated plasmids 327H and 330H, respectively. HIV gp145dID (plasmid 332H, 341H and 340H) are based on HIV-1 clade B strain BaL, 6101 and clade C 1086, respectively, and express HIV gp145 Env (corresponding to AA 1–727 of HXB2) lacking the extracellular gp41 immunodominant region (corresponding to AA 590–608 of HXB2). All HIV Env proteins were produced from an RNA/codon optimized genes cloned into pCMVkan. Endotoxin-free DNAs (Qiagen, Valencia, CA) were prepared according to the manufacturer's protocol.
HIV Env CE DNA expression upon transient transfection
HEK293T cells seeded in 60 mm plates at a density of 106 cells/plate and were transfected by the Calcium Phosphate DNA co-precipitation procedure using 0.5 µg HIV env CE or 0.1 µg HIV gp145 env plasmid DNA together with 7 µg Bluescript as carrier DNA. Six hours after transfection the medium was replaced with 3 ml of complete DMEM. After 2 days, supernatants and cells were harvested, and the cells were lysed in 1 ml of hypertonic N1 lysis buffer (20 mM HEPES pH7.9, 10% glycerol, 1 mM MgCl2, 400 mM NaCl, 0.5 mM DTT, 0.5% Triton X-100), sonicated briefly for 2 × 6 seconds and centrifuged at 14 000 rpm for 15 min at 4°C. Endoglycosidase treatments were performed using 20 microliter cell lysates from HEK283 transfected cells. The cells were transfected with 1 µg DNA CE DNA and 0.2 µg env DNA, respectively. Extracts were untreated or digested for 1 hr at 37 °C with endoglycosidase H (EndoH) or N-Glycosidase F (PNGase F) (New England Biolabs, Inc., Ipswich, MA), respectively, following the manufacturer's instructions. Protein expression was analyzed by Western immunoblots using 12% sodium dodecyl sulfate polyacrylamide gels (Nu-Page Bis-Tris, NuPAGE, Invitrogen, Life Technologies Corp., Carlsbad, CA) and blotted onto nitrocellulose membranes which were probed with a rabbit anti-HIV gp120 antibody (dilution 1:2000), followed by anti-Rabbit IgG-HRP labeled antibody (1:10,000 dilution, GE Healthcare, Piscataway, NJ). As control, the membranes were probed with anti-actin antibody (clone C4, EMD Millipore, Billerica, MA) at a dilution of 1:10,000. The bands were visualized using the enhanced chemiluminescence (ECL) Prime Western blotting detection system (GE HealthCare, Piscataway, NJ).
Immunofluorescence assay
The HeLa-derived HLtat cells (2 × 105 cells/35 mm glass-bottomed plate) were transfected with 200 ng of HIV Env CE1-FLAG (plasmid 327H) and Env CE2-FLAG (330H). After 24 h, the cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.5% Triton-X 100 in PBS, incubated with mouse anti-FLAG antibody (#F1804, Sigma, St. Louis, MO), followed by incubation with anti-mouse antibody conjugated with Alexa-Fluor 488 (Life Technologies, Carlsbad, CA, at 1:750 dilution each) as secondary antibodies. The nuclei were stained with DAPI (Life Technologies, Carlsbad, CA). Cells were visualized on a Zeiss Observer Z1 fluorescent microscope using Zeiss Axiovision software (Carl Zeiss Microimaging GmbH, Göttingen, Germany).
Vaccination of Rhesus macaques
This study was performed in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Indian rhesus macaques were housed and handled in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care International at the Advanced BioScience Laboratories Inc., MD, and were approved by the Institutional Animal Care and Use Committee (OLAW assurance number A3467–01 and USDA Certificate number 51-R-0059). All DNA vaccine mixtures contained 0.2 mg of expression-optimized macaque IL-12 DNA (plasmid AG157).
Four macaques (L985, P574, R067 and R288) received 3 DNA priming vaccinations using a mixture containing 1 mg (vaccination 1 and 2) or 2 mg (vaccination 3) of each HIV Env CE1 amd Env CE2 plasmids. The env pDNA booster vaccination used 1 mg of each of the 3 HIV gp145dID plasmids (BaL, 6101, 1086). The pDNA vaccine was formulated in water and delivered via i.m. injection at 2 different sites (0.3 ml each site) followed by in vivo electroporation (IM/EP) using the Elgen 1000 device (Inovio Pharmaceuticals Inc., Plymouth Meeting, PA).
