ABSTRACT
We previously developed optimized DNA vaccines against both Lassa fever and Ebola hemorrhagic fever viruses and demonstrated that they were protective individually in guinea pig and nonhuman primate models. In this study, we vaccinated groups of strain 13 guinea pigs two times, four weeks apart with 50 µg of each DNA vaccine or a mock vaccine at discrete sites by intradermal electroporation. Five weeks following the second vaccinations, guinea pigs were exposed to lethal doses of Lassa virus, Ebola virus, or a combination of both viruses simultaneously. None of the vaccinated guinea pigs, regardless of challenge virus and including the coinfected group, displayed weight loss, fever or other disease signs, and all survived to the study endpoint. All of the mock-vaccinated guinea pigs that were infected with Lassa virus, and all but one of the EBOV-infected mock-vaccinated guinea pigs succumbed. In order to determine if the dual-agent vaccination strategy could protect against both viruses if exposures were temporally separated, we held the surviving vaccinates in BSL-4 for approximately 120 days to perform a cross-challenge experiment in which guinea pigs originally infected with Lassa virus received a lethal dose of Ebola virus and those originally infected with Ebola virus were infected with a lethal dose of Lassa virus. All guinea pigs remained healthy and survived to the study endpoint. This study clearly demonstrates that DNA vaccines against Lassa and Ebola viruses can elicit protective immunity against both individual virus exposures as well as in a mixed-infection environment.
Introduction
Viral hemorrhagic fevers are among the most acute and deadly diseases known. Lassa virus (LASV) and Ebola virus (EBOV), two hemorrhagic fever viruses that previously caused outbreaks in distinct geographic ranges, are now known to occur in an overlapping geographic area of West Africa. The unprecedented West African human outbreak of EBOV that occurred 2014 through 2015 was the first time the disease had been observed in the well-known Lassa Fever endemic areas of Liberia, Sierra Leone, Nigeria and Guinea.Citation1,Citation2 With over 30,000 cases of Ebola virus disease occurring over the period, it was the largest outbreak of EBOV on record, but still pales by comparison to the estimated annual burden of 300,000-500,000 cases of LASV in the endemic area per year.Citation2,Citation3 There is critical need for safe and effective medical countermeasures for both of these viruses, and development of vaccines or therapeutics that could target both viruses simultaneously would be ideal to not only protect military and peace-keeping personnel deployed to the now EBOV/LASV endemic area, but also as an important tool in improving the overall public health in the region. Also, it is possible that people in the endemic area could become infected with both LASV and EBOV simultaneously, thus a dual-agent countermeasure strategy is an important consideration.
Our laboratory is engaged in development of DNA-based vaccines against biodefense targets, and we reasoned that development of a vaccine that could induce protective immunity simultaneously to both LASV and EBOV could fill a critical need. DNA vaccines are a good match for the target deployment area because they are stable and do not require rigorous cold-chain management, they are adaptable and can be made quickly and in large volumes in response to outbreak situations, and have been shown to be safe and effective when paired with optimal delivery devices.Citation4–7 Several DNA vaccine candidates are currently in clinical trials, such as viruses of biodefense concern, including vaccines against hantaviruses and Venezuelan equine encephalitis virus.Citation8–10 We have previously screened DNA-based viruses against EBOV and LASV individually in guinea pig and nonhuman primate disease models with success.Citation11–13 In the study presented here, we assessed the ability of the previously-tested DNA vaccines against LASV and EBOV, administered as subcutaneous injections followed by dermal electroporation, to protect guinea pigs from lethal infection with LASV, EBOV, or from simultaneous infection with both viruses. We also performed an additional cross-challenge experiment in which the guinea pigs vaccinated with both DNA vaccines that survived exposure to individual viral agents (LASV or EBOV) were held in the BSL-4 lab for approximately 90 days after the end of the primary exposure study, then were exposed to the opposite virus to simulate if the multi-agent vaccine could protect against each virus if exposures were separated temporally. To our knowledge, this study is the first report of a LASV/EBOV coinfection model, so in addition to the vaccine proof-of-concept studies, we report novel basic descriptive and pathologic parameters of coinfected guinea pigs.
Results
Antibody response to vaccination
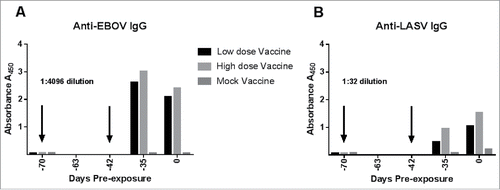
In order to trace the development of LASV-specific and EBOV-specific IgG in vaccinated guinea pigs during the vaccination phase and subsequent virus exposures, and to determine what effect vaccine dose on subsequent antibody production, we performed glycoprotein-based ELISAs of samples collected prior to the first vaccination, one week after the second vaccination, and just prior to virus exposure (Day 0). Guinea pigs received either a low dose of vaccine (50 µg DNA per vaccination) or a high dose of vaccine (100 µg DNA per vaccination). It was necessary to conserve the small sample volumes collected during the pre-exposure phase, so samples from guinea pigs receiving the low dose vaccine were pooled together, as were those from guinea pigs receiving the high dose vaccine. Plasma from the mock-vaccinated guinea pigs was also pooled. For the post-challenge samples, plasma from guinea pigs within each of the nine exposure groups was pooled for the analysis. As shown in , there was no measureable IgG specific for EBOV (1A) or LASV (1B) in the samples collected just prior to the first vaccination, which is indicated by the first downward arrow. The second vaccination was administered four weeks after the first one (Day -42), and one week after the second vaccination (Day -35), samples were again examined for LASV- and EBOV-specific IgG. Antigen specific IgG was detected at high levels for EBOV. LASV-specific IgG was also detected after the second vaccination, but at a much lower level than what was observed for EBOV. In our previous experiments with the LASV DNA vaccine, we have also observed a modest humoral response to vaccine prior to virus exposure.Citation16 The last sample prior to virus exposure was collected approximately five weeks after the second vaccination, immediately prior to virus exposure (Day 0). Levels of EBOV-specific IgG were beginning to decline on Day 0, but levels of LASV-specific IgG were higher on Day 0 than in the samples collected after the second vaccination. For both EBOV and LASV, the animals that received the high dose vaccine mounted a stronger antibody response than those that received the low dose vaccine. The mock-vaccinated guinea pigs did not have measureable EBOV- or LASV-specific IgG at any of the sampling timepoints.
