ABSTRACT
In the current study, an improved NGS approach was developed to study the B-cell repertoire evolution in a simple mouse immunization model including only two DNA immunizations. The combination of 5′RACE and Ion Torrent long reads enabled unbiased immunoglobulin repertoire analysis even from small amounts of peripheral mouse blood. The B-cell population expanded by the vaccine displayed a relatively strong clonality. Upon priming with the first vaccine dose, we observed a consistent pattern of V-segment gene and CDR3 usage (public specificities). Interestingly, this pattern diversified with the second dose of immunization —it was relatively different in individual mice in spite of having received the same vaccine regimen (private specificities). Nevertheless, there were several instances in which the same public V-segment genes and CDR3s that were expanded after the first dose were further amplified after the second immunization. Taken together, it appears that the major clonotypes expanded by vaccination were originally a homogeneous subset that later diversified after a second dose leading to diverse “private” clonal compositions in different mice. These results established a new platform valuable to perform longitudinal analyses of the Ig germline gene usage and clonotype evolution throughout an immunization regimen in a commonly used animal model.
Introduction
Traditionally the humoral responses elicited by vaccines are mainly monitored by the levels and quality of polyclonal antibodies in serum and other specific tissues at various time points after immunization. The introduction of monoclonal antibody (mAb) technology enabled examining the quality of individual antibody components in an immune serum including the immunoglobulin (Ig) germline gene usage and sequence. However the process of isolating mAbs is selective and does not necessarily reflect the full picture of antibody responses. Next generation sequencing (NGS) technology with its high-throughput analysis of a large pool of genes at the same time, provides a powerful tool to investigate the antibody repertoire with unprecedented efficiency and depth, although the application of NGS to the study of antibody responses following immunization needs to be further refined.
B-cell repertoire NGS has been reported in the past few years. In many instances, the samples are of human origin,Citation1–4 although there have also been reports of primateCitation5 and rabbit.Citation6,7 In these studies, a multiplex PCR approach has proven sufficient to provide enough coverage of the different V-segment genes without significantly skewing the original repertoire as a PCR artifact. In mice however, the number of different V-gene segments is greater,Citation8 and designing a primer set covering all the possibilities is more challenging. In the current report, 5′RACE,Citation9 an appealing alternative to multiplex PCR, was used to eliminate the need for bioinformatics corrections and also simplify the PCR reactions required to obtain the amplicons to be sequenced. We optimized the Ion TorrentCitation10 platform which enabled us to read over 400 bases from mouse 5′RACE amplicons in a single reverse read from the junction between variable and constant region, thus covering the V-region from the end of framework region 4 to the beginning of framework region 1. We used HIV gp120 as a model antigen, since we have previously shown gp120 DNA immunization of mice to be very effective.Citation11 Using this method, B-cell repertoire NGS was conducted to monitor the dynamics of Ig germline gene usage and clonotype expansion following immunizations in a mouse model.
Results
5′RACE has been utilized to clone antibody genes from mouse hybridomas,Citation12 but it is yet to be established whether it would be applicable to immunoglobulin repertoire sequencing by NGS. In the current study, upon 5′RACE and amplicon generation by PCR, barcoded samples were run on the Ion Torrent PGM platform (ThermoFisher Scientific). A significant number of reads covering the entire variable region after quality score trimming was observed. A summary of the quality-filtered reads corresponding to each cell population and individual is provided in Table S1. This method was then used in the current study to monitor the B cell repertoire evolution in a mouse gp120 DNA immunization model.
The DNA vaccine was chosen in the current study because our previous data demonstrated that DNA immunization with an HIV-1 gp120-expressing plasmid leads to a measurable antibody response, and the avidity of the resulting antibodies increases with subsequent doses.Citation11 Furthermore, the gp120 DNA vaccine was more effective in priming germinal center (GC) B cell responses than a recombinant gp120 protein vaccine with the same amino acid sequence.Citation13 The immunization schedule, sample collection and processing are depicted in . There are only two groups in this simple pilot study. Mice in Group A received two gp120-expressing DNA vaccine immunizations on Days 0 and 21, followed with one recombinant gp120 protein immunization 4 days before splenocytes collection. Mice in Group B received the empty DNA vaccine vector twice and one time gp120 protein immunization in the same schedule as in Group A. No adjuvant was included for either DNA or protein vaccines to minimize the impact of non-antigen specific immune stimulations.
Figure 1. Schematic of the study design. Two DNA immunizations were administered by gene gun followed by a protein boost without adjuvant.