Two groups of macaques (total 16 animals) were vaccinated with intact env DNA as part of other studies and PBMC collected after the last vaccination were analyzed for Env CE and total Env-specific responses for this study. Seven macaques (T134 through T151) were vaccinated (V1, V2 and V3) with a mixture of 2 mg of the 3 HIV gp145dID pDNA (mixture of HIV-1 clade B BaL and 6101, and clade C 1086) formulated in phosphate-buffered saline solution. The pDNA vaccine was delivered via i.m. injection at one site (0.5 ml) followed by in vivo electroporation with the CELLECTRA® 5P device (Inovio Pharmaceuticals, Inc., Plymouth Meeting, PA). Nine macaques (P909 through P941 starting by the letter “P”) were vaccinated with a mixture of 3 mg of HIV expressing BaL, 6101, 1086 and EnvC (GenBank accession number AAD12112.1) as gp160 (plasmids 217H, 98H, 284H, 158H) and as gp140 (plasmids 229H, 228H, 285H, 246H) and the full-length single-chain (FLSC) protein, a CD4-BaL gp120 fusion protein (plasmid 203H). The pDNA mixtures was formulated in phosphate-buffered saline solution and delivered via i.m. injection at 2 different sites (0.5 ml each site) followed by in vivo electroporation (IM/EP) using the ICHOR device (ICHOR Medical Systems, San Diego, CA).
Intracellular cytokine staining
Ficoll-hypaque isolated PBMC were cultured in 96-well plates in the presence of various peptide pools from HIV, at a final concentration of 1 μg/ml for each peptide. Two peptide pools, combining 15-mer peptides overlapping by 11 AA and 10-mer peptides overlapping by 9 AA (Infinity Biotech Research & Resource, Inc.) were prepared to cover all the CE. Pool 1 contains 126 peptides and covers CE1, 7, 8, 9, 10, 13 and 16 and pool 2 contains 125 peptides and covers CE6, 11, 12, 14, and 15. The Env CE-specific results are presented as the sum of the 2 pools. Analysis of total Env-specific responses was performed using pools of 15-mer peptides spanning gp120 (using BaL peptides) and gp41 (PTE pool). Antigen-specific T cells were measured by intracellular cytokine staining followed by polychromatic flow cytometryCitation18,22 using the following cocktail of cell surface antibodies: CD3-APCCy7 (clone SP34–2), CD4-V500 (clone L200), CD8-Alexa Fluor-405 (clone 3B5, Invitrogen, Carlsbad, CA), CD28-PerCP Cy5.5 (clone CD28.2, BioLegend, San Diego, CA) and CD95-FITC (clone DX2) (BD PharMingen, San Diego, CA). Ten minutes after addition of peptides, the CD107a-eFluor 660 (clone eBioH4A3, eBioscience San Diego, CA) or CD107a-PE antibody (clone eBioH4A3, eBioscience San Diego, CA) was added. After cell permeabilization, intracellular staining was performed using IFN-γ-PE Cy7 (clone B27, BD PharMingen), TNF-α-Alexa Fluor 700 (clone Mab11, BD PharMingen) and Granzyme B-PE antibodies or Granzyme B-APC antibodies (clone GB12, Invitrogen). As negative and positive controls, PBMC were cultured in medium without peptide pools or stimulated with PMA and calcium ionophore (Sigma, St. Louis, MO). Peptide-stimulated samples were considered positive if the responses were 2-fold higher than that of unstimulated medium only control and greater than 0.01 after subtracting the medium control value. Samples were acquired on a LSR II or Fortessa flow cytometer (BD Biosciences, San Jose, CA), and the data were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR).
Humoral immune response analysis of DNA vaccinated macaques
Plasma samples were heat-inactivated for 30 minutes at 56°C. Binding antibodies to HIV env CE immunogen and intact Env were detected by Western immunoblot using cell extracts from HEK293 cells transfected with 1 μg of env CE DNA and 0.5 μg of env DNA (BaL gp145dID; 1086 gp145dID, respectively), separated on 12% SDS-PAGE, and the membranes were probed with pooled plasma (at a 1:100 dilution) and the bands were visualized with anti-monkey IgA, M, G-HRP antibody (1:10,000 dilution; cat# 43R-IG050hrp; Fitzgerald Industries International Inc., MA).Citation22 The binding titers to HIV IIIB gp120 were determined by standard ELISA using serial dilutions of plasma samples (Advanced Bioscience Laboratory, Rockville, MD), measuring optical absorbance at 450 nm. Pepscan analysis was performed using 20-mer peptides overlapping by 14 AA derived from HIV BaL.