Figure 1. Antigen-specific IgG response to multi-agent vaccination prior to virus exposures. The x axis defines study day relative to Day 0, which is virus exposure. The vaccination days are represented as negative numbers indicating that they occurred prior to study Day 0. EDTA-treated plasma from guinea pigs vaccinated with the low-dose (50 µg) multiagent vaccine, High-dose (100 µg) multiagent vaccine or the mock vaccine were pooled into their respective groups for analysis of IgG response via antibody capture ELISA. The arrows indicated when the vaccinations were administered. Blood was collected just prior to the first vaccination timepoint, which was study day -70, and serves as the zero control for this experiment. A second vaccination was administered to all guinea pigs on study day -42. Blood samples collected just prior to the first vaccination, one week after the second vaccination, and on Day 0 (samples collected just prior to virus exposure) were selected for analysis by ELISA. A) The antibody response to EBOV antigen was strong in the vaccinated guinea pigs, as indicated by the EBOV ELISA. Plasma dilutions of 1:4096 were required to obtain a reading below the dynamic range limit of the plate reader. B) Vaccination with the multi-agent vaccine induced lower levels of Anti-LASV antibodies than those observed on the EBOV plates. A plasma dilution of 1:32 was graphed for the LASV antigen ELISAs.
Protective efficacy of the DNA vaccines against viral challenges
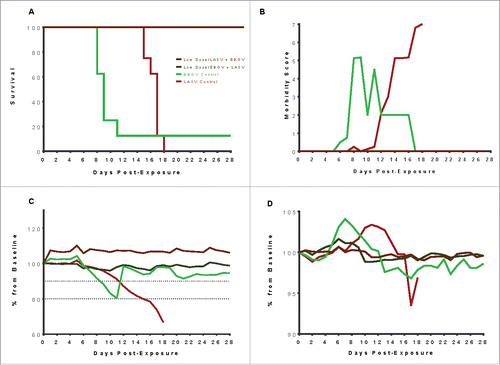
All vaccinated guinea pigs survived to the study endpoint (day 28 post exposure) regardless of the initial dose of vaccine (high or low), or challenge virus, including those exposed to LASV and EBOV simultaneously, compared to the guinea pigs in the control groups that all succumbed (). The typical lethal window for EBOV infection in guinea pigs is between days 8 and 12 post-exposure whereas the normal disease course for LASV in guinea pigs is typically longer, with animals reaching euthanasia criteria between 14 and 18 days post-exposure. In this study, the mock-vaccinated control guinea pigs exposed to LASV alone succumbed between days 14 and 19 post exposure; control guinea pigs exposed to EBOV alone succumbed between days 8 and 14 post exposure, and LASV/EBOV co-exposed guinea pigs succumbed between days 8 and 15 post exposure. The co-exposed control group succumbed in three distinct phases, which will be referred to as early (Days 8–9), mid (Days 10–11), or late (Days 12–19).
Figure 2. Outcomes of the primary vaccine study following exposure to 1000 PFU EBOV, 1000 PFU LASV, or 1000 PFU each of LASV and EBOV simultaneously. (A) All vaccinated guinea pigs survived, regardless of whether they received high (100 µg) or low dose (50 µg) multi-agent vaccine. The mock-vaccinated control groups succumbed in three distinct phases: in the typical window for EBOV (days 7–14), in the typical window for LASV (days 14–19) or an interim window. (B) Morbidity scores for mock-vaccinated guinea pigs increased as disease signs were observed until euthanasia criteria were met. None of the vaccinated GPs experienced observable disease signs after virus exposure. (C) Vaccinated guinea pigs maintained their body weights postexposure. In contrast, mock-vaccinated guinea pigs lost weight steadily, starting approximately 6 days postexposure and continuing to euthanasia. Dashed lines indicate 10% and 20% weightloss estimates. (D) Normal body temperature was maintained by all vaccinated guinea pigs while mock-vaccinated controls experienced a febrile state starting approximately day 5 postexposure in guinea pigs exposed to EBOV alone and EBOV + LASV, and starting approximately day 9 post exposure in guinea pigs exposed to LASV alone. The temporal development of fevers in the mock-vaccinated animals mimics the tri-phasic morbidity observed in the survival curve.
