ABSTRACT
Our previous preclinical studies and a Phase I clinical trial DP6-001 have indicated that a polyvalent Env formulation was able to elicit broadly reactive antibody responses including low titer neutralizing antibody responses against viral isolates of subtypes A, B, C and AE. In the current report, a panel of 62 gp120 immunogens were screened in a rabbit model to identify gp120 immunogens that can elicit improved binding and neutralizing antibody responses and some of them can be included in the next polyvalent formulation. Only about 19% of gp120 immunogens in this panel were able to elicit neutralizing antibodies against greater than 50% of the viruses included in a high throughput PhenoSense neutralization assay when these immuongens were tested as a DNA prime followed by a fixed 5-valent gp120 protein vaccine boost. The new polyvalent formulation, using five gp120 immunogens selected from this subgroup, elicited improved quality of antibody responses in rabbits than the previous DP6-001 formulation. More significantly, this new polyvalent formulation elicited higher antibody responses against a panel of gp70V1/V2 antigens expressing V1/V2 sequences from diverse subtypes. Bioinformatics analysis supports the design of a 4-valent or 5-valent formulation using gp120 immunogens from this screening study to achieve a broad coverage against 16 HIV-1 subtypes.
Introduction
After close to three decades' effort, it is becoming clear that the heterologous prime-boost vaccination approach is the one showing the greatest promise among a long list of vaccination approaches tested for HIV vaccine development. RV144 trial using an ALVAC vector prime and recombinant gp120 protein boost is the only trial of an HIV vaccine where a modest reduction in the risk of HIV acquisition was observed.Citation1 Almost at the same time, a Phase I clinical trial DP6-001 conducted in healthy adult volunteers, using the DNA prime- recombinant gp120 protein boost strategy developed by our team, showed robust immunogenicity including 100% response rate of high level HIV-1 antigen specific antibody and T cell immune responses.Citation2,3 Sera from immunized human sera had low titer neutralizing antibody responses against relatively difficult to neutralize viruses from subtypes A, B, C and E although the Nab titers were poor against the standard Tier 2 viral isolates.Citation2 It is possible that DP6-001 formulation using a polyvalent Env formulation in both DNA prime and protein boost immunizations was able to elicit broad antibody responses confirming findings from our preclinical study that the polyvalent formulations were more effective than monovalent formulations in eliciting broader neutralizing antibody responses.Citation4
However, five gp120 immunogens included in DP6-001 were randomly selected based on the available Env sequences from primary HIV-1 isolated in the mid-1990s. With more primary HIV-1 Env sequences available in the last 10–15 years, it is an important question to understand whether these Env proteins have the same level of immunogenicity in eliciting antibody responses, especially the ability to elicit neutralizing antibody responses, and such information can guide the rational selection of next generation polyvalent Env formulations.
In the current study, we first conducted a high-throughput immunogenicity study in the rabbit model to compare the abilities of individual gp120 immunogens delivered in the form of DNA vaccines to elicit neutralizing antibody responses when the DNA-immunized animals received a fixed protein boost which includes five recombinant gp120 proteins. Our previous studies have demonstrated that recombinant Env proteins alone are not effective in eliciting reliable neutralizing antibodies yet the inclusion of a DNA priming step was able to greatly enhance the immunogenicity of Env proteins to elicit improved Nab.Citation5-8
Our data indicated not every gp120 has the same immunogenicity and only about 10–15% Env was capable of eliciting immune sera neutralizing more than 50% viral isolates in a study panel. Based on this analysis, a new polyvalent Env formulation was produced consisting of five new gp120 immunogens selected from the current screening study. This new polyvalent gp120 formulation showed enhanced potency and improved breadth of antibody responses, in comparison with the previous DP6-001 polyvalent formulation. The new polyvalent gp120 formulation was also able to elicit broader anti-V2 antibody responses which may be involved in ADCC functions. Data presented in this report support the production and testing of the next generation polyvalent Env formulation in a human study.
Result
Immunogenicity testing of a panel of 62 gp120 DNA vaccines in a DNA prime-protein boost design to produce immune rabbit sera for neutralizing activity analysis
In our previous DP6-001 Phase I clinical study in healthy adult volunteers, the polyvalent Env formulation delivered by the DNA prime-protein boost approach was highly immunogenic including the potent and broadly cross reactive Env-specific antibody responses in 100% of the volunteers based on solid phase antibody assays including ELISA and Western Blot analyses.Citation2 Furthermore, immune sera from this study demonstrated weak neutralizing antibody responses in a high-throughput TZM-bl neutralization assay against a diverse panel of primary HIV-1 isolates from subtypes A, B, C and AE which are not easy to neutralize.Citation9-11 However, the titers of Nab activities were relatively low against this panel of primary viruses and the same set of human immune sera was not able to neutralize the standard Tier 2 viruses.Citation2
In the current study, we investigated the possibility of using a new polyvalent Env formulation to generate improved protective antibody responses. The original DP6-001 formulation included four primary gp120 immunogens (92UG037.8 of subtype A, 92US715.6 of subtype B, 96ZM651 of subtype C and 93TH976.17 of subtype AE) and one TCLA gp120 immunogen (Bal) (Supplement Table 1). They were included in the DP6-001 formulation randomly because there were limited intact gp120 molecular clones available at the time. In contrast, the current study included 62 HIV-1 gp120 immunogens covering subtypes A, B, C, D, F, G and CRF01_AE (AE) including one consensus AE (). These Env sequences were originally cloned from HV-1 infected patients in various geographic regions including Africa, North America, South America, Europe and Asia and from different tissues including plasma, PBMCs, lymph nodes, and brainsCitation9,10,12-27 (). The number of gp120 immunogens included in this panel is not same among different subtypes because several rare subtypes have less available molecular clones than other subtypes. More importantly, it is recognized that HIV genotypes could not well predict the neutralizing antibody subtypes.Citation28
Table 1. List of the original HIV-1 viruses from which gp120 DNA vaccines were produced.
Because the central objective of the current study is to screen among a large panel of gp120 immunogens and understand whether any particular gp120 has improved ability in eliciting antibody responses than the previous DP6-001 formulation, the study was designed to focus on gp120 immunogens with such potential. Individual gp120 sequences were cloned into the same DNA vaccine vector pJW4303 and used as the priming vaccine in a standard DNA prime-protein boost strategy in rabbits (), which we have shown to be superior to DNA or protein immunization alone.Citation2,4-8,29 The reason to include a fixed 5-gp120 protein as the boost has multiple considerations: 1) it will take significant time and resources to produce over 60 individual recombinant gp120 proteins, and the variation on the quality and purity of these proteins may further impact the immunogenicity results; and 2) a polyvalent Env formulation is more capable to expand the breadth of antibody responses than a single Env protein boost. A polyvalent Env formulation may also minimize the bias if only one fixed protein is used as the boost. The inclusion of these five gp120 proteins in the boost was based on the availability of highly purified quality and a large quantity supply.
Figure 1. The rabbit immunization and serum collection schedules for individual gp120 immunogen screening study.