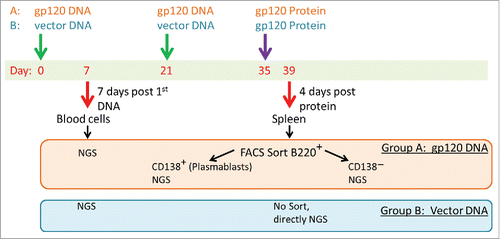
The first sample collection on Day 7 consisted of peripheral blood cells from a small volume of blood and did not require euthanasia, so that the same animals could be subsequently sampled. The protein inoculation was intended as a stimulus for pre-existing memory cells elicited by DNA priming vaccination to migrate to the spleen and potentially differentiate into plasmablasts. The second sample collection was 4 days after the protein boost, and the differences in the immunoglobulin (Ig) repertoire between the two groups could be attributed to the expansion of B-cells mediated by two gp120 DNA immunizations and not to protein. It is not expected that a single small dose of protein without adjuvant 4 days before the final sample collection could be immunogenic in vector control-immunized mice. Indeed, there was no detectable gp120-specific antibody response in the serum of Group B mice, whereas the gp120-specific titer in Group A was high at the same time point (Fig. S1). Both groups received the same gp120 protein dose and serum was collected 4 days after protein inoculation.
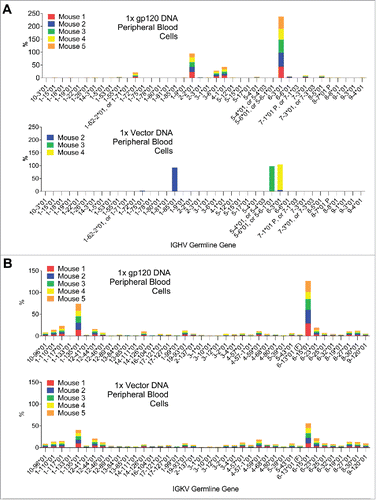
The gp120 DNA-immunized mice in Group A showed much greater diversity in their IgG heavy chain repertoires: 11 different germline genes were utilized (). For kappa reads the pattern is more similar between vector and gp120-immunized (). Interestingly, the germline gene distribution after the first gp120 DNA immunization displayed a “public” pattern in which germline gene usage was remarkably consistent among five different mice in the same group. In contrast, empty vector-immunized mice in Group B displayed an extremely polarized heavy V-segment gene usage repertoire in peripheral blood cells after the first DNA immunization. Only 3 germline genes were observed at more than 1% of the total reads in gamma chain amplicons, and the vast majority of the reads were assigned to a single germline gene per mouse.
Figure 2. A) V-gene segment usage in gamma reads from peripheral blood cells of vector control-immunized mice and gp120-immunized mice. The distribution in vector control-immunized mice is oligoclonal and very different from that of gp120-immunized mice. B) Same as A, but in kappa reads.
Splenocytes were harvested on day 4 after protein vaccine boost, and cells were sorted by FACS into two populations: B220+ CD138− and B220+ CD138+. CD138 is a known plasmablast marker and its expression in B-cells is indicative of differentiation into antibody secreting cells.Citation14 A small number of plasmablasts were observed in splenocytes from Group A mice (Fig. S2 and Table S1), but no plasmablasts were observed in Group B and therefore no sorting was performed on Group B samples. Overall, fewer plasmablasts are expected in a vaccine setting than after a viral infection.
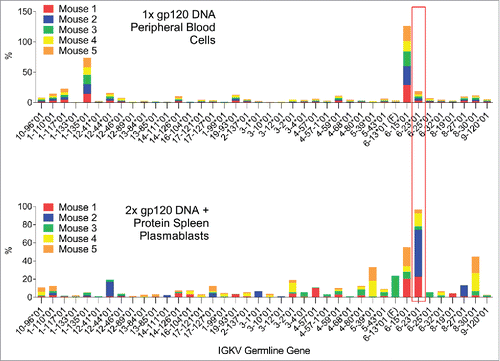
It is interesting to observe that the gamma chain V-segment gene usage was greatly diversified after the boost immunization. Although the dominant V-segment genes observed after the first immunization were still heavily used by some mice after the 2nd dose, the appearance of “private” V-segment gene usage patterns was observed in certain individual mice, especially in plasmablasts (). A very similar scenario was observed for the kappa chain V-segment gene usage (). There was manifest overlap between the repertoires of the individual mice after the 1st DNA immunization in peripheral blood cells; whereas with the 2nd DNA immunization and boost, the repertoires were more diversified and “private” V-segment gene usage emerged.
Figure 3. Comparison of the gamma chain V-segment germline gene usage by IgG expressing B-cells in peripheral blood cells 7 days after the first DNA immunization or in spleen plasmablasts 4 days after the 2nd DNA and boost. Germline genes which are dominant in both time points are highlighted.
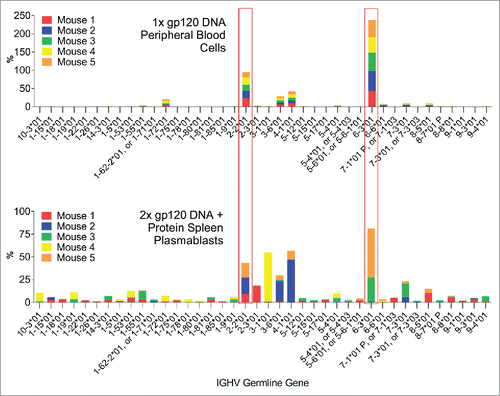
Figure 4. Comparison of the V-segment kappa germline gene usage by B-cells in peripheral blood cells 7 days after the first DNA immunization or in spleen plasmablasts 4 days after the 2nd DNA and boost. Germline genes which are dominant in both time points are highlighted.