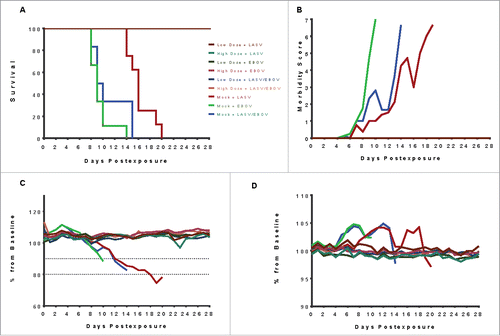
Daily observations of the guinea pigs were performed after virus exposures and the development of disease signs in all groups were observed and recorded. Each guinea pig was assigned a morbidity score daily, based on signs of illness including piloerection, anorexia and weight loss, activity, rash development, neurologic deficit, dyspnea, hypothermia, respiratory distress, and non-ambulatory state (). Each of these disease signs was given a number from 1 to 7 according to severity with a score of 7 corresponding to a non-ambulatory state in which euthanasia criteria was met. Guinea pigs were assigned a score corresponding to the most severe disease sign they were experiencing at each observation period. In addition to disease signs, weight and temperatures were also recorded daily ( and ). The body weights and temperatures of vaccinated guinea pigs were stable throughout the study period, whereas guinea pigs assigned to the control groups experienced severe weight loss and became febrile prior to succumbing to disease.
Changes in IgG levels after virus exposure
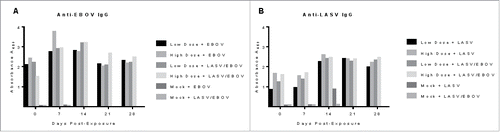
EBOV-specific IgG levels increased in the vaccinated guinea pigs after virus exposure starting at the Day 7 sample collection (, ). Guinea pigs receiving the high dose vaccine and exposed to EBOV alone and EBOV + LASV experienced the highest increases. IgG levels remained high at Day 14 before beginning to decline slightly in all vaccinated survivors by Day 21. The disease course for LASV is extended compared to that of EBOV in guinea pigs, thus there were some animals in the mock-vaccinated control group who were still alive at the Day 14 sampling and some LASV-specific IgG was detected in those animals. None of the EBOV-only or LASV+EBOV-exposed control animals developed a detectable IgG response prior to succumbing to disease. Samples collected from the vaccinated survivors at the end of the study (Day 28) had IgG levels comparable to those observed for the Day 21 samples.
Figure 3. Antigen-specific IgG responses against EBOV and LASV in vaccinated guinea pigs receiving high dose (100 µg DNA) or low dose (50 µg DNA) multi-agent vaccine and exposed to EBOV alone, LASV alone, or EBOV and LASV in a simultaneous infection. (A) Levels of anti-EBOV IgG increased from the pre-exposure levels by day 7, then declined slightly by Day 21 and Day 28. (B) Levels of LASV IgG increased in all vaccinated animals by Day 14, and remained increased through the end of the study (Day 21). The Mock-vaccinated group that were exposed to LASV alone began to mount a LASV-specific IgG response as observed on Day 14, but these animals succumbed before Day 21.
Cross challenge of guinea pigs
At the conclusion of the in-life portion of the primary animal study, the guinea pigs that received the low dose vaccine prior to being exposed to LASV or EBOV alone were retained in the BSL-4 lab and were held for approximately 90 days pending a cross-challenge experiment in which the guinea pigs exposed to LASV alone in the primary study were then exposed to EBOV, and the guinea pigs exposed to EBOV alone in the primary experiment were then exposed to LASV. Naïve age and weight-matched guinea pigs were also used to populate control groups for each virus. The cross-challenge experiment was performed in the same way as the primary study except that blood samples were not collected post exposure. All of the vaccinated guinea pigs survived cross-challenge exposure, without development of disease signs. All LASV-exposed control guinea pigs succumbed and all but one of the guinea pigs exposed to EBOV succumbed to lethal disease ().
Figure 4. Outcomes of a cross-challenge study in which exposures to EBOV and LASV were temporally separated. Guinea pigs receiving the low dose vaccine and exposed to either LASV alone or EBOV alone were held in the BSL-4 laboratory for approximately 90 days following the end of the Primary exposure study, pending a cross-challenge experiment. Age and weight-matched control guinea pigs were assigned to virus exposure groups, and vaccinated guinea pigs that survived LASV-only exposure in the primary experiment were exposed to 1000 PFU EBOV by the subcutaneous route, while vaccinated guinea pigs that survived LASV-only exposure in the primary experiment were exposed to 1000 PFU EBOV by the subcutaneous route. All vaccinated guinea pigs survived the cross-challenge, indicating that the multi-agent LASV/EBOV DNA Vaccine can protect when exposures are temporally separated. (A) Survival curve for the cross-challenge study shows that all LASV-exposed, and all but one of the EBOV-exposed control guinea pigs succumbed, while the vaccinated guinea pigs survived to the endpoint. (B) Vaccinated guinea pigs remained well following virus exposure, whereas the control guinea pigs developed disease signs typical of LASV or EBOV exposure. The surviving EBOV-exposed control animal that survived developed disease signs following exposure, but recovered and had no observable signs of disease after day 17 post-exposure. (C) Vaccinated guinea pigs maintained their bodyweights following exposure, compared to control guinea pigs that lost weight steadily until euthanasia. The jump in weight in the EBOV control group occurring between days 11 and 12 is due to the last critically ill guinea pig succumbing, leaving the lone surviving control guinea pig. (D) Body temperatures remained stable for the vaccinated guinea pigs. Control guinea pigs experienced a febrile state similar to that seen in the primary study.
Pathologic analysis of co-exposed mock-vaccinated guinea pigs
Mock vaccinated guinea pigs exposed to LASV had mild histologic lesions that could not be classified as LASV-specific. Despite the lack of convincing histologic lesions, guinea pigs in this group succumbed of typical LASV disease and demonstrated positive immunohistochemistry staining for LASV antigen in all tissues examined (). Non-specific binding of antibodies used for analysis to endothelial tissue of guinea pigs was observed, but antigen-specific staining in the hepatocytes, pneumocytes, tissue leukocytes, and adrenal cortical cells was characteristic of previously reported LASV infections,Citation14 confirming that these animals were exposed to LASV.