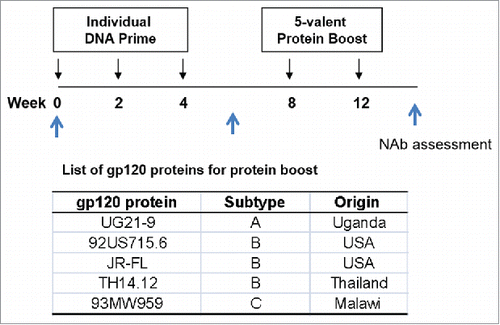
New Zealand White (NZW) rabbits were first immunized by one of 62 individual DNA vaccines three times by a gene gun followed by two boost immunizations with a fixed 5-gp120 protein mix (UG21-9, 92US715.6, JR-FL, TH14-12 and 93MW959) delivered subcutaneously along with the Incomplete Freund's Adjuvant (IFA). IFA was used in the current study because it was used in previous published studies from our group so the data from the current study can be compared with the previous data. Sera were collected before immunization and after either DNA prime or protein boost. The neutralizing antibody analysis was done by a high throughput commercial “PhenoSense Assay” which included a target panel of 13 primary viral isolates (92RW020 and 94UG103 of subtype A, AC10.0.29, PVO.4, QH0692.42, SC422661.8, and JRCSF of subtype B, 93IN905 and 98CN006 of subtype C, 92UG046 and 94UG114 of subtype D, and 92TH021 and CMU02 of subtype AE) and 3 TCLA strains (MN, NL43 and SF162).
Neutralizing activities of immune rabbit sera elicited with different gp120 DNA prime
Among 62 gp120 immunogens tested in this single DNA prime and 5-valent gp120 protein boost standard design, they can be divided into three subgroups based on the breadth of neutralizing activities (). The first subgroup of 12 gp120 immunogens (about 19.3% of this group) elicited rabbit immune sera neutralized more than 50% of the viral isolates in this testing panel, albeit at low titers. The second subgroup of 10 (16.1%) neutralized between a quarter to up to one half of the viral isolates (25–49%). The remaining 40 (64.5%) gp120 immunogens elicited much narrow neutralizing activities (6–24% of the viral isolates).
Table 2. The neutralizing antibody breadth induced by various gp120 DNA vaccine prime in rabbits.
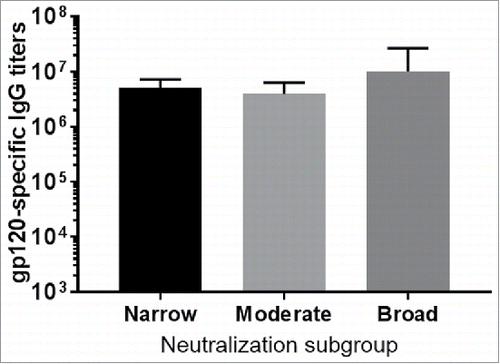
To evaluate whether stronger neutralizing antibody responses correlated with higher binding antibody responses, titers of gp120-specific antibody responses for the above three subgroups were measured. Overall there was no significant difference in titers of binding antibody responses among three neutralization subgroups (). Therefore, gp120 immunogens in the best neutralizing antibody subgroup did not elicit unusually higher antibody responses than gp120 immunogens in other subgroups, and the level of binding antibody responses was not responsible for neutralizing antibody responses.
Figure 2. The gp120-specific antibody titers as measured by ELISA. Rabbit sera from individual gp120 screening study were tested against the fixed five gp120 proteins use in the boost. The end titration titers shown are the averages with standard deviation of each neutralization subgroup (narrow, moderate or broad).
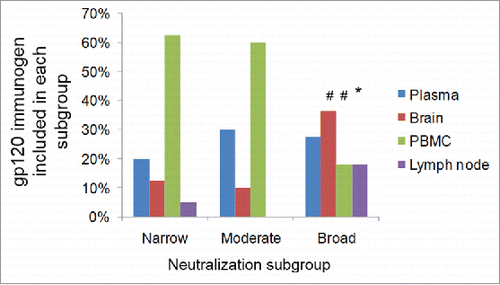
The gp120 immunogens that were best able to elicit Nab responses are from different subtypes and do not appear to be clustered closely based on the gene sequence analysis (). The neutralizing antibody profile was further analyzed for its relationship to the sources of gp120 immunogens (). In the best Nab subgroup, there are similar numbers of gp120 immunogens from all four major tissue sources (plasma, brain, PBMC and lymph node), with no indication that any particular source may be responsible for the improved breadth of Nab responses. When different sources are analyzed, PBMC represented a higher percentage of gp120 immunogens (60%) in both moderate and narrow Nab subgroups. But this may only reflect the fact that the PBMC source gp120 immunogens are the majority of gp120 immunogens included in the current study (). On the other hand, most of brain source and lymph node source gp120 immunogens in the current study are found in the best Nab subgroup (). Due to the small sample size, this observation will need future studies to establish any true correlation.
Figure 3. The phylogenetic tree analysis of the amino acid sequences of 62 gp120 DNA immunogens included in the screening study. The arrows indicate those gp120 immunogens in the broad NAb subgroup.
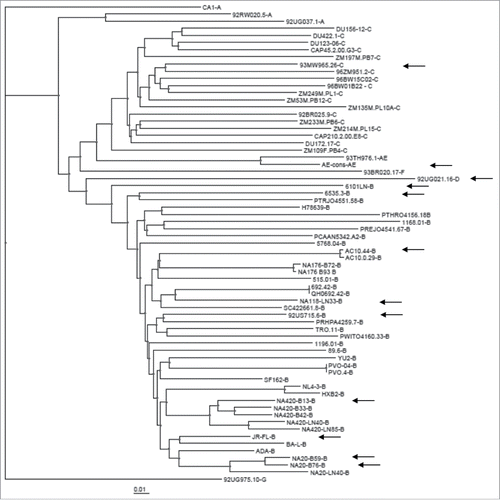
Figure 4. The tissue sources of gp120 immunogens identified in different Nab subgroups. The numbers shown are percentages of each source in a particular subgroup. The sum of all sources within one subgroup equals to 100%. The statistical significance is calculated using Fisher exact test for the difference of one source between broad and moderate subgroups, or between broad and narrow subgroups, # indicating p < 0.01 and * indicating p < 0.05.
The V1/V2 loop of HIV-1 Env proteins is the region with the highest sequence variation including the length of V1/V2 as previously published.Citation30 Lengths of the V1 and V2 loops among the gp120 did not correlate with breadth of neutralization (supplement Fig. 1).