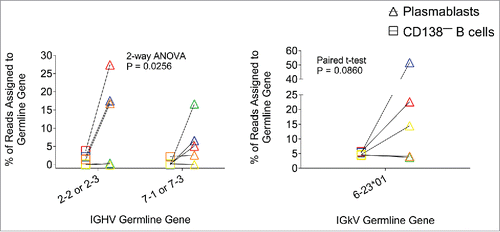
When comparing the V-segment gene usage between plasmablasts and the B220+ CD138− population after boost immunization, we observed very different patterns (). These samples were taken 4 days after protein inoculation, at which time we expected the plasmablast expansion to reach its peak. Thus, the B220+ CD138− subpopulation represents B-cells that have not been activated and may serve as a baseline, while the B220+ CD138+ plasmablasts should represent antigen-specific B-cells generated by the DNA priming vaccination that have been recently recalled to the spleen and activated by the protein boost. There was a “public” gene usage pattern for gamma reads: IGHV2-2 or IGHV2-3, and IGHV7-1 or IGHV7-3 were exclusively over-represented in the plasmablast subpopulation and not in the B220+ CD138− B-cells or in the vector control-immunized mice ( and Fig. S4). In order to obtain statistical significance, two germline genes had to be clustered together. This may seem arbitrary, but indeed the sequence of the two genes clustered together is very similar, even at the CDR1 and CDR2 regions (Supplemental Figure 5). Similarly, for kappa reads IGKV6-23 was preferentially used by plasmablasts and not by B220+ CD138− B-cells or splenocytes from vector control group mice (Supplemental Figure 6).
Figure 5. Relative abundance of germline gene assignments in two subpopulations—plasmablasts (B220+CD138+) or other B cells (B220+CD138−)—from mice immunized with two doses of gp120 DNA. Each color represents an individual mouse. 4 out of 5 mice showed increased germline gene usage in plasmablasts for either one or both of the germline genes shown. P value denotes statistical significance of a 2-way ANOVA for the variable “subpopulation” (plasmablasts vs. CD138− B cells) for the gamma chain, or for a paired one-tailed t-test for the kappa chain.
The CDR3 sequence can be used as a clonotype marker. Cells expressing the same CDR3 and V-segment gene are very likely to have originated from a single precursor. When the ranking of unique CDR3 amino acid sequences was analyzed, we observed once more a pattern with shared “public” CDR3s after the first DNA prime for both gamma and kappa chain reads (). After the boost, we observed diversification and the appearance of dominant “private” CDR3s in the different mice (). Interestingly, there was only one mouse in which 3 of the 5 dominant CDRH3s after the first DNA immunization went on to expand even further after the boost (, blue).
Figure 6. A: Relative frequencies of the dominant CDRH3s from gamma reads after the 1st immunization. None of the dominant CDRH3s were observed in vector control-immunized mice. B: Relative frequencies of the dominant CDRL3s from kappa reads after the 1st immunization. All of the dominant CDRL3s in gp120-immunized mice were also present in vector control-immunized mice, albeit at lower frequencies (data not shown).
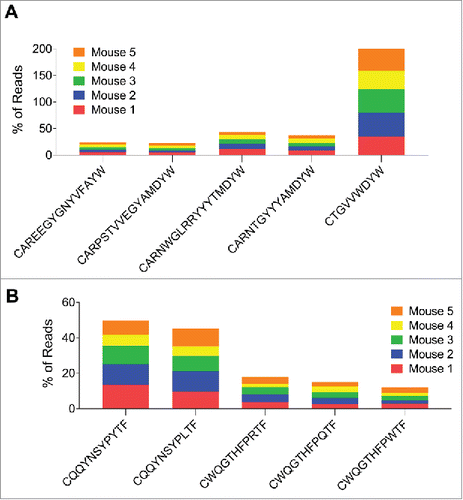
Figure 7. The relative frequency of CDR3s common to two or more mice (“public” CDR3s) after 1 or 2 gp120 DNA doses. Each color represents an individual mouse. Top: public CDRH3s for IgG-expressing B cell heavy chain. After the first immunization all public CDRH3s appeared at a very similar frequency in all 5 mice, whereas after the second DNA immunization the scatter was considerable—each individual mouse displayed different frequencies of the different public CDRH3s. Bottom: public CDRL3s for B-cells expressing kappa chain. CQQYSSYPLTF is a rare CDRL3 after the first immunization that goes on to become dominant and shared by 3 out of 5 mice.