Figure 5. Histology and immunohistochemistry of selected spleen and liver tissues from a co-infected guinea pig that succumbed in the early phase (Day 9). IHC staining for EBOV and LASV was also performed on spleen tissue from an uninfected guinea pig for comparison (insets). Staining for EBOV antigen was much more prominent in this animal than LASV in the early phase. The spleen H&E stain reveals lymphocytolysis and lymphoid depletion. The IHC reveals intense EBOV staining in the presence of lymphocytolysis and lymphoid depletion. LASV staining was light. The Liver H&E stain shows multifocal hepatocyte necrosis and sinusoidal fibrin thrombi. There is heavy focal staining of EBOV antigen in the liver. LASV IHC staining was not determined (ND).
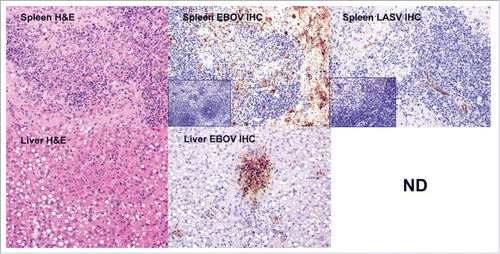
The most consistent histologic lesion seen in the mock-vaccinated guinea pigs exposed to EBOV was necrosis and/or loss of lymphocytes in lymph nodes at one or more locations and splenic white pulp, and is consistent with what is normally reported for EBOV disease in guinea pigs.Citation15 These lesions were particularly evident in the mesenteric lymph nodes. Positive immunohistochemistry (IHC) in leukocytes (predominantly macrophages) in affected follicles in the splenic white pulp correlates with the presence of EBOV antigen and lymphocyte necrosis. Other consistent histologic lesions in this group included multifocal random hepatocyte necrosis +/- sinusoidal fibrin thrombi, and deposition of fibrin and necrotic debris and reticular cell hyperplasia in the red pulp of the spleen. Positive IHC staining in hepatocytes, pneumocytes, and leukocytes in the liver, splenic red pulp, and lung was co-located with the histologic lesions described above (data not shown).
Three distinct waves of mortality occurred in the LASV+EBOV co-exposed control group. The animals that died early (days 8–9) had histologic lesions similar to the animals in the EBOV-only group, including lymphocytolysis in multiple lymphoid tissues, hepatocyte necrosis, hepatic sinusoidal fibrin thrombi, splenic red pulp fibrin and necrotic debris, and interstitial inflammation in the lung. Positive IHC staining for EBOV antigen was seen in leukocytes of multiple tissues, hepatocytes, pneumocytes, adrenal cortical cells, renal tubular epithelium, and scattered fibroblasts and endothelial cells. Interestingly, positive IHC staining for LASV antigen was also seen in multiple tissues, but was limited to the endothelium, thus we suspect this is due to non-specific staining of the LASV monoclonal antibody that has been described in previous studies.Citation14,Citation16,Citation17 The guinea pigs that died later (days 14–19) had limited histologic lesions, similar to the animals in the LASV-only group. Minimal to mild lymphocytolysis in lymphoid tissues, and minimal necrosis in the liver were the most consistent findings. Minimal to mild interstitial inflammation was present in the lungs of these animals. It is significant that there was no IHC staining for EBOV antigen in any of the tissues of the guinea pigs that died in the late stage, suggesting that these guinea pigs were able to clear EBOV virus prior to the time of death. However, these guinea pigs had positive IHC staining for LASV in multiple tissues, to include hepatocytes, adrenal cortical cells, and endothelium of multiple tissues. As mentioned previously, there was also non-specific endothelial staining in these tissues. Co-infected guinea pigs that succumbed during the mid phase (days 10–11) showed characteristics observed in those that died early and late in the disease course. A summary of histologic lesions present in the co-exposed guinea pigs is included as . A selection of IHC results are presented in .
Table 1. Summary of Pathologic Findings in guinea pigs exposed to LASV and EBOV simultaneously by the subcutaneous route.
Detection of viral antigens in tissues of mock-vaccinated guinea pigs
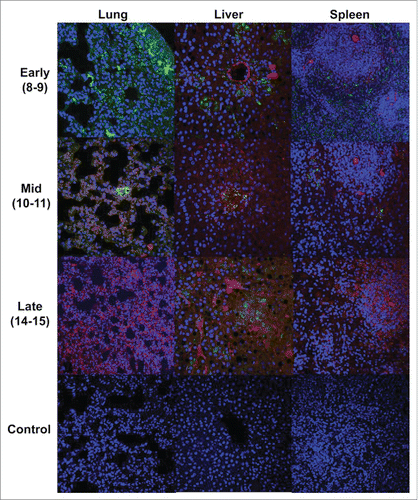
We performed immunofluorescence analysis (IFA) on the tissue samples from the mock-vaccinated, coinfected guinea pigs in order to determine if EBOV and LASV target the same cells. Slides of lung, liver and spleen tissues were stained for LASV and EBOV antigens and examined on a confocal microscope. For this analyses, polyclonal rabbit sera derived by injection of rabbits with LASV recombinant GP proteins was used instead of the monoclonal antibody that was used for IHC. Although background staining was apparent, antigen specific staining was well defined, and when the fluorescent fields were merged, there was no evidence of co-staining of cells, indicating that LASV and EBOV target different cell types in tissues ().