Disclosure of potential conflicts of interest
G.N.P., and B.K.F. are inventors on US Government-owned patents related to DNA vaccines and gene expression optimization. G.N.P., B.K.F., A.V. and J.I.M are inventors US Government- and Washington University- co-owned patent applications on the Conserved Element technology. K.E.B. and N.Y.S. are full time employees of Inovio Pharmaceuticals and as such receive compensation in the form of salary and stock options. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
X.H. performed experiments, analyzed results, and contributed to drafting the paper; M.R., C.A., B.C., J.B. performed experiments; K.E.B. and N.Y.S. contributed essential methods; J.I.M., S.M., S.L G, A.V., G.N.P., and B.K.F. designed the research, analyzed results, and wrote the paper.
Acknowledgments
We thank D. Weiss, J. Treece, H. Anderson and staff at Bioqual and I. Kalisz (ABL) for excellent support, C. LaBranche and D. Montefiori (Duke University) for the neutralization assay, N. Miller (DAIDS/NIAID) for support, and T. Jones for editorial assistance. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 PTE Env Peptide Set (Cat #11551).
Funding
This work was support by the Intramural Research Program of the National Cancer Institute (to G.N.P and B.K.F.). This work was supported in part thorough the Simian Vaccine Evaluation Unit SVEU P185 (DAIDS, NIAID).
References
- Carlson JM, Brumme CJ, Martin E, Listgarten J, Brockman MA, Le AQ, Chui CK, Cotton LA, Knapp DJ, Riddler SA, et al. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J Virol. 2012;86:13202-16. doi:10.1128/JVI.01998-12. PMID:23055555
- Carlson JM, Le AQ, Shahid A, Brumme ZL. HIV-1 adaptation to HLA: a window into virus-host immune interactions. Trends Microbiol. 2015;23:212-24. doi:10.1016/j.tim.2014.12.008. PMID:25613992
- Keane NM, Roberts SG, Almeida CA, Krishnan T, Chopra A, Demaine E, Laird R, Tschochner M, Carlson JM, Mallal S, et al. High-avidity, high-IFNgamma-producing CD8 T-cell responses following immune selection during HIV-1 infection. Immunol Cell Biol. 2012;90:224-34. doi:10.1038/icb.2011.34. PMID:21577229
- Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, Sanders-Buell E, Heath L, Magaret CA, Bose M, Bradfield A, et al. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med. 2011;17:366-71. doi:10.1038/nm.2316. PMID:21358627
- Christie NM, Willer DO, Lobritz MA, Chan JK, Arts EJ, Ostrowski MA, Cochrane A, Luscher MA, MacDonald KS. Viral fitness implications of variation within an immunodominant CD8+ T-cell epitope of HIV-1. Virology. 2009;388:137-46. doi:10.1016/j.virol.2009.03.003. PMID:19368950
- Brander C, Hartman KE, Trocha AK, Jones NG, Johnson RP, Korber B, Wentworth P, Buchbinder SP, Wolinsky S, Walker BD, et al. Lack of strong immune selection pressure by the immunodominant, HLA-A*0201-restricted cytotoxic T lymphocyte response in chronic human immunodeficiency virus-1 infection. J Clin Invest. 1998;101:2559-66. doi:10.1172/JCI2405. PMID:9616227
- Herbeck JT, Rolland M, Liu Y, McLaughlin S, McNevin J, Zhao H, Wong K, Stoddard JN, Raugi D, Sorensen S, et al. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J Virol. 2011;85:7523-34. doi:10.1128/JVI.02697-10. PMID:21593162
- Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253-72. doi:10.1084/jem.20090365. PMID:19487423
- Mullins JI, Nickle DC, Heath L, Rodrigo AG, Learn GH. Immunogen sequence: the fourth tier of AIDS vaccine design. Expert Rev Vaccines. 2004;3:S151-9. doi:10.1586/14760584.3.4.S151. PMID:15285713
- Nickle DC, Jensen MA, Gottlieb GS, Shriner D, Learn GH, Rodrigo AG, Mullins JI. Consensus and ancestral state HIV vaccines. Science. 2003;299:1515-8; author reply −8. doi:10.1126/science.299.5612.1515c. PMID:12624248
- Doria-Rose NA, Learn GH, Rodrigo AG, Nickle DC, Li F, Mahalanabis M, Hensel MT, McLaughlin S, Edmonson PF, Montefiori D, et al. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J Virol. 2005;79:11214-24. doi:10.1128/JVI.79.17.11214-11224.2005. PMID:16103173