Antibody responses elicited with the new 5-valent gp120 DNA prime-protein boost formulation
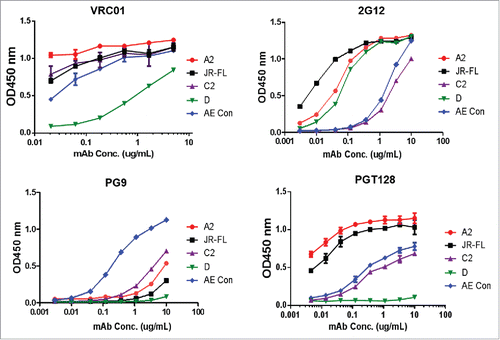
Next, the new polyvalent vaccine was formulated with five gp120 immunogens selected from the broad Nab group to examine whether the new formulation is able to elicit better neutralizing antibody responses than the previous DP6-001 formulation. One gp120 immunogen from each of five major subtypes (subtypes A, B, C, D and AE) was selected from the broad Nab group () to expand the coverage of HIV-1 viral Env spectrum. The preliminary antigenicity analysis using ELISA with four representative mAbs demonstrated that these five gp120 immunogens preserved broad epitopes such as those recognized by the CD4bs mAb VRC-01, glycan specific mAbs 2G21 and PGT128. The subtype AE gp120 immunogen even showed good binding by PG9, a well-known trimer specific mAb PG9. The overall high levels of binding were observed except subtype D gp120 immunogen which had much lower binging with VRC-01 and PGT128 ().
Table 3. New 5-valent gp120 immunogens.
Figure 5. The antigenicity analysis (binding antibody) of the 5 gp120 immunogens included in the new polyvalent formulation by gp120-specific bnAbs VRC01, 2G12, PG9 and PGT128 in serial dilutions. ELISA was run with individual gp120 proteins (A2, JR-FL, C2, D or AE con) coated on the microtiter plates as indicated. Curves are shown as the average of two duplicate wells.
The more complete binding affinities of these five gp120 immunogens were measured with a bigger panel of gp120-specific human and rabbit mAbs covering a wide range of epitopes (including C1, C2, C5, V2, V3, CD4bs and glycan) and the IgG-CD4 fusion protein, showing an overall high level of binding affinities () except a few epitopes on gp120 immunogen from subtypes D which is not unexpected given the significant difference between subtype D and other HIV-1 M group subtypes.
Table 4. Antigenicity analysis of new 5-vlaent gp120 immunogens.
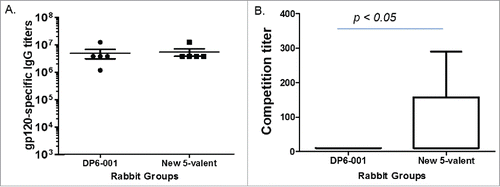
The relative immunogenicity between DP6-001 and the new 5-valent was compared in rabbits using the standard three-time DNA immunizations followed with twice protein immunizations using the matched five gp120 immunogens. The levels of gp120-specific binding antibodies as measured by standard ELISA were very similar between two polyvalent formulations. Both formulations are highly immunogenic in eliciting high titer gp120-specific IgG responses (). However, further studies identified the difference in quality of antibody responses. Our previous studies in rabbits and human trial demonstrated that the DNA prime-protein boost approach is capable of eliciting CD4bs antibodies. In the current study, the new polyvalent formulation was able to induce higher levels of CD4bs antibody responses than the previous DP6-001 formulation based on the competition of CD4bs mAb b12 ().
Figure 6. Analyses of antibody responses in rabbit immune sera elicited by either DP6-001 or the new polyvalent formulation. (A) The gp120-specific antibody titers were measured by ELISA against the autologous 5 gp120 proteins included in either DP6-001 or new polyvalent formulation. (B) The ability of rabbit sera to outcompete binding of CD4bs-specific mAb b12 to JR-FL & VSV-G pseudotyped viruses. Competition titer is defined as the serum dilution preventing 50% of pseudoviral binding to the ELISA plate. The statistical analysis was done with t-test.
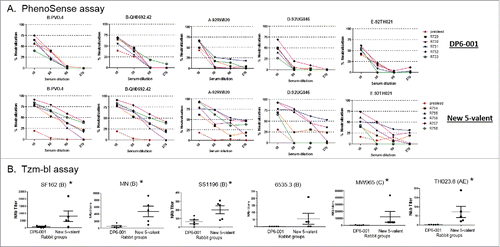
The neutralizing activities of rabbit immune sera were analyzed by two different types of assays. The first is the high throughput commercial PhenoSense assay by Monogram. Individual serum titration curves showed an upward shift of new polyvalent rabbit sera when compared with that of the DP6-001 sera against 5 viruses in this assay (). The second neutralization study was conducted by the TZM-bl assay conducted at Duke University. Two Tier 1A TCLA and four Tier 2 viruses were included in the TZM-bl assay. In both assays, the neutralizing activities of the new polyvalent Env formulation rabbit sera were clearly higher than the rabbit sera elicited by the DP6-001 formulation ().
Figure 7. Neutralizing activity in rabbit sera elicited by ether DP6-001 and new polyvalent formulation. (A) Percent of inhibition by rabbit immune sera in the PhenoSense assay. Each curve represent serum from one rabbit. (B) Neutralizing titers in rabbit immune sera as tested by TZM-bl assays. Five rabbit sera were tested for each formulation. The mean titers with standard deviations are shown. NAb titer is defined as the serum dilution capable of inhibiting 50% of viral infection. Rabbit sera collected two weeks after the final boost immunization were tested for their ability to neutralize various viruses. The statistical significance (p < 0.05) is indicated with “*”, using one way ANOVA.
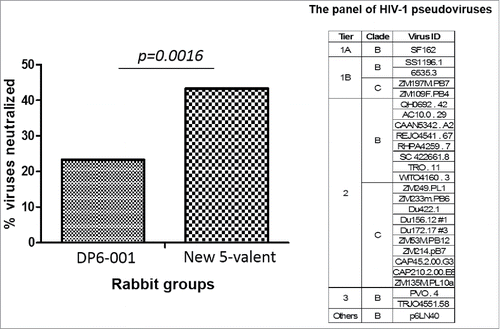
An additional neutralization assay was conducted against a panel of selected viruses including those from different standard tiers (). At 1:10 serum dilution, rabbit sera elicited by the new polyvalent formulation neutralized about 40% of viruses in this panel while the previous DP6-001 vaccine rabbit sera neutralized only slightly above 20% of viruses.
Figure 8. Neutralizing breadth of rabbit sera elicited by DP6-001 and new 5-valent gp120 vaccines against a panel of 26 HIV-1 pseudoviruses as shown in TZM-bl assay. The percentage of virus neutralized was calculated based on the positive neutralization events of 5 rabbits/each group against 26 viruses. The positive NAb is defined as the serum capable of inhibiting = />50% of viral infection at > = 1:10 serum dilution. The statistical significance is indicated and analyzed using Fisher exact test. Rabbit sera collected two weeks after the final boost immunization were used for the assay.
The profile of epitope-specific antibody responses in rabbit sera elicited by the new polyvalent formulation was analyzed by peptide array binding. It showed a pattern of antibody responses against epitopes spreading to all constant and variable domains including the V2 domain in multiple gp120 immunogens (). Further detailed analysis was done using the gp70V1/V2 chimeric antigen system expressing V1/V2 domains from gp120 immunogens included in both DP6-001 and the new polyvalent formulations. Sera elicited by the new polyvalent formulation had higher antibody responses against this group of gp70V1/V2 antigens than the sera elicited by DP6-001 formulation (). Because ADCC activities against V2 region was found to be an immune correlate of protection in RV144 studies, the ability of new 5-valent formulation to elicit higher antibody responses against diverse V1/V2 immunogens is highly significant.