Figure 6. Merged immunofluorescent images of lung, liver and spleen tissues from Co-infected GPs stained for EBOV (green) and LASV (red) antigens. EBOV staining was more prominent in tissues at the early stage. Both EBOV and LASV antigens are present in tissues at mid phase, and predominantly LASV antigens are present in the late phase samples. The absence of yellow color in these images indicates that LASV and EBOV did not infect the same cell populations in the target tissues. Background fluorescence is apparent, as is nonspecific staining of the LASV antibody in endothelial tissue, which is a previously observed issue with the antibody used.
To enable the detection of tissue differences that might relate to physical changes observed in the three-phased mortality (early, mid and late), we used a novel in-situ hybridization (ISH) technique employing nucleic acid probes instead of antibodies. We examined lung, liver, and spleen tissues from guinea pigs that succumbed in the early, mid, or late disease stage and differentially stained for the presence of LASV or EBOV genomes (). In the guinea pigs that succumbed early (Day 8–9), we were able to show the presence of moderate levels of EBOV genomes in the lung, and higher levels in the liver and spleen. LASV genomes were not observed in the lungs in co-infected animals that succumbed early, but were present in the liver and spleen at lower levels than EBOV. Much lower levels of EBOV antigen were observed in guinea pigs that succumbed in the mid stage (Days 10–11) Notably, LASV genomes still appear to be absent in the lungs but they were present in liver and spleen. LASV antigen stained more intensely in the spleen than EBOV in coinfected animals that succumbed in the mid phase. The guinea pigs that succumbed in the late phase (Day 14–15) were negative for EBOV staining in all tissues. It is important to point out that the brown color present in the spleen tissue of the Late EBOV and the negative control is thought to be deposits of hemosiderin, and are not considered antigen-positive staining in this assay. The late phase tissues were absent of EBOV-specific staining but were intensely positive for LASV antigen in the lung and spleen, and moderately positive in the liver.
Figure 7. A newly adapted ISH technique using nucleic acid probes on formalin fixed- tissues reveals more specific staining patterns for LASV and EBOV antigens. Staining patterns differ in the co-infected GPs dependent upon disease progression (early, mid, and late). Tissues from animals that succumbed early had prominent staining of EBOV antigen in lung, liver and spleen. Moderate staining of LASV was also present in liver and spleen in the early sample. Both viruses stained with relatively equal intensity in the mid phase samples in liver and spleen, but only EBOV was detected in lung at the mid phase. EBOV antigen was still detected in the spleen at low levels during the late phase, but LASV antigen was detected at a much higher level in this tissue, as well as being present in high levels in the lung and liver. The uninfected control samples demonstrate the specificity of the nucleic acid probe staining technique.
Discussion
In this study, we were able to show that a dual-agent DNA vaccination strategy was effective against LASV and EBOV exposure individually, and in a coinfection model. In the primary experiment, we assessed the ability of the optimized LASV-GPC and EBOV-GP DNA vaccines to elicit protective immunity against both viruses in the same animal. We were able to observe a brisk humoral response to the vaccine against EBOV, and a much lower but clearly measureable humoral response to LASV after the second vaccination timepoint. We demonstrated that guinea pigs receiving both vaccines at different sites via dermal electroporation, were completely protected against exposure with each virus individually, and were also protected against simultaneous exposure to both viruses. Our ELISA experiment shows that the guinea pigs that received the high dose vaccine had a higher antibody response during the vaccination phase and after virus exposure than those that received the low dose vaccine, but there were no observable differences between these vaccinated groups in terms of protection from disease and death. In addition, we were able to show in the cross-challenge experiment that the multi-agent vaccine was able to protect against LASV and EBOV when exposure to the second virus was delayed for an extended period of time after the initial exposure. We believe this indicates that the multiagent vaccine can provide durable protection against each virus when exposures are spread apart in time.
Although the vaccinated guinea pigs survived infection, the mock-vaccinated guinea pigs that succumbed to infection provided an opportunity for a first-of-its-kind analysis of a LASV/EBOV coinfection animal model. Our initial observational and pathologic study of this coinfection model revealed that despite showing three distinct phases of morbidity, the coinfected guinea pigs tended to experience disease signs typical of either LASV or EBOV exposure. There were no unique disease signs observed in the coinfected guinea pigs, and histology and IHC staining patterns did not reveal unique disease pathology. We were able to show that although LASV and EBOV are capable of infecting the same myeloid cell types in the periphery,Citation18-21 the ultimate target cell ranges of these viruses in the tissues do not overlap in the coinfected guinea pigs. Due to the high background staining and non-specific binding that we observed with traditional antibody-based staining techniques, we adapted an in situ hybridization (ISH) technique using virus-specific nucleic acid probes to analyze the tissues from coinfected guinea pigs. We found this method to be more sensitive and accurate for differentiating coinfected samples than IHC and IFA, and it allowed us to more precisely detect virus in a variety of infected tissues. Using the ISH technique, we were able to track the dynamics of EBOV and LASV antigen in selected tissues over time in the coinfected animals, revealing that EBOV was the dominant antigen present in animals that succumbed in the early phase, that both viruses were present in animals succumbing in the mid phase, and that LASV was the predominant antigen present in animals that succumbed in the late phase.
While the results of this multiagent vaccine efficacy study in guinea pigs is quite encouraging, our ability to fully investigate the host immune response to the vaccine and virus exposure, especially in the co-infected group, is hampered by the general lack of availability of guinea-pig specific immune assays and reagents. In order to more directly assess the durability of protection against each virus in our multiagent DNA vaccine formulation, a study in which guinea pigs are vaccinated and held for a longer period of time prior to virus exposures would be advantageous. We are currently attempting to secure funding to continue to refine this work in the guinea pig model prior to screening in primates.
