ABSTRACT
Since the first confirmed case of H7N9 infection was reported in China, there have been five epidemic waves of human H7N9 infections between 2013 and 2017. The fifth wave differed from the previous four waves in that highly pathogenic avian influenza (HPAI) H7N9 viruses with multiple basic amino acids at the cleavage site were detected in humans, poultry and environmental samples. The HPAI H7N9 viruses were genetically and antigenically distinct from previous H7N9 viruses. Therefore, a new candidate vaccine virus(CVV) derived from a HPAI A/Guangdong/17SF003/2016-like virus was proposed by the World Health Organization(WHO). According to the WHO recommendations, we constructed a new CVV using reverse genetic technology, with a (6+2) gene constitution. The (6+2) reassortant virus possessed hemagglutinin(HA) with multiple basic amino acids removed and the neuraminidase from A/Guangdong/SF003/2016 in a high-yield A/Puerto Rico/8/34 virus backbone. Sequence analysis confirmed that no mutations had occurred in the HA of V1E1(the initial CVV rescued in Vero cells and followed by passage in eggs), but a mixture of arginine (R)/glycine (G)/isoleucine (I) was detected at position 220 (H3 numbering) in the HA of V1E2 to V1E5 with different percentages. Furthermore, V1E5 showed improved growth characteristics and immunogenicity compared with V1E1, and retained low pathogenicity in chickens and chicken embryos, but the mutation changed its antigenicity. Our study indicates that antigenic changes should be closely monitored during the development of H7N9 CVV in eggs. Additionally, although V1E5 changes the antigenicity, the antisera had some reactivity to previous H7N9 CVVs.
Introduction
Human infections with avian influenza A (H7N9) viruses were first reported to World Health Organization (WHO) on 31 March 2013.Citation1 Five waves of outbreaks have occurred since then. During the first four waves, all of the detected viruses belonged to low pathogenicity avian influenza virus and these viruses were silently circulating in poultry.Citation2,Citation3 However, recent epidemiological studies have shown that mild human A (H7N9) infections are more common than H5N1 infections,Citation4 supporting the experimental findings that H7N9 is unusually well adapted to the human respiratory tract.Citation5 During the fifth wave, more human infections with avian A (H7N9) were reported with a wider geographic spread in China.Citation6 It is particularly important to note that highly pathogenic avian influenza (HPAI) A(H7N9) viruses were detected and resulted in fatal outcomes in 2016.Citation7,Citation8 Fatality rate is the proportion of deaths within a designated population of “cases” (people with a medical condition) over the course of the disease. Since 2013, a total of 1564 laboratory-confirmed cases of human infected with avian influenza A(H7N9) viruses, including at least 612 deaths. Compared with another subtype avian influenza A(H5N1) virus, the fatality rate(612/1564) of A(H7N9) viruses is lower than that of A(H5N1) virus(454/860) (Updated on September 27,2017).Citation9 However, since the virus continues to be detected in animals and environments, further human cases can be expected.Citation9The HPAI A (H7N9) viruses were genetically and antigenically distinct from other A(H7N9) viruses and the current candidate vaccine viruses (CVVs).Citation10A previous report, which employed well-established immunoinformatic tools, showed that H7N9 virus had poor immunogenicity.Citation11In addition, an H7 subtype influenza has never before infected humans to any extent, meaning that the human population has little or no immunity, which could lead to a high mortality rate.Citation12 Avian influenza A (H7N9) virus therefore posed a significant threat to public health, as confirmed by this virus ranking as the influenza virus with the highest potential pandemic risk when a Risk Assessment Tool was applied.Citation13 Therefore, a new CVV derived from a HPAI A/Guangdong/17SF003/2016-like virus (SF003), with multiple basic amino acids (KRTA) at the cleavage site, was proposed by the WHO.Citation10
A new reassortant CVV could be generated either by conventional reassortment or by reverse genetics (RG), including, in the latter case, those viruses derived from synthetic nucleic acid.Citation14 For the HPAI influenza viruses, (6+2) reassortant viruses with hemagglutinin (HA) and neuraminidase (NA) derived from the wild-type virus in a high-yield A/Puerto Rico/8/34 (PR8) virus backbone were generally obtained as CVV using reverse genetics technologies.Citation15
In this study, according to the WHO guidance on the development of influenza vaccine reference viruses,Citation10 we constructed a (6+2) reassortant virus, RG-SF003 as a CVV, with HA and NA derived from A/Guangdong/17SF003/2016 in the PR8 backbone by RG. We evaluated the genetic stability after multiple passages in eggs, growth ability in eggs or MDCK cells, viral protein yields, antigenic identity with the original virus and pathogenicity in vivo.