The dominant CDRH3s after the 1st gp120 DNA immunization () accounted for 77% of the total reads, with the top 5 accounting for 66%. This is indicative of a highly polarized repertoire, probably antigen-specific, since the peripheral blood cell sample was taken at the expected peak time of the B-cell response. Further supporting the hypothesis that the dominant CDRH3s observed after the 1st gp120 DNA immunization are antigen-specific is the fact that none of the top 10 CDRH3s found in this sample were observed in the vector control mice (data not shown). However, a different scenario was observed for the kappa chain CDRL3s: the top 5 CDRL3s only accounted for 28% of the reads on average (). In addition, all five dominant CDRL3s were observed in the vector control mice, albeit at lower frequencies (data not shown). This discrepancy between gamma and kappa chain is probably explained by the fact that the gamma chain reads are undoubtedly derived from cells expressing IgG, as the reverse primers were specifically designed to amplify IgG heavy chain cDNA and not any other subclasses. On the other hand, the kappa reverse primers would also amplify kappa chain cDNA originating from cells expressing any subclass from their heavy chain locus. Therefore, while the heavy chain reads are derived exclusively from IgG-expressing cells, the kappa chain reads reflect a combination of B-cells from every subclass, and thus the enrichment in antigen-specific kappa reads is diluted by contributions from naïve IgM/IgD-expressing cells. After the boost immunization, IgM-expressing B-cells were removed from the splenocyte cell suspension using magnetic beads, and therefore most kappa reads should derive from IgG-expressing cells.
When looking at the dominant CDR3 repertoire in plasmablasts after boost, we observed “private” patterns for both gamma and kappa as opposed to the “public” patterns seen after the 1st immunization (). This is consistent with the patterns observed for V-segment gene usage described above.
Discussion
Prior to NGS, B-cell repertoires have been studied by traditional sequencing techniques with isolated B-cells of interest or hybridoma cells, but these techniques were insufficient for creating a high-resolution profile of the Ig gene usage. It is important to monitor the immune cell population dynamics after antigen stimulation and to establish patterns of variable gene utilization, and to correlate them with the nature of the antigen design. In the current study, we established the feasibility of using NGS to conduct B-cell repertoire analysis with peripheral blood samples in addition to splenocytes. Reddy and others have been able to obtain immunoglobulin repertoires from mouse bone marrow and spleen.Citation15–18 These two sources are no doubt very relevant, however sampling these organs is a terminal procedure and in order to establish B-cell dynamics, repetitive sampling is required. To the best of our knowledge this is the first report of a methodology that enables sequencing the immunoglobulin repertoire from a small volume of mouse peripheral blood. This is of great importance, since variation between individuals is very high after several instances of exposure to antigen. Thus, our methodology enables establishing a time course of the B-cell repertoire evolution in a mouse model.
One key finding was that upon the first dose of DNA immunization, there was a remarkable overlap in the IgG repertoire of the 5 different mice. Other studies have shown much lower degrees of overlap in cells from different individual mice.Citation15 This is probably due to the fact that in those studies spleen or bone marrow samples were analyzed after multiple immunizations, whereas in the present report we were able to analyze peripheral blood samples shortly after the first immunization. We believe that both the sampling timing and the nature of the vaccine can greatly influence the composition of the B-cell repertoire. The peak of the plasmablast response has been observed before at 4 days post-immunization in secondary lymphoid organs.Citation19 We expected the expansion of vaccine-induced B-cells to be maximal at that time. We hypothesized that by sampling the repertoire at that time, any variations from an unimmunized control would likely reflect the selection and expansion of antigen-specific cells.
In contrast with the observations after one dose, upon boost we ceased to observe such a large degree of overlap between individuals, consistent with published observations.Citation15 This transition from a public repertoire after one dose to mostly private repertoires after the second dose was quite remarkable and unexpected. It is possible that DNA vaccines produce a very homogeneous form of antigen and therefore the B-cells in the naïve repertoire that can be selected through binding are more limited than if a more heterogeneous antigen were presented, such as a recombinant protein or virus-based vaccine, which may include denatured or partially degraded forms of antigen, or mixed antigens. In the current study, spleens were harvested only 4 days after protein boost, and therefore we do not anticipate that the protein boost had a significant effect on remodeling the B-cell repertoire, rather, it was intended as a means to recall to the spleen the memory B-cells generated after the 2nd dose of DNA.
Alternatively, we can speculate that upon a first encounter with antigen there are only a limited number of unique naïve B-cell clones that are stimulated to expand, and even fewer antigen-specific clones undergo class switching and expand enough to be above our detection threshold. There could be other clones that are initially undetected due to their small numbers, who upon re-encounter with antigen accumulate more somatic mutations that enable them to compete with the clones that expanded more strongly after the first dose. This later process of somatic mutation and selection may be more stochastic and thus lead to the observed private repertoires when comparing among individuals after the second DNA dose. Further studies and larger numbers of mice and sampling time points will be needed in order to confirm this hypothesis.