Conclusion
Our results provide a foundation for future studies on understanding and preventing disease in regions of the world where both LASV and EBOV are present. In future experiments, we intend to further assess the samples collected during this study, and to characterize the immune responses elicited by the vaccines. Although we can measure antibody responses using ELISA or neutralization tests, there is limited data to suggest a correlation with antibody development and disease protection. Cell mediated immunity is difficult to evaluate in the guinea pig model due to the lack of available reagents; however, we aim to gain some basic insights into how the multi-agent vaccine was able to stimulate strong, virus-specific, and lasting immune responses to both viruses simultaneously by planned proteomic and transcriptomic analysis of samples collected during this study. In addition, samples collected for the mock-vaccinated guinea pigs will allow us to elucidate the natural history and immune response to coinfection with LASV and EBOV and potentially identify common pathways that could be exploited for further vaccine or therapeutics development. Finally, we plan to explore the ability to protect against other routes of exposure, including aerosol or intranasal, and to include other viruses or other strains of LASV and EBOV in vaccine formulations. The success of this initial proof-of-concept study indicates that multiagent DNA-based vaccine strategies represent a viable approach for development of medical countermeasures against biodefense targets.
Materials and methods
DNA vaccines
The construction of the codon-optimized LASV-GPC DNA vaccine is described elsewhere.Citation16 Briefly, the published sequence for LASV GPC gene, Josiah strain, (Genbank Accession number AY628203.1) was used as the basis of the vaccine construct. The sequence was optimized for expression in Cavia Porcellus by GeneArt The optimized sequence was synthesized and cloned into the NotI/BglII site of expression vector pWRG7077 (Powdermed) to create the LASV-GPC vaccine plasmid. The sequence of the EBOV (Kikwit 1995 strain) glycoprotein gene was also optimized to yield the same GP expression product as previously reported. (Genbank accession number AAQ55048) was also codon-optimized, and the optimization process resulted in the ablation of the 7U/8U editing site thus this plasmid does not express the soluble version of the EBOV GP. The optimized gene was cloned into the pWRG7077 expression plasmid, and is described elsewhere.Citation12,Citation22 Research grade LASV and EBOV vaccine plasmids used in this study were manufactured by Aldevron (Fargo, ND). The pWRG7077 empty plasmid was used for all Mock vaccinations.
Virus strains
Guinea pig-adapted LASV, Josiah strain, was derived from a human clinical sample obtained from the CDC (CDC # 800789) and received at USAMRIID in August of 1978. Prior to shipping to USAMRIID, the serum sample had been passaged three times in VERO cells at the CDC. At USAMRIID, the virus was passaged once in VERO cells, then 1000 plaque forming units (PFU) of this stock were subcutaneously injected into Strain 13 guinea pigs which were then euthanized 7 days postinfection. Clarified spleen homogenates were quantified by plaque assay, then spleen homogenate containing 1000 PFU of LASV was passaged into another set of guinea pigs. On the eighth such passage, the virus was uniformly lethal in guinea pigs, and a clarified spleen homogenate from this eighth passage was used to infect monolayers of VERO cells. Supernatants from this VERO passage were collected, centrifuged to clarify and then placed in 1 ml aliquots for storage at -80ºC. One additional passage of this stock occurred in 2010, which is the source of LASV used in this study.Citation14,Citation16 A target dose of 1000 PFU was used for the experiments.
Guinea pig-adapted EBOV was derived by passage of a fatal human clinical isolate of EBOV, Mayinga strain, four times through guinea pig spleens as described above for guinea pig adapted LASV. The clarified spleen homogenate from the fourth passage was then passaged once through Vero E6 cells. One additional passage of this stock through VERO E6 cells occurred in 2009.Citation15 A target dose of 1000 PFU was used for the experiments.
Guinea pig studies
Research was conducted under an IACUC approved protocol in compliance with the Animal Welfare Act, PHS Policy, and other Federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011. Strain 13 guinea pigs (8 per group) received two intradermal vaccinations immediately followed by electroporation, spaced four weeks apart and consisting of either 50 µg of each (low dose) or 100 µg of each (high dose) DNA vaccine or a mock vaccine at discrete sites on the shaved abdomens. Five weeks following the second vaccination, guinea pigs were exposed to a 1000 PFU target dose of Lassa virus via subcutaneous (SQ) exposure, EBOV via intraperitoneal (IP) exposure, or 1000 PFU target doses of both viruses simultaneously (LASV via SQ and EBOV via IP). Guinea pigs were monitored daily and assessed for development of disease signs. Weight and temperature data was collected and recorded daily. Guinea pigs that became moribund were humanely euthanized in accordance with IACUC-approved criteria. In order to understand the pathology of LASV/EBOV coinfection in guinea pigs, a subset of LASV-exposed, EBOV-exposed and LASV+EBOV-exposed guinea pigs were selected for pathologic analysis.
Strain 13 guinea pigs that received the low dose vaccine and survived the primary LASV only or EBOV only challenge were held for approximately 90 days in BSL-4 pending a cross-challenge experiment. In this experiment, guinea pigs that initially were exposed to LASV alone and survived to the primary study endpoint were exposed to a target subcutaneous dose of 1000 PFU EBOV, and guinea pigs that initially were exposed to EBOV alone and survived were exposed to a target subcutaneous dose of 1000 PFU of LASV. Guinea pigs were monitored daily and assessed for development of disease signs. Weight and temperature data was collected and recorded daily. Guinea pigs that became moribund were humanely euthanized in accordance with IACUC-approved criteria.