Results
Generation of reassortant viruses by reverse genetics
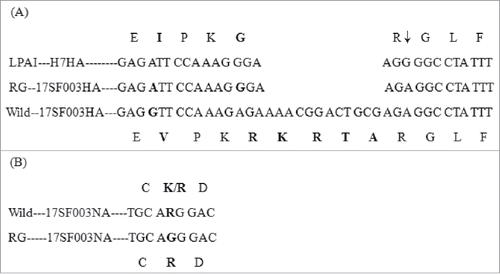
The reassortant virus RG-SF003 was obtained using the (6+2) RG method in Vero cells, in which the six backbone fragments were derived from PR8, whereas the HA and NA genes were synthesized based on the sequence of the SF003 virus from our laboratory. To confirm the reassortant virus, we sequenced the first-generation rescued virus RG-SF003 V1E1 (initial growth in Vero cells followed by one passage in eggs). Sequence analysis for V1E1 indicated that the cleavage site of the HA sequence was consistent with that of the low pathogenic H7 virus with the desired four amino acid (KRTA) deletion in the multibasic cleavage motif () and no other mutations were detected compared with the parental strain SF003. Wild-type virus SF003 had either a K or R at position 292(N2 numbering)Citation16 and we synthesized R at position 292. The sequence of NA was identical to our synthesized sequences (). Six internal gene segments were derived from the high-yield PR8. The sequencing results for RG-SF003 V1E1 virus indicated that the gene was identical to that carried on the plasmids used in its creation.
Figure 1. Modified sequence in HA and NA of reassortant viruses. (A) HA cleavage site sequence. LAPI–HA7: consensus sequence of low pathogenic avain influenza viruses H7 (reference to the sequence in NCBI gene bank, Accession Number: APR73185, AJU15335, AKI82233, APW83929, AHL21384, AJE61993, ANH96462, ALR82230, AJU15321, APW83918); RG-17SF003HA: sequence of (6+2) reassortant H7N9 virus by reverse technology with deletion of KRTA; Wild-17SF003HA: sequence of wild-type virus A/Guangdong/17SF003/2016; ↓: proteolytic cleavage site. (B) Part of amino acid sequence associated with resistance to neuraminidase inhibitors (N2 numbering: 291–293 position). Wild—17SF003NA: wild-type 17SF003 virus containing NA-292K/R; RG-17SF003NA: (6+2) reassortant 17SF003 virus containing NA-292R. Change sequences were in bold.
Stability of the reassortant virus during passage
To determine the genetic stability of the HA and NA genes, rescued V1E1 virus were propagated in chicken embryos for an additional four passages and these were designated as V1E2, V1E3, V1E4 and V1E5, respectively. At each passage, the embryonated eggs were checked and dead embryos would be recorded. The allantoic fluid of each passage was tested for the HA titer. No dead eggs were observed at any virus passage. The HA titer for passages V1E1 to V1E5 were 512–1024 hemagglutination units (), similar to that of PR8.
Table 1. Amino acid difference and virus titers of reassortant viruses.
The CVVs of different passages were sequenced and compared with V1E1. The HA sequence remained the low pathogenic motif in the cleavage site from V1E1 to V1E5, but there was one variation at position 220 in the HA gene of V1E2 toV1E5 (H3 numbering, ). Sequencing results revealed a mixture(R/G/I) at amino acid position 220(H3 numbering) in the HA of V1E2 to V1E5 compared with a homozygous R at the same amino acid position of V1E1. The amino acid I firstly appeared in V1E3. The percentage of G increased with the increasing of passage and reached the highest percentage (70%) in V1E5 (). Sequence analysis of the NA gene from V1E2 to V1E5 revealed that no mutations had occurred compared with the NA gene of V1E1. During the preparation of this manuscript, in order to learn which amino acid would the dominant one, RG-SF003 viruses were propagated in eggs for additional five times. The sequence results showed I gradually became the dominant amino acid from V1E6 to V1E10 (data not shown).