However, some bias may be introduced by this method due to the small sample size. It is known that terminally differentiated B-cells produce more immunoglobulin RNA than naive or memory cells.Citation20 Since NGS repertoire analysis measures cDNA, the fact that some cells in a mixed population express more RNA than others could confound the relationship between the fraction of reads in the NGS data set and the cellular composition of the original repertoire. We have attempted to minimize this problem by sorting cells that are known to produce larger amounts of immunoglobulin RNA, such as plasmablasts. This allowed for a more detailed analysis of the B cell repertoire in our study. Unfortunately, sorting cells from a small volume of mouse peripheral blood is not technically feasible at this point and requires further methodology development. This led to the shortcoming of the current study that plasmablast samples were collected only once (after 2nd immunization) and they were compared against B-cells collected from peripheral blood after the first immunization. More systematic longitudinal analyses of the B-cell repertoire in peripheral blood after immunization in a vaccine regimen with more doses are warranted. In addition, the data collected from the current study may be affected by using gp120 as the immunogen or plasmid DNA plasmid as the format of vaccines. It would also be interesting to see whether the initial public and subsequent private repertoire patterns observed here also applied to other antigens and vaccine modalities.
In future studies, monoclonal antibodies (mAbs) can be isolated from the same study design with known sequence and serve as leads to mine the immune repertoire in search of reads with similar V-genes and CDRs to identify new functional antibodies. This would also enable the assessment of any novel vaccine's capability to elicit antibodies similar to lead mAbs of interest. In addition, extensive analysis of somatic mutation at different time points also enables tracing evolutionary pathways for final functional antibodies. In this way, novel vaccines can be designed and studied with much greater precision to achieve the desired immune responses and protection.
Materials and methods
Animals
C57BL/6 female 6–8 week old mice were purchased from the Jackson Laboratory and maintained in the Department of Animal Medicine at the University of Massachusetts Medical School in accordance with the “Guide for the Care and Use of Laboratory Animals” (National Research Council). All protocols have been reviewed and approved by the Institutional Animal Care and Use Committee (IACUC).
DNA immunization
DNA immunizations were performed using a Helios Gene Gun (Bio-Rad) as described beforeCitation21 following the schedule shown in . 3.6 ug of plasmid DNA encoding gp120-JRFL or empty expression vector (pJW4303) were administered per mouse and per immunization.
Protein inoculation
HIV-1 gp120 JR-FL protein was produced by transient transfection of 293F cells with the vaccine construct using the 293Freestyle system (Thermo Fisher Scientific). Supernatants were collected 72 hours post-transfection and gp120 JR-FL was purified over a lentil lectin agarose affinity column (GE Healthcare). 50 ug per mouse of purified protein dissolved in PBS were administered intraperitoneally in the absence of adjuvant 4 days prior to euthanasia and spleen collection.
Peripheral blood cell collection
Seven days after the 1st DNA vaccination, peripheral blood cells were isolated from whole blood as follows. Whole blood was obtained by facial vein puncture: 150 uL were collected into potassium EDTA microtainers (BD Biosciences) and passed through heparinized hematocrit capillary tubes at the time of collection. Blood was transferred to 1.5 mL polypropylene centrifuge tubes and after centrifugation at 1,000 xg for 10 min @ 4°C, the supernatant (plasma) was stored at −20°C to be later assayed for the presence of anti-gp120 antibodies by ELISA. The soft cell pellet was resuspended in 50 uL rat normal serum and 900 uL Hanks Balanced Salt Solution (HBSS, Mediatech) with 2% FCS, 1mM EDTA, without Ca2+ or Mg2+. After a 5-minute incubation period at room temperature, 2.5 ug (5 uL) of biotinylated TER-119 (BD biosciences) was mixed in and incubated for 10 minutes at room temperature. Next, 25 uL of Streptavidin RapidSpheres (Stemcell Technologies) were added, mixed in, and incubated 3 minutes at room temperature. Tubes were placed on a magnetic rack (Thermo Fisher Scientific) and let stand for 3–4 minutes until the beads were visibly clumped on the side. Collected the supernatant and centrifuged at 1,000 xg for 10 min at 4°C. Resuspended in 200 uL of red blood cell lysis buffer (Sigma) and incubated for 3 minutes on ice. Quenched with 1 mL HBSS 2% FCS 1mM EDTA and pelleted cells at 1,000 xg for 10 min at 4°C. Removed the supernatant and lysed the pellet with 0.5 mL of TRIzol reagent (Thermo Fisher Scientific) for 5 minutes at room temperature. Added 100 uL of chloroform, mixed vigorously, and centrifuged at 12,000 xg for 15 min at 4°C. Moved the aqueous phase to a new tube and added 0.5 volumes of 200-proof ethanol and mixed. Loaded the mix into a Quick-RNA MicroPrep column (Zymo Research) and centrifuged for 30 seconds at 12,000 xg at room temperature. Washed column with 400 uL RNA prep buffer, 700 uL RNA wash buffer, once again with 400 uL RNA wash buffer, and spun at 12,000 xg for 2 minutes. Transferred the column into a new 1.5 mL RNase-free collection tube and pipetted 10 uL RNase-free water onto the membrane. After 3 minutes at room temperature, spun down at 12,000 xg for 1 minute. Stored at −80°C until 5′RACE was performed.