Detection of antigen-specific IgG
ELISAs were performed on plasma from blood samples collected prior to the first vaccination, one week after the second vaccination, and weekly starting on the day of virus exposure. ELISA kits for EBOV-GP were custom prepared by Zalgen, Inc. Antigen for LASV-GP ELISA kit was generously provided by Erica Ollmann Saphire of The Scripps Research Institute. The antigen was coated onto plates by Zalgen, Inc. and provided to the investigators for use in this study. Manufacturer's instructions were followed, with the exception of the anti-human IgG HRP-conjuaged antibody being substituted for a goat-anti-guinea pig IgG-HRP conjugate (Novex, Life Technologies). Briefly, plasma from guinea pigs were pooled within each treatment/exposure group for each blood collection timepoint. Two-fold serial dilutions were performed on these samples and incubated on the antigen-coated ELISA plates at room temperature, followed by a 4x wash, and a second incubation with a 1:10000 dilution of the HRP-conjugate antibody. After a final wash, the plates were developed with the substrate solution provided in the kits, a stop solution was applied, then the plates were read on a plate reader at 450 nm. There was no positive control available to use in order to quantify the antibody response, so changes in absorbance values over time compared to the first pre-vaccination blood sample for each pooled group was used to determine the development of antigen-specific antibodies.
Analysis of tissues
Necropsies were performed on a subset of guinea pigs immediately following euthanasia in the USAMRIID BSL 4 laboratory. Tissues from major organ systems were collected, immersion fixed in 10% neutral buffered formalin and held in biocontainment for a minimum of 21 days. Histopathology samples were routinely processed, embedded in paraffin, sectioned and stained with hematoxylin and eosin. Immunohistochemistry was performed on replicate tissue sections for both partial and full necropsies using an Envision kit (Dako). A mouse monoclonal antibody specific for Lassa virus GP1 (USAMRIID clone #52-2074-7A) was used at a dilution of 1:15000. After deparaffinization and peroxidase blocking, an antigen retrieval step was performed using a TRIS/EDTA buffer in a steamer for 30 minutes. Tissue slides were covered with primary antibody, incubated at room temperature for 30 minutes and rinsed. The secondary antibody, a peroxidase-labeled polymer, was applied for 30 minutes and the slides rinsed again. Substrate-chromogen solution (DAB, Dako) was applied for five minutes; the slides were rinsed in distilled water, counterstained with hematoxylin for two minutes, dehydrated, cleared with xyless and then coverslipped. Slides were examined with a Nikon Eclipse 600 light microscope.
For immunofluorescent antibody staining, the formalin-fixed paraffin embedded guinea pig tissue sections were deparaffinized and rehydrated through a series of graded ethanol. After 0.1% Sudan black B (Sigma) treatment to quench autofluoresence, the sections were boiled in citrate buffer (pH 6.0) for 15 minutes to unmask antigen. After rinses with PBS (pH 7.4), the section were blocked with PBS containing 5% normal goat serum overnight at 4°C. Then the sections were incubated with mouse monoclonal anti-LASV (USAMRIID clone #52-2074-7A) and rabbit polyclonal anti-EBOV GP (USAMRIID) antibodies for 2 hours at room temperature. After rinses with PBS, the sections were incubated with secondary Alexa Fluor 488 conjugated goat anti-rabbit antibody and Alexa Fluor 561 conjugated goat anti-mouse antibody for 1 hour at room temperature. Sections were cover slipped using the Vectashield mounting medium with DAPI (Vector Laboratories). Images were captured on a Zeiss LSM 780 confocal system and processed using ImageJ software.
In situ hybridization (ISH) analysis was performed using RNAscope® 2.5 HD RED kit according to the manufacturer's instructions (Advanced Cell Diagnostics, Hayward, CA). Briefly, 20 ZZ probes set targeting to 466–1433 of Lassa genome with GenBank accession number KM821901.1 and 20ZZ probes set targeting to the compliment strand of 1673–2598 of EBOV Zaire genome with GenBank accession number J04337.1 were synthesized. After deparaffinization and peroxidase blocking, the sections were heated in antigen retrieval buffer and then were digested by proteinase. The section were covered with ISH probes and incubated at 40°C in hybridization oven for two hours. They were rinsed and the ISH signal is amplified by applying Pre-amplifier and Amplifier conjugated with HRP. A red substrate-chromogen solution was applied for 10 minutes at room temperature. The slides were further stained with hematoxylin, air dried, and mounted.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors thank personnel of the Pathology Division at USAMRIID for careful blocking and cutting and staining of tissues slides for analysis. We would also like to acknowledge the generous gift of LASV-GP antigen from Dr. Erica Ollmann Saphire of The Scripps Research Institute that enabled us to measure the LASV-specific response to vaccination and virus exposure.
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number 1R01AI105383 awarded to Drs. Cashman and Schmaljohn. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the U.S. Army.
References
- Baize S. Ebola virus in West Africa: new conquered territories and new risks-or how I learned to stop worrying and (not) love Ebola virus. Curr Opin Viro. 2015;10:70–6. doi:10.1016/j.coviro.2015.01.008.
- Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, et al. Emergence of Zaire Ebola virus disease in Guinea. N Eng J Med. 2014;371:1418–25. doi:10.1056/NEJMoa1404505.
- Ogbu O, Ajuluchukwu E, Uneke CJ. Lassa fever in West African sub-region: an overview. J Vector Borne Diseases. 2007;44:1–11.
- Dupuy LC, Schmaljohn CS. DNA vaccines for biodefense. Expert Rev Vaccines. 2009;8:1739–54. doi:10.1586/erv.09.132.
- Broderick KE, Khan AS, Sardesai NY. DNA vaccination in skin enhanced by electroporation. Methods Mol Biol. 2014;1143:123–30. doi:10.1007/978-1-4939-0410-5_8.