Antigenic analysis of the reassortant virus
To examine the effect of the HA amino acid differences on the antigenicity of V1E1 and V1E5, we determined the reactivity of ferret antisera to V1E1 and V1E5 (). Post-infection(p.i.) ferret sera samples were collected on day 17 and antibody titers were evaluated by a hemagglutination inhibition (HI)assay against the homologous virus and the reference virus using turkey erythrocytes.V1E1 and V1E5 were immunogenic and induced HI antibody titers (80 and 1280, respectively) against homologous viruses. Antisera to V1E1 inhibited wild-type SF003 virus and V1E1 virus with the same HI titer of 80 but reacted well with V1E5 at a HI titer of 640. Antisera to V1E5 inhibited hemagglutination of the wild-type SF003 and V1E1viruses to a 16 fold lower (HI titer 80) than the homologous virus, V1E5 (HI titer 1280), indicating that antigenicity varied with HA mutation at position 220. We also used the WHO recommended CVVs and antisera to perform further tests, and results showed that both antisera to V1E1 and V1E5 were cross-reactive with the CVVs, but the antisera to the CVVs showed an eight-fold different HI titer to V1E1 compared with homologous viruses. The antisera to the CVVs showed a similar HI titer to V1E5 compared with homologous viruses, with the exception of A/Shanghai/2/2013 (RG), which showed four-fold different HI titer. Wild-type SF003 virus and V1E1 exhibited the same reactivity to the reference antisera indicating that V1E1 retained similar antigenicity to wild-type SF003 and distinct antigenicity from A/Anhui/1/2013(wild-type), A/Anhui/1/2013 (RG), A/Shanghai/2/2013(wild-type), A/Shanghai/2/2013 (RG) and A/Hunan/2650/2016 (RG).
Table 2. Antigenic analysis of candidate vaccine viruses by hemagglutination inhibition.
Growth characterization of reassortant viruses in vitro
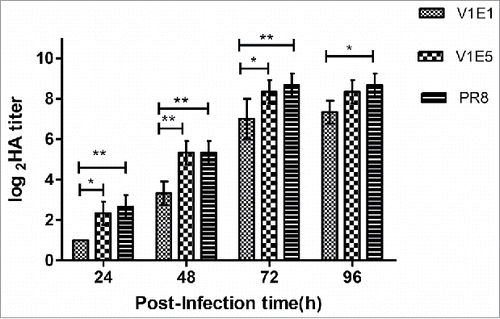
To analyze the replication efficiency of viruses possessing dominant G variants at position 220 in the HA gene, we tested the viral titer in eggs and MDCK cells. The results showed that the V1E5 titers were higher than those of V1E1 in eggs and MDCK cells (). Next, viral growth characteristics were assessed in MDCK cells. MDCK cells were infected with virus (V1E1 and V1E5) at a multiplicity of infection(M.O.I.) of 0.001 and supernatants were collected at the indicated times and tested using hemagglutination assays, PR8 was included as a control virus.. The results () showed that the HA titer of V1E5 was significantly higher than that of V1E1 at 24, 48 and 72 h p.i. (*p < 0.05, **p < 0.01 and *p < 0.05, respectively). There was no significant different between V1E5 and V1E1 at 96h p.i. The HA titer of V1E1 was significantly lower than that of PR8 at each time point (24, 48, 72h p.i. **p < 0.01, and 96h p.i. *p < 0.05). We noticed that there was no significant different of HA titer between V1E5 and PR8 from 24 to 96h p.i. indicating V1E5 had similar growth characteristics to that of PR8.
Figure 2. Growth characteristics of different viruses in MDCK cells. V1E1: RG-17SF003V1E1 with initial rescued in Vero cells followed by 1 passage growth in eggs; V1E5: RG-17SF003V1E5 with initial rescued in Vero cells followed by 5 passages growth in eggs; PR8: A/Puerto Rico/8/34 generated by RG. MDCK cell monolayers were infected at a multiplicity of infection of 0.001 of different viruses. The data was the results of three independent tests and analyzed by two-way ANOVA using GraphPad Prism 5 software package (version 5.0). ** represents p < 0.01; * represents p < 0.05.
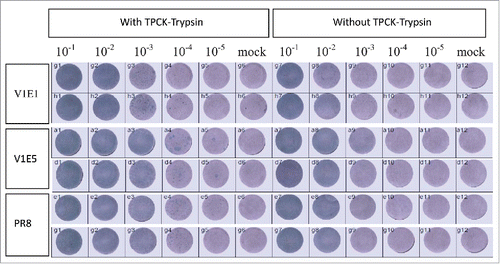
Trypsin-dependent plaque formation in MDCK cells
A previous study showed that HPAI SF003 had comparable ability to replicate both in the presence and absence of N-p-tosyl-L-phenylalanine chloromethyl ketone-treated (TPCK)-trypsin.Citation16 The CVV should maintain an HA cleavage site consistent with low pathogenic phenotype upon multiple passage in embryonated eggs.Citation14 We tested whether RG-SF003 V1E1 and V1E5 maintained low pathogenic phenotype using a trypsin-dependent plaque formation assay in MDCK cells. The results showed that V1E1 and V1E5 could not form plaques in the absence of TPCK- trypsin, whereas in the presence of TPCK- trypsin plaques could be clearly identified (). The ability of V1E1 and V1E5 to form plaques was comparable to that of the control virus PR8 in the presence or absence of TPCK- trypsin.