ELISA
ELISA for the detection of gp120-specific antibody in plasma or serum samples was conducted as previously described.Citation11 Plates were coated with the same gp120-JR-FL used for protein inoculation.
Cell staining, magnetic separation and FACS sorting
Spleens were removed after mouse euthanasia and placed on petri dishes with 10 mL RPMI 1640 10% FCS (Thermo Fisher Scientific). Single cell suspensions were obtained by pressing the tissue through a 70 micron nylon mesh into 50 cc conical tubes. The suspension was topped off to 50 mL with RPMI 1640 10% FCS, and centrifuged at 350 xg for 5 minutes at 4°C. The cell pellet was resuspended in Hanks Balanced Salt Solution (HBSS, Mediatech) with 2% FCS, 1mM EDTA, without Ca2+ or Mg2+ (FACS buffer), moved to a 15 mL conical tube, and cells were counted and their density adjusted. For every 100 million cells resuspended in 1 mL the following were added: 8 ug (2 uL) 2.4G2 monoclonal antibody (rat anti-mouse FcγRII/III, BioXcell), and 50 uL normal rat serum. Incubated at room temperature for 3 minutes. Added the following biotinylated antibodies and incubated samples 10 minutes at room temperature: 5 ug (10 uL) TER-119 (BD Biosciences), 5 ug (10 uL) Hamster anti-mouse CD3epsilon (BD Biosciences) and 5 ug (10 uL) Goat anti-mouse IgM (Southern Biotech). Added 125 µL of Streptavidin RapidSpheres (Stemcell Technologies) and incubated at room temperature for 2.5 minutes. Topped up to 5 mL with column buffer, mixed and placed the tube into “the Big Easy silver EasySep magnet” (Stemcell Technologies) for 2.5 minutes. Supernatants containing the depleted samples were decanted into new 50 mL tubes and topped up to 20 mL with ice-cold FACS buffer and kept on ice. Group B samples contained 5–8 million cells, which were pelleted and lysed in TRIzol and frozen at −80°C until sorting of group A samples was finished and RNA extraction was performed. Group A samples were resuspended in FACS buffer at 30 million cells/mL and the following antibodies were added: Biotin-anti-mouse CD138 (BD Biosciences; 100 ng/million cells), V450 Rat Anti-Mouse CD45R (B220) (clone RA3-6B2, BD Biosciences; 40 ng/million cells). Cell suspensions were incubated for 30 min on ice in the dark, topped up to 50 mL with ice-cold PBS, spun down at 4°C and 400 × g for 5 min supernatants were discarded. Cell pellets were again resuspended as above and Neutravidin-PE was added (Thermo Fisher Scientific; 100 ng/million cells). Cells were incubated on ice overnight. Topped up to 50 mL with ice-cold FACS buffer, spun down at 4°C and 400 × g for 5 minutes and discarded supernatants. Repeated once. Resuspended the cell pellets in 1 mL staining buffer and moved to 12 × 75 mm tubes; kept at 4°C until sorting time. Cells were sorted into B220+CD138+ (plasmablasts) and B220+CD138− (B-cells) by FACS using a FACS Aria II (BD Biosciences). After sorting, plasmablast samples contained 1 to 2,000 cells, whereas B220+CD138− B cell samples contained 2 to 5 million cells. Sorted samples were centrifuged and the cell pellets were lysed in TRIzol and chloroform was added according to the manufacturer's recommendations (Thermo Fisher Scientific). 0.2 mL Chloroform were added per 1 mL of TRizol lysate and vigorously mixed. After centrifugation at 12,000 xg for 15 min at 4°C, moved the aqueous phase to a new tube and added 0.5 volumes of 200-proof ethanol and mixed. Group A B220+CD138− B cell samples and group B samples were loaded into an RNEasy Mini Kit column (Qiagen), washed according to manufacturer's instructions, and RNA was eluted in 25 uL of water. Group A plasmablast samples were loaded into a Quick-RNA MicroPrep column (Zymo Research) and processed as described above for peripheral blood samples. RNA was stored at −80°C until 5′RACE was performed.
5′RACE and amplicon generation
5′RACE was performed using the template switch method.Citation22 Single-stranded cDNA (sscDNA) synthesis was carried out using the Mint-2 5′RACE Kit (Evrogen) according to the manufacturer's instructions starting from 3 uL of RNA. sscDNA was purified using 1.8x volumes of AMPure XP beads (Beckman Coulter), washed once with 200 uL 70% Ethanol and eluted in 45 uL water.