- Broderick KE, Shen X, Soderholm J, Lin F, McCoy J, Khan AS, Yan J, Morrow MP, Patel A, Kobinger GP, et al. Prototype development and preclinical immunogenicity analysis of a novel minimally invasive electroporation device. Gene Ther. 2011;18:258–65. doi:10.1038/gt.2010.137.
- Diehl MC, Lee JC, Daniels SE, Tebas P, Khan AS, Giffear M, Sardesai NY, Bagarazzi ML. Tolerability of intramuscular and intradermal delivery by CELLECTRA((R)) adaptive constant current electroporation device in healthy volunteers. Hum Vaccin Immunother. 2013;9:2246–52. doi:10.4161/hv.24702.
- Hannaman D, Dupuy LC, Ellefsen B, Schmaljohn CS. A Phase 1 clinical trial of a DNA vaccine for Venezuelan equine encephalitis delivered by intramuscular or intradermal electroporation. Vaccin. 2016;34:3607–12. doi:10.1016/j.vaccine.2016.04.077.
- Hooper JW, Moon JE, Paolino KM, Newcomer R, McLain DE, Josleyn M, Hannaman D, Schmaljohn C. A Phase 1 clinical trial of Hantaan virus and Puumala virus M-segment DNA vaccines for haemorrhagic fever with renal syndrome delivered by intramuscular electroporation. Clin Microbiol Infect. 2014;20(Suppl 5):110–7. doi:10.1111/1469-0691.12553.
- Schmaljohn CS, Spik KW, Hooper JW. DNA vaccines for HFRS: laboratory and clinical studies. Virus Re. 2014;187:91–6. doi:10.1016/j.virusres.2013.12.020.
- Cashman KA, Broderick KE, Wilkinson ER, Shaia CI, Bell TM, Shurtleff AC, Spik KW, Badger CV, Guttieri MC, Sardesai NY, et al. Enhanced efficacy of a Codon-optimized DNA vaccine encoding the glycoprotein precursor gene of Lassa virus in a guinea pig disease model when delivered by dermal electroporation. Vaccines (Basel. 2013;1:262–77. doi:10.3390/vaccines1030262.
- Grant-Klein RJ, Altamura LA, Badger CV, Bounds CE, Van Deusen NM, Kwilas SA, Vu HA, Warfield KL, Hooper JW, Hannaman D, et al. Codon-optimized filovirus DNA vaccines delivered by intramuscular electroporation protect cynomolgus macaques from lethal Ebola and Marburg virus challenges. Hum Vaccin Immunother. 2015;11:1991–2004. doi:10.1080/21645515.2015.1039757.
- Grant-Klein RJ, Van Deusen NM, Badger CV, Hannaman D, Dupuy LC, Schmaljohn CS. A multiagent filovirus DNA vaccine delivered by intramuscular electroporation completely protects mice from ebola and Marburg virus challenge. Hum Vaccin immunother. 2012;8:1703–6. doi:10.4161/hv.21873.
- Bell TM, Shaia CI, Bearss JJ, Mattix ME, Koistinen KA, Honnold SP, Zeng X, Blancett CD, Donnelly GC, Shamblin JD, et al. Temporal progression of lesions in guinea pigs infected with Lassa virus. Vet Pathol. 2017;54(3):549–562. doi:10.1177/0300985816677153.
- Twenhafel NA, Shaia CI, Bunton TE, Shamblin JD, Wollen SE, Pitt LM, Sizemore DR, Ogg MM, Johnston SC. Experimental aerosolized guinea pig-adapted Zaire ebolavirus (variant: Mayinga) causes lethal pneumonia in guinea pigs. Vet Pathol. 2015;52:21–5. doi:10.1177/0300985814535612.
- Cashman KA, Broderick KE, Wilkinson ER, Shaia CI, Bell TM, Shurtleff AC, Spik KW, Badger CV, Guttieri MC, Sardesai NY, et al. Enhanced efficacy of a codon-optimized DNA vaccine encoding the glycoprotein precursor gene of Lassa virus in a guinea pig disease model when delivered by dermal electroporation. Vaccine. 2013;1:262–77. doi:10.3390/vaccines1030262.
- Cashman KA, Smith MA, Twenhafel NA, Larson RA, Jones KF, Allen RD, 3rd, Dai D, Chinsangaram J, Bolken TC, Hruby DE, et al. Evaluation of Lassa antiviral compound ST-193 in a guinea pig model. Antiviral Res. 2011;90:70–9. doi:10.1016/j.antiviral.2011.02.012.
- Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol. 2003;163:2347–70. doi:10.1016/S0002-9440(10)63591-2.
- Walker DH, Wulff H, Lange JV, Murphy FA. Comparative pathology of Lassa virus infection in monkeys, guinea-pigs, and Mastomys natalensis. Bull World Health Organ. 1975;52:523–34.
- Baize S, Kaplon J, Faure C, Pannetier D, Georges-Courbot MC, Deubel V. Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells. J Immunol. 2004;172:2861–9. doi:10.4049/jimmunol.172.5.2861.
- Mahanty S, Hutchinson K, Agarwal S, McRae M, Rollin PE, Pulendran B. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J Immunol. 2003;170:2797–801. doi:10.4049/jimmunol.170.6.2797.
- Grant-Klein RJ, Van Deusen NM, Badger CV, Hannaman D, Dupuy LC, Schmaljohn CS. A multiagent filovirus DNA vaccine delivered by intramuscular electroporation completely protects mice from ebola and Marburg virus challenge. Hum Vaccin Immunother. 2012;8:1703–6. doi:10.4161/hv.21873.