Figure 9. Evaluation of serum antibodies binding in new 5-valent vaccine immunized rabbit to linear peptides spanning gp120 antigens by JPT peptide microarray. The peptides were 15-mer with 11-overlap. The peptide panels A, B, C, D and E represent autologous immunogens in the new 5-valent vaccine: 92UG037.8 (subtype A), JR-FL (subtype B), 93MW965.26 (subtype C), 92UG021.16 (Subtype D), and AE consensus (CRF_01 AE). Peptide panel M is the Group M consensus sequence. The peptide panels LN40 and B33 are two additional subtype B viruses. The signal intensities were the group mean of the normalized values with pre-bleed background subtracted. Rabbit sera collected two weeks after the final boost immunization were used for peptide microarray assay.
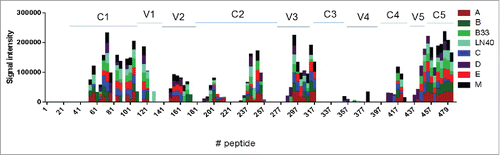
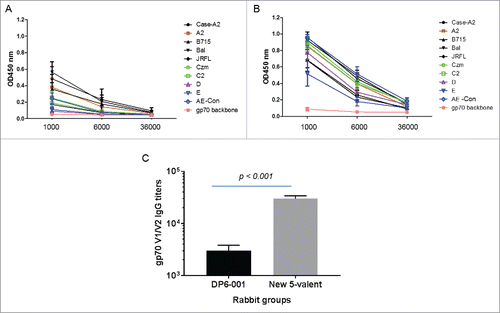
Figure 10. ELISA analysis of gp70V1/V2-specific antibody responses in rabbit sera induced by either DP6-001 (Panel A) or the new 5-valent vaccines (Panel B). Ten individual gp70V1/V2 scaffold proteins were used as coating antigens covering subtypes A, B, C, D and AE. The gp70V1/V2 antigens autologous to the DP6-001 were A2 (92UG037.8, subtype A), US715 (92US715.6, subtype B), Ba-L (subtype B), Czm (96ZM9651, subtype C) and AE (93TH967, CRF_01 AE). The gp70V1/V2 antigens autologous to the new 5-valent were A2 (92UG037.1, subtype A), JR-FL (subtype B), C2 (93MW965.26, subtype C), D (92UG021.16 Subtype D) and AE-con (AE consensus, CRF_01 AE). The Case-A2 gp70V1/V2 is a commonly used subtype B antigen as the reference control. The empty gp70 vector was used as the negative control. Rabbit sera collected two weeks after the final boost immunization were used in this study. Each curve represents one rabbit serum from each immunogen group against one gp70V1/V2 antigen. Panel C shows the mean antibody titers between A and B with statistical significance indicated using the t-test.
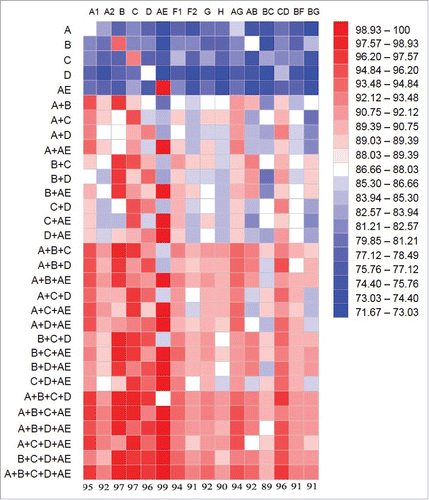
The current study further supports the value of an improved polyvalent gp120 formulation. Because of the extremely high mutation rate of HIV-1 viruses, it has been one of the key challenges in developing an HIV vaccine that can cover enough breadth of circulating HIV viral isolates. We used a bioinformatics model to see the impact of polyvalency to the coverage of circulating HIV-1 isolates (). In this model, the sequence homology was analyzed between gp120 sequences included in a polyvalent formulation and the consensus sequences from each of the 16 key subtypes of HIV-1 M group. The heat map method was used to show the level of homology. It is clear that the homology is very low when only one gp120 immunogen is tested – only the matching subtype shows some degree of homology but not to any other subtypes. When the valency is increased from two-valent to five-valent, the homology increased to over 90% to any subtype. Actually, even four-valent formulation is already able to achieve high homology, especially the one without the subtype D (A+B+C+AE). The results from the bioinformatics model and the neutralizing data reported here provide the direction for future HIV vaccine development.
Figure 11. Bioinformatics analysis on the coverage between various gp120 immunogen formulations and the consensus sequences of HIV-1 subtypes. The A, B, C, D and AE indicated at the left represent the new 5-valent gp120 immunogens: 92UG037.8 (subtype A), JR-FL (subtype B), 93MW965.26(subtype C), 92UG021.16 Subtype D), and AE consensus (CRF_01 AE). Sixteen HIV-1 subtypes are indicated on the top, representing the consensus sequences of each subtype. Heat map analysis was used to indicate the level of coverage, with warmer color indicating higher levels of coverage. Numbers indicated at the bottom of heat map are the percentages of sequence coverage between the five-valent gp120 formulation and 16 different subtypes.
Discussion
In the last three decades, HIV vaccine development has put major emphasis on the optimal vaccine delivery approaches and the induction of broad neutralizing antibody responses. The unexpected protection results of RV144 trial suggested the heterologous gene-based vaccine prime and recombinant protein vaccine boost offers an alternative approach based on non-neutralizing antibodies. An improved polyvalent Env formulation offers potential value in augmenting the protection afforded in RV144 which only included Env immunogens from two subtypes. Our early rabbit immunogenicity study showed that randomly selected polyvalent gp120 formulations were able to elicit statistically significant higher neutralizing antibody responses in rabbit immune sera than any of the individual gp120 immunogens included in those polyvalent formulations.Citation4 The 5-valent DP6-001 formulation tested in a subsequent Phase I human clinical study elicited broadly reactive antibody responses against gp120 antigens from subtypes A to G and low titer but broad neutralizing antibody responses in a high throughput PhenoSense assay.Citation2 Recently, ALVAC/Pentavalent gp120 protein (B/E/E/E/E) vaccine with increased diversity of gp120 motifs in the immunogen elicited a broader antibody response and enhanced protection in a SHIV challenge model in macaques,Citation31 supporting the importance to develop polyvalent HIV-1 vaccines.
In the current study, we investigated the possibility to further improve the quality of antibody responses based on the optimal selection of gp120 immunogens in a new polyvalent formulation, Several important pieces of information were learned. First, it appears that not all the gp120 immunogens are created equal in their ability to elicit high quality antibody responses as measured by neutralizing activities. Second, the new polyvalent formulation with selected gp120 immunogens was able to elicit better neutralization antibodies than the original DP6-001 which was randomly formulated. While the sera elicited by the new polyvalent formulation still could not neutralize the standard Tier 2 panel of viral isolates, the improvement on neutralizing antibodies against other viruses was clear including both breadth and potency of Nab using the sera elicited by DP6-001 as the control. Third, the new polyvalent formulation was also able to elicit higher antibody responses against a diverse group of gp70V1/V2 antigens. As we learned from RV144 trial, ADCC mediated through V2 epitopes may correlate to the protection efficacy.Citation32 It is possible that a better gp120 immunogen may have the structure features that are beneficial to the induction of both Nab and ADCC responses. IgA may affect ADCC as learned from RV144 but there are many IgA subtypes in rabbit which makes the measurement of rabbit IgA more difficult.Citation33 Future human studies are needed to establish the improvement of ADCC with the new polyvalent formulation.