- Fischer W, Liao HX, Haynes BF, Letvin NL, Korber B. Coping with viral diversity in HIV vaccine design: a response to Nickle et al. PLoS Comput Biol. 2008;4:e15; author reply e25. doi:10.1371/journal.pcbi.0040015. PMID:18225944
- Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, Kuiken C, Haynes B, Letvin NL, Walker BD, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100-6. doi:10.1038/nm1461. PMID:17187074
- Barouch DH, O'Brien KL, Simmons NL, King SL, Abbink P, Maxfield LF, Sun YH, La Porte A, Riggs AM, Lynch DM, et al. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med. 2010;16:319-23. doi:10.1038/nm.2089. PMID:20173752
- Santra S, Liao HX, Zhang R, Muldoon M, Watson S, Fischer W, Theiler J, Szinger J, Balachandran H, Buzby A, et al. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat Med. 2010;16:324-8. doi:10.1038/nm.2108. PMID:20173754
- Nickle DC, Jojic N, Heckerman D, Jojic V, Kirovski D, Rolland M, Kosakovsky Pond S, Mullins JI. Comparison of immunogen designs that optimize peptide coverage: reply to Fischer et al. PLoS Comput Biol. 2008;4:e25. doi:10.1371/journal.pcbi.0040025. PMID:18463692
- Nickle DC, Rolland M, Jensen MA, Pond SL, Deng W, Seligman M, Heckerman D, Mullins JI, Jojic N. Coping with viral diversity in HIV vaccine design. PLoS Comput Biol. 2007;3:e75. doi:10.1371/journal.pcbi.0030075. PMID:17465674
- Hu X, Valentin A, Dayton F, Kulkarni V, Alicea A, Rosati M, Chowdhury B, Gautam R, Broderick KE, Sardesai NY, et al. DNA Prime-boost Vaccine Regimen to Increase Breadth, Magnitude, and Cytotoxicity of the Cellular Immune Responses to Subdominant Gag Epitopes of SIV and HIV. J Immunol. 2016;197:3999-4013. doi:10.4049/jimmunol.1600697. PMID:27733554
- Kulkarni V, Rosati M, Valentin A, Ganneru B, Singh AK, Yan J, Rolland M, Alicea C, Beach RK, Zhang GM, et al. HIV-1 p24gag derived conserved element DNA vaccine increases the breadth of immune response in mice. PLos One. 2013;8:e60245. doi:10.1371/journal.pone.0060245. PMID:23555935
- Kulkarni V, Valentin A, Rosati M, Alicea C, Singh AK, Jalah R, Broderick KE, Sardesai NY, Le Gall S, Mothe B, et al. Altered response hierarchy and increased T-cell breadth upon HIV-1 conserved element DNA vaccination in macaques. PLoS One. 2014;9:e86254. doi:10.1371/journal.pone.0086254. PMID:24465991
- Kulkarni V, Valentin A, Rosati M, Rolland M, Mullins JI, Pavlakis GN, Felber BK. HIV-1 Conserved elements p24CE DNA vaccine induces humoral immune responses with broad epitope recognition in macaques. PLoS One. 2014;9:e111085. doi:10.1371/journal.pone.0111085. PMID:25338098
- Mothe B, Hu X, Llano A, Rosati M, Olvera A, Kulkarni V, Valentin A, Alicea C, Pilkington GR, Sardesai NY, et al. A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques. J Transl Med. 2015;13:60. doi:10.1186/s12967-015-0392-5. PMID:25879820
- Letourneau S, Im EJ, Mashishi T, Brereton C, Bridgeman A, Yang H, Dorrell L, Dong T, Korber B, McMichael AJ, et al. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One. 2007;2:e984. doi:10.1371/journal.pone.0000984. PMID:17912361
- Ondondo B, Murakoshi H, Clutton G, Abdul-Jawad S, Wee EG, Gatanaga H, Oka S, McMichael AJ, Takiguchi M, Korber B, et al. Novel conserved-region T-cell mosaic vaccine with high global HIV-1 coverage is recognized by protective responses in untreated infection. Mol Ther. 2016;24:832-42. doi:10.1038/mt.2016.3. PMID:26743582
- Kran AM, Sorensen B, Nyhus J, Sommerfelt MA, Baksaas I, Bruun JN, Kvale D. HLA- and dose-dependent immunogenicity of a peptide-based HIV-1 immunotherapy candidate (Vacc-4x). AIDS. 2004;18:1875-83. doi:10.1097/00002030-200409240-00003. PMID:15353973
- Mothe B, Llano A, Ibarrondo J, Zamarreno J, Schiaulini M, Miranda C, Ruiz-Riol M, Berger CT, Herrero MJ, Palou E, et al. CTL responses of high functional avidity and broad variant cross-reactivity are associated with HIV control. PLoS One. 2012;7:e29717. doi:10.1371/journal.pone.0029717. PMID:22238642