The fragments to be sequenced were generated by PCR using specific reverse primers and a universal forward primer which anneals to the sequence introduced by template switch during 5′RACE. In addition, the PCR primers used were fused to the adapters necessary for Ion Torrent template preparation (Adapter full-length P1 and adapter A, Thermo Fisher Scientific). All primers were added from a stock solution at 10 uM. Amplicons were generated by PCR using the Kapa HotStart HiFi kit (Kapa Biosystems) following manufacturer's instructions. Template volumes, annealing temperatures, and extension times were optimized for the different reactions. For kappa V-region, 1.3 uL amplification primers FLP1-M1 and mKappa-AdA with an IonXpress barcode were used, along with 5 uL template (purified sscDNA) in a final volume of 40 uL. The annealing temperature was 62°C; extension for 20 s at 72°C; 30 cycles were performed. The PCR reaction was loaded in a 1.5% agarose gel and the amplicon band at approximately 600 bp was cut and purified with a NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Bethlehem, PA) eluting in 15 uL water. Due to the diversity of the gamma chain constant region sequence in the vicinity of the variable region for the different IgG subclasses, two reverse primers were necessary: the IgG2b reverse primer covered IgG1, IgG2a, and IgG2b sequences; while the IgG3 reverse primer was specific for IgG3. Thus, two separate PCR reactions were conducted to amplify gamma chain V-region amplicons: one with IgG2b and one with IgG3 reverse primer. Gamma chain V-region amplicons were amplified in two phases: first, adding 1.7 uL M1 primer (from Mint-2 kit, Evrogen) and mIgC-179 reverse (which covered all IgG subclasses), along with 10 uL template (purified sscDNA) in a final volume of 50 uL. The annealing temperature was 62°C, extension for 20 s at 72°C; 10 cycles. The entire PCR reaction volume was purified with 0.8x volumes (40 uL) of SPRI size select beads (Beckman Coulter), washed once with 200 uL 85% Ethanol and eluted in 20 uL water. 19 uL of this eluate was used in a second round of PCR with primer FLP1-M1 and mIgG2b-AdA or IgG3-AdA with an IonXpress barcode, in a final volume of 40 uL. The annealing temperature was 62°C, extension for 20 s at 72°C, 25 cycles. The PCR reaction was loaded in a 1.5% agarose gel and the amplicon band at approximately 650 bp was cut and purified with a NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel), eluting in 15 uL water. Primer sequences (5′ to 3′): M1: AAGCAGTGGTATCAACGCAGAGT, FLP1-M1: CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGATAAGCAGTGGTATCAACGCAGAGT, mIgC-179 reverse: CAGACTGCAGKASAGMTG; mIgG2b-AdA: CCATCTCATCCCTGCGTGTCTCCGACTCAGctaaggtaacGATAGGGGCCAGTGGATAGACTGATGG (the lowercase letters represent the IonXpress barcode sequence, in this case IonXpress_001); mIgG3-AdA: CCATCTCATCCCTGCGTGTCTCCGACTCAGtaaggagaacGATAGGGACCAAGGGATAGACAGA (the lowercase letters represent the IonXpress barcode sequence, in this case IonXpress_002); mKappa-AdA: CCATCTCATCCCTGCGTGTCTCCGACTCAGaagaggattcGATGGATACAGTTGGTGCAGCAT (the lowercase letters represent the IonXpress barcode sequence, in this case IonXpress_003).
Library preparation
Purified amplicons were subjected to 2 rounds of size selection using 0.6x volumes of SPRI size select beads (Beckman Coulter) and eluted in 15 uL of water. Concentrations were measured using the Qubit HS dsDNA kit (Thermo Fisher Scientific). IgG1/2a/2b amplicons (amplified with IgG2b-AdA reverse primer) and IgG3 amplicons (amplified with IgG3-AdA reverse primer) were mixed 1:1, and the mix was in turn mixed in equimolar amounts with the kappa chain amplicons. Concentrations were adjusted to 50 pM and the mix was used to template IonSphere Particles using the Ion PGM Template IA 500 Kit (Thermo Fisher Scientific) following manufacturer's instructions. Templated particles were enriched using an Ion OneTouch Enrichment System (Thermo Fisher Scientific).
Sequencing
Enriched templated Ion Sphere Particles prepared as shown above were annealed to the sequencing primer and bound to the sequencing polymerase using an Ion PGM Hi-Q™ OT2 kit (Thermo Fisher Scientific), and loaded into Ion 318 chips and sequenced using an Ion Torrent PGM instrument (Thermo Fisher Scientific) following the manufacturer's instructions but increasing the flow number to 1,350 flows.
Data analysis
Fastq files were downloaded from the Ion Torrent server (Thermo Fisher Scientific) after running the file generator plugin and reads over 400 bases long and with phred quality score >20 were selected using the AbMining toolbox suite.Citation23 The output fasta file was split into blocks of 500,000 reads and submitted to the IMGT High V-quest server.Citation24 The output data were clustered into clonotypes based on unique CDR3 and V-gene usage. Lists of unique V-genes and CDR3s were generated using the advanced filter function in Microsoft Excel and their relative frequencies were calculated with the COUNTIF function. BLAST+Citation25 was utilized to generate pairwise alignments of the unique CDR3s from different mice. Shared CDR3s were identified in this way and their relative proportions for each individual mouse and time point were plotted. All plotting and statistical analyses were carried out in GraphPad Prism.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Material.zip
Download Zip (867 KB)Funding
The study reported here was supported in part by NIH grants 5 U19 AI082676, 5 P01AI082274 and 5R21/R33AI087191, and by Bill and Melinda Gates Foundation grant OPP1033112.