The current study was unable to identify the exact factors contributing to those 19% gp120 immunogens that are able to prime the immune system to elicit better neutralizing antibody responses. It is possible that the tissue sources from which the original HIV-1 primary viruses were isolated may play a role because gp120 immunogens isolated from the brain and lymph are mainly discovered from this subgroup. It was reported in literature that HIV-1 viruses isolated from different tissues may have differences in target cell type preference (T cell tropic vs. macrophage tropic), affinity to CD4 and sensitivity to different neutralizing mAbs.Citation34-36 We further observed that a pair of two primary gp120 immunogens isolated from the same patients with high sequence homology but different CD4 affinity and profile of Nab mAb sensitivity showed different immunogenicity in eliciting Nab responses.Citation37 Future studies need to understand the impact of tissue sources to the selection of viral Env products for viral fitness and/or structure modifications such as type of glycosylation.
The bioinformatics model analysis conducted in the current report pointed to the theoretical basis for a polyvalent Env HIV vaccine. While the current analysis is oversimplified by using a consensus gp120 sequence to represent a full subtype of gp120 sequences, the collection of 16 consensus gp120 sequences does cover the diverse scope of HIV-1 gp120 immunogens. It is quite interesting to see that the coverage by the new 5-valent gp120 formulation against these 16 HIV-1 subtypes can reach greater than 90. However, the sequence homology between any one of these five gp120 immunogens to these 16 subtype consensus gp120 sequences was very low unless to its autologous subtype. With the increase on the numbers of gp120 immunogens in a formulation, the coverage to 16 subtypes was improved. These data validated the polyvalent formulation as a key principle in vaccinology and provided strong argument to the use of polyvalent formulation design even to a highly mutated pathogen like HIV-1. A number of recent studies also demonstrated the effectiveness of polyvalent Env formulations even such formulations only including multiple Env immunogens from 1–2 subtypes.Citation31,38
The bioinformatics analysis also showed that if subtype D is not considered a major target viral subtype, then the four-valent formulation including four gp120 immunogens selected from the current study will also achieve a very high percentage of coverage against different subtypes. A clinical formulation based on these four gp120 immunogens (subtypes A, B, C and AE) has been successfully produced from CHO cells under GMP and is scheduled to be tested as a matched polyvalent DNA prime – protein boost vaccine in a clinical trial to be run by the HIV Vaccine Trial Network (HVTN). Data from future clinical studies will further validate the findings learned from the current rabbit immunogenicity study.
Materials and methods
DNA vaccines
Sixty-two individual plasmid DNA vaccines expressing HIV-1 gp120 proteins from subtypes A, B, C, D, AE, F and G () were constructed utilizing the same vector pJW4303 at NheI and BamHI sites under the tPA leader.Citation39 The expression of gp120 proteins from each individual DNA vaccine was verified by transient transfection of 293T cells and Western blot analysis as previously described.Citation4 The DNA vaccines were purified using QIAGEN Mega DNA prep kit (Valencia, CA) for rabbit immunization studies.
Recombinant gp120 proteins
Nine recombinant gp120 proteins, HIV-1 subtypes A (92UG037.8, UG21-9), B (92US715.6, JR-FL, TH14-12), C (93MW959, 93MW965.26), D (92UG021.16) and AE (consensus) (Supplement Table 1), included in the current study were produced from stably transfected CHO cell lines as previously described.Citation2,4 CHO cell cultures were harvested and gp120 proteins were purified over a lectin column, and further polished by the ion-exchange and size-exclusion chromatography as needed. The quality of purified gp120 proteins was verified by SDS-PAGE and Western blot analysis. These gp120 proteins were used as the protein boost immunizations in rabbit studies, and as antigens to evaluate the Env-specific antibody responses by ELISA, and the binding affinity analysis by using the forteBio Octet QKe system.
gp70V1/V2 scaffold proteins
Nine recombinant gp70V1/V2 proteins were generated in the current study. The V1/V2 regions from HIV-1 subtypes A (92UG037.8), B (92US715.6, JR-FL, Case-A2), C (96ZM965, 93MW965.26), D (92UG021.16) and AE (93TH967, consensus) was individually PCR-amplified from relevant gp120 DNA vaccines, and subcloned into the mammalian expression vector pJW4303 containing the MLV gp70 backboneCitation40 at the KpnI and BamHI cloning sites. The V1/V2 was fused at the C-terminal of the gp70 protein with a 6xHis tag at N-terminus of the fusion protein. After DNA sequencing verification of the construct and large DNA prep using QIAGEN kit, the 293T cells were transfected with each gp70V1/V2 plasmid DNA to express the gp70V1/V2 proteins. The transfected 293T cell supernatant was harvested at 72 hours after the transfection and gp70V1/V2 proteins were purified by using a nickel affinity column. The purity of gp70V1/V2 proteins were verified by SDS-PAGE and their reactivity was analyzed by Western-blot analysis using a homemade anti-V1/V2 polyclonal rabbit serum.
Env-specific antibodies
The CD4 binding site specific monoclonal antibody, VRC01,Citation41 was obtained from the AIDS Research & Reference Reagent Program. The glycan specific mAb PGT128Citation42 was provided by International AIDS Vaccine Initiative (IAVI) and the V2 quaternary mAb PG9Citation43 was provided as a gift from Dr. Dennis Burton at Scripps Institute. The glycan mAb, 2G12,Citation44 were purchased from Polymun (Klosterneuburg, Austria). The V2-specific mAb 2158Citation45 was produced by Dr. Susan Zolla-Pazner's group at Mount Sinai School of Medicine. The rabbit mAbs R15 (C1), R20 (V3), R53 (C4) and R13 (C5) were generated in-house from gp120 immunized rabbit.Citation46,47
Rabbit immunization
New Zealand White (NZW) rabbits of 6–8 weeks' old were used in the current study with two DNA prime-protein boost study designs. The rabbits were purchased from Harlan Laboratories (Indianapolis, IN) and housed in the animal facility managed by the Department of Animal Medicine at the University of Massachusetts Medical School (UMMS) in accordance of the protocol approved by UMMS' Institutional Animal Care and Use Committee (IACUC).
In the first gp120 antigen screening study, each rabbit received one individual DNA vaccine prime followed by a fixed five gp120 protein mix boost consisting of gp120 proteins from HIV-1 subtypes A (UG21-9), B (92US715.6, JR-FL, TH14-12) and C (93MW959). In the first study, each group included three NZW rabbits. In the second polyvalent formulations comparison study, each rabbit received either the DP6-001 or the new polyvalent DNA prime followed by the boost of gp120 proteins matching the polyvalent gp120 immunogens used in the DNA priming. In the second study, each group included five NZW rabbits.