- Rolland M, Jensen MA, Nickle DC, Yan J, Learn GH, Heath L, Weiner D, Mullins JI. Reconstruction and function of ancestral center-of-tree human immunodeficiency virus type 1 proteins. J Virol. 2007;81:8507-14. doi:10.1128/JVI.02683-06. PMID:17537854
- Rolland M, Manocheewa S, Swain JV, Lanxon-Cookson EC, Kim M, Westfall DH, Larsen BB, Gilbert PB, Mullins JI. HIV-1 conserved-element vaccines: relationship between sequence conservation and replicative capacity. J Virol. 2013;87:5461-7. doi:10.1128/JVI.03033-12. PMID:23468488
- Rolland M, Nickle DC, Mullins JI. HIV-1 group M conserved elements vaccine. PLoS Pathog. 2007;3:e157. doi:10.1371/journal.ppat.0030157. PMID:18052528
- Khan AS, Broderick KE, Sardesai NY. Clinical development of intramuscular electroporation: providing a “boost” for DNA vaccines. Methods Mol Biol. 2014;1121:279-89; PMID:24510832
- Felber BK, Valentin A, Rosati M, Bergamaschi C, Pavlakis GN. HIV DNA Vaccine: Stepwise improvements make a difference. Vaccines. 2014;2:354-79. doi:10.3390/vaccines2020354. PMID:26344623
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G –>A hypermutation. Bioinformatics. 2000;16:400-1. doi:10.1093/bioinformatics/16.4.400. PMID:10869039
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi:10.1186/1471-2105-5-113. PMID:15318951
- Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. J Clin Invest. 2007;117:3563-75. doi:10.1172/JCI32047. PMID:17975674
- Zhang SC, Martin E, Shimada M, Godfrey SB, Fricke J, Locastro S, Lai NY, Liebesny P, Carlson JM, Brumme CJ, et al. Aminopeptidase substrate preference affects HIV epitope presentation and predicts immune escape patterns in HIV-infected individuals. J Immunol. 2012;188:5924-34. doi:10.4049/jimmunol.1200219. PMID:22586036
- Rosati M, von Gegerfelt A, Roth P, Alicea C, Valentin A, Robert-Guroff M, Venzon D, Montefiori DC, Markham P, Felber BK, et al. DNA vaccines expressing different forms of simian immunodeficiency virus antigens decrease viremia upon SIVmac251 challenge. J Virol. 2005;79:8480-92. doi:10.1128/JVI.79.13.8480-8492.2005. PMID:15956591
- Kopycinski J, Cheeseman H, Ashraf A, Gill D, Hayes P, Hannaman D, Gilmour J, Cox JH, Vasan S. A DNA-based candidate HIV vaccine delivered via in vivo electroporation induces CD4 responses toward the alpha4beta7-binding V2 loop of HIV gp120 in healthy volunteers. Clin Vaccine Immunol. 2012;19:1557-9; PMID:22837097
- Harari A, Bart PA, Stohr W, Tapia G, Garcia M, Medjitna-Rais E, Burnet S, Cellerai C, Erlwein O, Barber T, et al. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J Exp Med. 2008;205:63-77; PMID:18195071
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209-20; PMID:19843557
- de Souza MS, Ratto-Kim S, Chuenarom W, Schuetz A, Chantakulkij S, Nuntapinit B, Valencia-Micolta A, Thelian D, Nitayaphan S, Pitisuttithum P, et al. The Thai phase III trial (RV144) vaccine regimen induces T cell responses that preferentially target epitopes within the V2 region of HIV-1 envelope. J Immunol. 2012;188:5166-76; PMID:22529301
- Orentas RJ, Hildreth JE, Obah E, Polydefkis M, Smith GE, Clements ML, Siliciano RF. Induction of CD4+ human cytolytic T cells specific for HIV-infected cells by a gp160 subunit vaccine. Science. 1990;248:1234-7; PMID:2190315
- Li J, Valentin A, Kulkarni V, Rosati M, Beach RK, Alicea C, Hannaman D, Reed SG, Felber BK, Pavlakis GN. HIV/SIV DNA vaccine combined with protein in a Co-immunization protocol elicits highest humoral responses to envelope in mice and macaques. Vaccine. 2013;31:3747-55
- Patel V, Jalah R, Kulkarni V, Valentin A, Rosati M, Alicea C, von Gegerfelt A, Huang W, Guan Y, Keele BF, et al. DNA and virus particle vaccination protects against acquisition and confers control of viremia upon heterologous SIV challenge. Proc Natl Acad Sci U S A. 2013;110:2975-80