References
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593-602. doi:10.1126/science.1207532
- DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, Varadarajan N, Giesecke C, Dörner T, Andrews SF, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31:166-9. doi:10.1038/nbt.2492
- Tan Y-C, Blum LK, Kongpachith S, Ju C-H, Cai X, Lindstrom TM, Sokolove J, Robinson WH. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin Immunol Orlando Fla. 2014;151:55-65.
- DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2015;21:86-91.
- Sundling C, Zhang Z, Phad GE, Sheng Z, Wang Y, Mascola JR, Li Y, Wyatt RT, Shapiro L, Karlsson Hedestam GB. Single-cell and deep sequencing of IgG-switched macaque B cells reveal a diverse Ig repertoire following immunization. J Immunol Baltim Md 1950. 2014;192:3637-44.
- Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G. Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PloS One. 2014;9:e101322. doi:10.1371/journal.pone.0101322
- Kodangattil S, Huard C, Ross C, Li J, Gao H, Mascioni A, Hodawadekar S, Naik S, Min-debartolo J, Visintin A, et al. The functional repertoire of rabbit antibodies and antibody discovery via next-generation sequencing. mAbs. 2014;6:628-36. doi:10.4161/mabs.28059
- Lefranc M-P, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, Duvergey H, Houles A, Paysan-Lafosse T, et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015;43:D413-422. doi:10.1093/nar/gku1056
- Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558-60. doi:10.1093/nar/27.6.1558
- Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434-9. doi:10.1038/nbt0612-562f
- Wang S, Farfan-Arribas DJ, Shen S, Chou T-HW, Hirsch A, He F, Lu S. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine. 2006;24:4531-40. doi:10.1016/j.vaccine.2005.08.023
- Ruberti F, Cattaneo A, Bradbury A. The use of the RACE method to clone hybridoma cDNA when V region primers fail. J Immunol Methods. 1994;173:33-9. doi:10.1016/0022-1759(94)90280-1
- Hollister K, Chen Y, Wang S, Wu H, Mondal A, Clegg N, Lu S, Dent A. The role of follicular helper T cells and the germinal center in HIV-1 gp120 DNA prime and gp120 protein boost vaccination. Hum Vaccines Immunother. 2014;10(7):1985-92. doi:10.4161/hv.28659
- Racine R, McLaughlin M, Jones DD, Wittmer ST, MacNamara KC, Woodland DL, Winslow GM. IgM Production by Bone Marrow Plasmablasts Contributes to Long-Term Protection against Intracellular Bacterial Infection. J Immunol Baltim Md 1950. 2011;186:1011-21.
- Wang B, Kluwe CA, Lungu OI, DeKosky BJ, Kerr SA, Johnson EL, Jung J, Rezigh AB, Carroll SM, Reyes AN, et al. Facile Discovery of a Diverse Panel of Anti-Ebola Virus Antibodies by Immune Repertoire Mining. Sci Rep. 2015;5:13926. doi:10.1038/srep13926
- Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010;28:965-9. doi:10.1038/nbt.1673
- Haessler U, Reddy ST. Using next-generation sequencing for discovery of high-frequency monoclonal antibodies in the variable gene repertoires from immunized mice. Methods Mol Biol Clifton NJ. 2014;1131:191-203.
- Friedensohn S, Khan TA, Reddy ST. Advanced Methodologies in High-Throughput Sequencing of Immune Repertoires. Trends Biotechnol. 2017;35:203-14. doi:10.1016/j.tibtech.2016.09.010
- Sze DM, Toellner KM, García de Vinuesa C, Taylor DR, MacLennan IC. Intrinsic constraint on plasmablast growth and extrinsic limits of plasma cell survival. J Exp Med. 2000;192:813-21. doi:10.1084/jem.192.6.813
- Calame KL, Lin K-I, Tunyaplin C. Regulatory mechanisms that determine the development and function of plasma cells. Annu Rev Immunol. 2003;21:205-30. doi:10.1146/annurev.immunol.21.120601.141138
- Wang S, Lu S. DNA immunization. Curr Protoc Microbiol. 2013;31:18.3.1-18.3.24.
- Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558-60. doi:10.1093/nar/27.6.1558
- D'Angelo S, Glanville J, Ferrara F, Naranjo L, Gleasner CD, Shen X, Bradbury ARM, Kiss C. The antibody mining toolbox: an open source tool for the rapid analysis of antibody repertoires. MAbs. 2014;6:160-72.
- Li S, Lefranc M-P, Miles JJ, Alamyar E, Giudicelli V, Duroux P, Freeman JD, Corbin VDA, Scheerlinck J-P, Frohman MA, et al. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat Commun. 2013;4:2333. doi:10.1038/ncomms3333
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi:10.1186/1471-2105-10-421