In both studies, rabbits received three DNA immunization at Weeks 0, 2, and 4, followed with the protein boosts delivered at Weeks 8 and 12. Sera were collected two weeks prior to the first immunization and two weeks after each animal immunization. DNA vaccines were delivered by a Bio-Rad Helios gene gun using 1-micron gold beads at a ratio of 2 µg of DNA per milligram of gold beads. Each animal received 36 µg of DNA at each immunization time point. For boost immunization, 50 µg of gp120 proteins (10 µg for each gp120 protein) was diluted in 500 µL PBS and mixed with 500 µL Incomplete Freund's Adjuvant (IFA). The 1 mL adjuvanted protein solution was then injected subcutaneously into the back of rabbits at multiple sites.
Enzyme linked immunosorbent assay (ELISA)
ELISA was run to detect gp120 protein or gp70-V1/V2 protein specific IgG responses in 96 well microtiter plates as previous described.Citation4,8 Serial diluted rabbit immune sera were added to plates coated with either gp120 or gp70-V1/V2 proteins, followed by incubation with biotinylated anti-rabbit IgG and a streptavidin horseradish peroxidase subsequently. For gp120 antigenicity analysis, the plates coated with various gp120 proteins were incubated with Env-specific human mAbs at different concentrations. Then the plates were incubated with biotinylated anti-human IgG and a streptavidin horseradish peroxidase subsequently.
The plates were finally developed for 3 min in 100 µL of a 3,3′5,5′-tetramethylbenzidine substrate. The OD values were read after stopping the reaction with 2N H2SO4. Endpoint titers of rabbit sera were defined as the last dilution of a serially diluted serum sample with greater than double the background optical density of a preimmune serum sample.
CD4bs-specific mAb b12 competitive binding assays
Competitive binding assays were performed as previously described.Citation48,49 JR-FL pseudovirus and Vesicular Stomatitis Virus (VSV) glycoprotein were produced with the pSG3ΔEnv backbone in 293T cells. Briefly, after coating the microtiter plates with 50 µL of mAb b12 at 5 µg/mL for 1 hr at room temperature, the serially diluted rabbit sera was incubated with pseudovirus correlating to 2.5 ng of p24/well for 1 hr. Then, the virus/sera mixture was incubated on the ELISA wells for 3 hrs at room temperature. Plates were then overlayed with 10,000 TZM-bl cells per well and incubated for 48 hrs at 37°C. Luciferase activity was determined per the manufacturer's instruction (Promega). Data is reported as the serum dilution at which the luciferase signal is reduced by 50% compared to a serum negative control.
HIV-1 neutralization assays
Two types of HIV-1 neutralization assays were performed in this study. The first neutralization assay (PhenoSense Assay, Monogram, Inc) used recombinant viruses pseudotyped with HIV-1 envelope proteins and a firefly luciferase indicator gene.Citation2,50,51 The panel of HIV-1 pseudoviruses included three TCLA strains (MN, NL43 and SF162), and 13 primary isolates including subtype A (92RW020 and 94UG103), B (AC10.0.29, PVO.4, QH0692.42, SC422661.8, and JRCSF), C (93IN905 and 98CN006), D (92UG046 and 94UG114), and AE (92TH021 and CMU02). Rabbit sera elicited from either one gp120 DNA prime – 5-valent protein boost, or 5-valent DNA prime and 5-valent protein boost (DP6-001 or the new polyvalent formulation) were tested in this high throughput neutralization assay with starting serum dilution at 1:10. The rabbit prebleed was used as the negative control and human HIV-1 infected patient serum N16 was used as the positive control. Neutralizing activity was calculated as the percent inhibition of viral replication (luciferase activity) at each antibody dilution compared with the prebleed-negative control: % inhibition = (1 – (luciferase + Ab/luciferase – Ab)) × 100. The specificity control was composed of a virus pseudotyped with an aMuLV envelope. An HIV-serum combination was considered to have positive neutralization if the inhibition of HIV was at least 50% and >3X higher IC50 than the same plasmas tested with aMuLV while the prebleed was not scored positive.
The second TZM-bl cell based neutralization assay was done as previously described,Citation6,11 either at Dr. Montefiori's lab at Duke University or in house at UMMS. The assays done at Duke titrated the neutralizing antibody titers against HIV pseudoviruses from subtype B (SF162, MN, SS1196, 6536.3), C (MW965) and AE (TH023.6). The in house neutralization assays were done against a panel of 14 subtype B pseudoviruses and 12 subtype C pseudoviruses as listed (), covering Tiers 1A, 1B, 2 and 3, using DP6-001 or new polyvalent immunized rabbits at 1:10 serum dilution. Neutralization was calculated as the percent change in luciferase activity in the presence of preimmune sera versus that of luciferase activity in the presence of immune sera ((Preimmune RLUs – Immune RLUs)/(Preimmune RLUs))*100.
ForteBio Octet QKe assay
The binding kinetics of mAbs to gp120 protein were studied using a ForteBio Octet QKe instrument in a 96-well format following the manufacturer's instructions. Protein A coated sensors (ForteBio) were loaded with Env-specific rabbit mAbs R15 (C1), R20 (V3), R53 (C4) and R13 (C5), or protein G coated sensors were coated with IgG-CD4, Env-specific mAbs VRC01 (CD4bs), PGT128 (Glycan), 2G12 (Glycan) or 2158 (V2), at 10 µg/ml diluted in the kinetics buffer (PBS/0.1%BSA/0.002%Tween 20). After capture, a 1-min wash in loading buffer removed excess unbound Ab to establish a new baseline signal. The biosensor tip was then put into wells containing the selected gp120 proteins at three-fold serially diluted concentrations from 100 to 11 nanomolar (nM), in loading buffer. The mAb-Env protein association on-rate and off-rate were measured. KD values were calculated as the ratio of off-rate/on-rate. The sensorgrams were corrected with the blank reference and fit with the software ForteBio Data Analysis package 7.0 using a 1:1 binding model with the global fitting function.
Linear epitope mapping by peptide microarray
To map the linear epitope recognition in immune rabbit sera, the rabbit sera collected at 2 weeks after the second protein boost and pre-bleed as control were analyzed against a library of HIV-1 Env linear peptides using JPT peptide microarray as previously reported.Citation52,53 Peptide libraries consisting of 15-mers overlapped by 12 amino acids were printed on glass slides, covering the full length of consensus gp120 Env from HIV-1 group M consensus, subtypes A, B, C, D and CRF01_AE including gp120 derived from vaccine strains. Briefly, the peptide microarray was performed using a Tecan HS4000 Hybridization WorkStation. All arrays were blocked with Superblock T20 PBS blocking buffer for 0.5 hour at 30°C with heat inactivated plasma diluted 1:100 in SuperBlock T20. The arrays were incubated for 45 minutes at 30°C with anti-IgG Cy5 secondary antibody (1.5 ug/ml final concentration). PBS containing 0.1% Tween was used for each of the washes between all steps. Arrays were scanned at 635 nm using an Axon GenePix 4300 Scanner (Molecular Devices, Sunnyvale, CA, USA) at a PMT setting of 600, 50% laser power. Images were analyzed using GenePix Pro 7 software (Molecular Devices).
Bioinformatics analysis
The coverage percentages of monovalent, two-valent, three-valent, four-valent, and five-valent gp120 matching current circulating HIV-1 isolates, including Clade A1, A2, B, C, D, E, F1, F2, G, H, AG, AB, BC, CD, BF and BG, were determined. The heatmap of coverage percentage was constructed by JColorGrid softwareCitation54 with input gp120 protein sequences.
Statistical analyses
To analyze the statistical significance of antibody responses induced by different vaccination regimens, One Way ANOVA, t-test and Fisher Exact tests were performed. The significance difference was considered when p < 0.05.
Abbreviations
| DP6-001 | = | an HIV-1 DNA prime-protein boost vaccine |
| Env | = | HIV-1 envelope glycoprotein |
| ELISA | = | enzyme-linked immunosorbent assay |
| HIV-1 | = | human immunodeficiency virus type 1 |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Material.zip
Download Zip (228.6 KB)Funding
This study was supported in part by NIH grants R01 AI065250, 5 U19 AI082676, 5 P01AI082274 and 5R21/R33AI087191, and Bill and Melinda Gates Foundation grant OPP1033112.
References
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209-20. doi:10.1056/NEJMoa0908492. PMID:19843557
- Wang S, Kennedy JS, West K, Montefiori DC, Coley S, Lawrence J, Shen S, Green S, Rothman AL, Ennis FA, et al. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine. 2008;26(31):3947-57. doi:10.1016/j.vaccine.2007.12.060. PMID:18724414
- Bansal A, Jackson B, West K, Wang S, Lu S, Kennedy JS, Goepfert PA. Multifunctional T-cell characteristics induced by a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine regimen given to healthy adults are dependent on the route and dose of administration. J Virol. 2008;82(13):6458-69. doi:10.1128/JVI.00068-08. PMID:18448544
- Wang S, Pal R, Mascola JR, Chou TH, Mboudjeka I, Shen S, Liu Q, Whitney S, Keen T, Nair BC, et al. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology. 2006;350(1):34-47. doi:10.1016/j.virol.2006.02.032. PMID:16616287
- Wang S, Arthos J, Lawrence JM, Van Ryk D, Mboudjeka I, Shen S, Chou TH, Montefiori DC, Lu S. Enhanced immunogenicity of gp120 protein when combined with recombinant DNA priming to generate antibodies that neutralize the JR-FL primary isolate of human immunodeficiency virus type 1. J Virol. 2005;79(12):7933-7. doi:10.1128/JVI.79.12.7933-7937.2005. PMID:15919951
- Vaine M, Wang S, Hackett A, Arthos J, Lu S. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine. 2010;28(17):2999-3007. doi:10.1016/j.vaccine.2010.02.006. PMID:20170767
- Vaine M, Wang S, Liu Q, Arthos J, Montefiori D, Goepfert P, McElrath MJ, Lu S. Profiles of human serum antibody responses elicited by three leading HIV vaccines focusing on the induction of Env-specific antibodies. PLoS One. 2010;5(11):e13916. doi:10.1371/journal.pone.0013916. PMID:21085486
- Vaine M, Wang S, Crooks ET, Jiang P, Montefiori DC, Binley J, Lu S. Improved induction of antibodies against key neutralizing epitopes by human immunodeficiency virus type 1 gp120 DNA prime-protein boost vaccination compared to gp120 protein-only vaccination. J Virol. 2008;82(15):7369-78. doi:10.1128/JVI.00562-08. PMID:18495775
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108-25. doi:10.1128/JVI.79.16.10108-10125.2005. PMID:16051804
- Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80(23):11776-90. doi:10.1128/JVI.01730-06. PMID:16971434
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84(3):1439-52. doi:10.1128/JVI.02108-09. PMID:19939925
- Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID networks for HIV isolation and characterization. J Virol. 1996;70(3):1651-67. PMID:8627686
- Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol. 2004;78(13):6915-26. doi:10.1128/JVI.78.13.6915-6926.2004. PMID:15194768
- Peters PJ, Duenas-Decamp MJ, Sullivan WM, Clapham PR. Variation of macrophage tropism among HIV-1 R5 envelopes in brain and other tissues. J Neuroimmune Pharmacol. 2007;2(1):32-41. doi:10.1007/s11481-006-9042-2. PMID:18040824
- Collman R, Balliet JW, Gregory SA, Friedman H, Kolson DL, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66(12):7517-21. PMID:1433527
- Montefiori DC, Pantaleo G, Fink LM, Zhou JT, Zhou JY, Bilska M, Miralles GD, Fauci AS. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J Infect Dis. 1996;173(1):60-7. PMID:8537683
- Ohagen A, Devitt A, Kunstman KJ, Gorry PR, Rose PP, Korber B, Taylor J, Levy R, Murphy RL, Wolinsky SM, et al. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J Virol. 2003;77(22):12336-45. PMID:14581570
- Ohagen A, Ghosh S, He J, Huang K, Chen Y, Yuan M, Osathanondh R, Gartner S, Shi B, Shaw G, et al. Apoptosis induced by infection of primary brain cultures with diverse human immunodeficiency virus type 1 isolates: Evidence for a role of the envelope. J Virol. 1999;73(2):897-906. PMID:9882290
- Schubert U, Clouse KA, Strebel K. Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J Virol. 1995;69(12):7699-711. PMID:7494279
- Ghorpade A, Nukuna A, Che M, Haggerty S, Persidsky Y, Carter E, Carhart L, Shafer L, Gendelman HE. Human immunodeficiency virus neurotropism: An analysis of viral replication and cytopathicity for divergent strains in monocytes and microglia. J Virol. 1998;72(4):3340-50. PMID:9525661
- McCutchan FE, Carr JK, Bajani M, Sanders-Buell E, Harry TO, Stoeckli TC, Robbins KE, Gashau W, Nasidi A, Janssens W, et al. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology. 1999;254(2):226-34. doi:10.1006/viro.1998.9505. PMID:9986789
- Ivey-Hoyle M, Culp JS, Chaikin MA, Hellmig BD, Matthews TJ, Sweet RW, Rosenberg M. Envelope glycoproteins from biologically diverse isolates of immunodeficiency viruses have widely different affinities for CD4. Proc Natl Acad Sci U S A. 1991;88(2):512-6. doi:10.1073/pnas.88.2.512. PMID:1899141
- Montefiori DC, Altfeld M, Lee PK, Bilska M, Zhou J, Johnston MN, Gao F, Walker BD, Rosenberg ES. Viremia control despite escape from a rapid and potent autologous neutralizing antibody response after therapy cessation in an HIV-1-infected individual. J Immunol. 2003;170(7):3906-14. doi:10.4049/jimmunol.170.7.3906. PMID:12646660
- Westervelt P, Gendelman HE, Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci U S A. 1991;88(8):3097-101. doi:10.1073/pnas.88.8.3097. PMID:2014229
- Theodore TS, Englund G, Buckler-White A, Buckler CE, Martin MA, Peden KW. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses. 1996;12(3):191-4. doi:10.1089/aid.1996.12.191. PMID:8835195
- Rodenburg CM, Li Y, Trask SA, Chen Y, Decker J, Robertson DL, Kalish ML, Shaw GM, Allen S, Hahn BH, et al. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res Hum Retroviruses. 2001;17(2):161-8. doi:10.1089/08892220150217247. PMID:11177395
- Novitsky VA, Montano MA, McLane MF, Renjifo B, Vannberg F, Foley BT, Ndung'u TP, Rahman M, Makhema MJ, Marlink R, et al. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: A set of 23 full-length clones from Botswana. J Virol. 1999;73(5):4427-32. PMID:10196340
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232-52. doi:10.1128/JVI.78.23.13232-13252.2004. PMID:15542675
- Lu S. Heterologous prime-boost vaccination. Curr Opin Immunol. 2009;21(3):346-51. doi:10.1016/j.coi.2009.05.016. PMID:19500964
- Huang Z, Chou A, Tanguay J, Shen S, Mboudjeka I, Chou TH, Lu S, Wang S. Levels of N-linked glycosylation on the V1 loop of HIV-1 Env proteins and their relationship to the antigenicity of Env from primary viral isolates. Curr HIV Res. 2008;6(4):296-305. doi:10.2174/157016208785132518. PMID:18691028
- Bradley T, Pollara J, Santra S, Vandergrift N, Pittala S, Bailey-Kellogg C, Shen X, Parks R, Goodman D, Eaton A, et al. Pentavalent HIV-1 vaccine protects against simian-human immunodeficiency virus challenge. Nat Commun. 2017;8:15711. doi:10.1038/ncomms15711. PMID:28593989
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275-86. doi:10.1056/NEJMoa1113425. PMID:22475592
- Burnett RC, Hanly WC, Zhai SK, Knight KL. The IgA heavy-chain gene family in rabbit: Cloning and sequence analysis of 13 C alpha genes. EMBO J. 1989;8(13):4041-7. PMID:2512120
- Duenas-Decamp MJ, Clapham PR. HIV-1 gp120 determinants proximal to the CD4 binding site shift protective glycans that are targeted by monoclonal antibody 2G12. J Virol. 2010;84(18):9608-12. doi:10.1128/JVI.00185-10. PMID:20610714
- O'Connell O, Repik A, Reeves JD, Gonzalez-Perez MP, Quitadamo B, Anton ED, Duenas-Decamp M, Peters P, Lin R, Zolla-Pazner S, et al. Efficiency of bridging-sheet recruitment explains HIV-1 R5 envelope glycoprotein sensitivity to soluble CD4 and macrophage tropism. J Virol. 2013;87(1):187-98. doi:10.1128/JVI.01834-12. PMID:23055568
- Peters PJ, Duenas-Decamp MJ, Sullivan WM, Brown R, Ankghuambom C, Luzuriaga K, Robinson J, Burton DR, Bell J, Simmonds P, et al. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology. 2008;5:5. doi:10.1186/1742-4690-5-5. PMID:18205925
- Vaine M, Duenas-Decamp M, Peters P, Liu Q, Arthos J, Wang S, Clapham P, Lu S. Two closely related Env antigens from the same patient elicited different spectra of neutralizing antibodies against heterologous HIV-1 isolates. J Virol. 2011;85(10):4927-36. doi:10.1128/JVI.00081-11. PMID:21411542
- Bricault CA, Kovacs JM, Nkolola JP, Yusim K, Giorgi EE, Shields JL, Perry J, Lavine CL, Cheung A, Ellingson-Strouss K, et al. A multivalent clade C HIV-1 Env trimer cocktail elicits a higher magnitude of neutralizing antibodies than any individual component. J Virol. 2015;89(5):2507-19. doi:10.1128/JVI.03331-14. PMID:25540368
- Wang S, Farfan-Arribas DJ, Shen S, Chou TH, Hirsch A, He F, Lu S. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine. 2006;24(21):4531-40. doi:10.1016/j.vaccine.2005.08.023. PMID:16140431
- Kayman SC, Wu Z, Revesz K, Chen H, Kopelman R, Pinter A. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J Virol. 1994;68(1):400-10. PMID:7504740
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856-61. doi:10.1126/science.1187659. PMID:20616233
- Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9(5):e1003342. doi:10.1371/journal.ppat.1003342. PMID:23658524
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466-70. doi:10.1038/nature10373. PMID:21849977
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100-8. PMID:8551569
- Spurrier B, Sampson J, Gorny MK, Zolla-Pazner S, Kong XP. Functional implications of the binding mode of a human conformation-dependent V2 monoclonal antibody against HIV. J Virol. 2014;88(8):4100-12. doi:10.1128/JVI.03153-13. PMID:24478429
- Chen Y, Vaine M, Wallace A, Han D, Wan S, Seaman MS, Montefiori D, Wang S, Lu S. A novel rabbit monoclonal antibody platform to dissect the diverse repertoire of antibody epitopes for HIV-1 Env immunogen design. J Virol. 2013;87(18):10232-43. doi:10.1128/JVI.00837-13. PMID:23864612
- Pan R, Chen Y, Vaine M, Hu G, Wang S, Lu S, Kong XP. Structural analysis of a novel rabbit monoclonal antibody R53 targeting an epitope in HIV-1 gp120 C4 region critical for receptor and co-receptor binding. Emerg Microbes Infect. 2015;4(7):e44. doi:10.1038/emi.2015.44. PMID:26251831
- Derby NR, Kraft Z, Kan E, Crooks ET, Barnett SW, Srivastava IK, Binley JM, Stamatatos L. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: Comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J Virol. 2006;80(17):8745-62. doi:10.1128/JVI.00956-06. PMID:16912322
- Crooks ET, Moore PL, Franti M, Cayanan CS, Zhu P, Jiang P, de Vries RP, Wiley C, Zharkikh I, Schülke N, et al. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology. 2007;366(2):245-62. doi:10.1016/j.virol.2007.04.033. PMID:17580087
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100(7):4144-9. doi:10.1073/pnas.0630530100. PMID:12644702
- Agwale SM, Forbi JC, Notka F, Wrin T, Wild J, Wagner R, Wolf H. Broad antibody mediated cross-neutralization and preclinical immunogenicity of new codon-optimized HIV-1 clade CRF02_AG and G primary isolates. PLoS One. 2011;6(8):e23233. doi:10.1371/journal.pone.0023233. PMID:21829720
- Imholte GC, Sauteraud R, Korber B, Bailer RT, Turk ET, Shen X, Tomaras GD, Mascola JR, Koup RA, Montefiori DC, et al. A computational framework for the analysis of peptide microarray antibody binding data with application to HIV vaccine profiling. J Immunol Methods. 2013;395(1–2):1-13. doi:10.1016/j.jim.2013.06.001. PMID:23770318
- 'Karasavvas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, Koup RA, Madnote S, Arworn D, Shen X, et al. The Thai Phase III HIV Type 1 Vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res Hum Retroviruses. 2012;28(11):1444-57. doi:10.1089/aid.2012.0103. PMID:23035746
- Joachimiak MP, Weisman JL, May B. JColorGrid: Software for the visualization of biological measurements. BMC Bioinformatics. 2006;7:225. doi:10.1186/1471-2105-7-225. PMID:16640789