Figure 3. Replication of reassortant viruses in MDCK cells with or without TPCK-trypsin. V1E1: RG-17SF003V1E1 with initial growth in Vero cells followed by 1 passage in eggs; V1E5: RG-17SF003V1E5 with initial growth in Vero cells followed by 5 passages in eggs; PR8: A/Puerto Rico/8/34 generated by RG. The original viruses were diluted from 10−1to 10−5 and each diluted viruses were inoculated in MDCK cells. Mock cells were inoculated with PBS.
Chicken embryo lethality test and pathogenicity testing in chickens
To verify whether pathogenicity varied with amino acid changes in the HA, we tested viral pathogenicity in chicken embryos and chickens. Nine-day-old specific pathogen free (SPF) chicken embryos were inoculated with 1:10 diluted titers of V1E1 and V1E5 respectively. As shown in , the 50% chicken embryo lethal dose (CELD50) for V1E1 was≥9.0log 50% egg infectious dose (EID50)/ml and that of V1E5 was ≥9.22logEID50/ml, at 48 h after inoculation. To determine the pathogenic potential of the reassortant virus in chickens, SPF chickens (n = 10) were inoculated with 1:10 diluted titer of V1E1 and V1E5 via the intravenous (i.v.) route with 8.0logEID50/ml and 8.22logEID50/ml, respectively. All chickens remained healthy throughout the 10-day observation period with no mortalities (). The intravenous pathogenicity index (IVPI) was calculated according to the standard of the World Organisation for Animal Health (OIE).Citation17 The IVPI for both of the viruses was zero indicating that both V1E1 and V1E5 had low pathogenicity in chickens. These results demonstrated that V1E1 and V1E5 exhibited similar pathogenic characteristics as low pathogenic avian influenza.
Table 3. Virulence assessment of RG-SF003 in chicken embryos and chickens.
Total protein and HA antigen yield analysis of RG-SF003
Although we had determined the growth characteristics of the viruses in MDCK cells, giving an indication of the total virus concentration, these parameters were not reliable indicators of the HA content of purified viruses. To examine the content of total viral protein to HA in the reassortant viruses, egg-propagated viruses were purified by ultracentrifugation through sucrose gradients and the total protein was analyzed (). The total protein content (per 10 eggs) of V1E1 virus was significant lower than that of V1E5 and PR8 viruses. The viral protein was deglycosylated using PNGase F at 37°C for 18 h, and then analyzed by SDS-PAGE under reducing conditions (). The proportion of HA protein in the corresponding band was determined using Image lab software 3.0 (deglycosylated HA1 and HA2 were combined to determine the total proportion of HA). The proportion of HA was determined using the following equation: HA1+HA2 /HA1+HA2+NP+M. The analysis showed that there were no significant differences between V1E1 and V1E5 with regard to the proportion of HA protein ().
Figure 4. A Total protein content (left Y axis) and HA proportion (right Y axis). Total protein content per 10 eggs of each virus. HA proportion was determined using the following equation: HA1+HA2 /HA1+HA2+NP+M. B SDS-PAGE profile of various influenza viruses using the same sample amount. M: Marker (KD). PR8: A/Puerto Rico/8/34 generated by RG; V1E1: RG-17SF003V1E1 with initial rescued in Vero cells followed by 1 passage growth in eggs; V1E5: RG-17SF003V1E5 with initial rescued in Vero cells followed by 5 passages growth in eggs. The data was the results of two independent tests and analyzed by two-way ANOVA using GraphPad Prism 5 software package (version 5.0).** represents p < 0.01; *** represents p < 0.001.
Discussion
In addition to the risk of infection with seasonal influenza viruses, avian influenza viruses have also been responsible for human infections in recent years. As a result of the genetic changes in influenza viruses caused by antigenic drift and, periodically, antigenic shift, the CVVs need to undergo annual reformulation. The WHO recommend that the CVVs match the circulating influenza strains of seasonal influenza viruses for vaccine manufacture and also of avian influenza viruses (H7N9, H5N1 and H5N6) for pandemic preparedness.Citation10 Novel strains of H7N9 avian viruses continue to be identified and have evolve both genetically and antigenically since the first confirmed case of human infection with this virus was reported in 2013.Citation1,Citation3,Citation10,Citation13,Citation18 A previous study showed that just a few additional mutations in the H7N9 virus may be sufficient for full adaptation to human-to-human transmission.Citation19 Currently, vaccination is the most effective strategy for the prevention and control of influenza and its associated morbidity and mortality.Citation20 Therefore, the development of HPAI H7N9 CVV for pandemic preparedness is necessary. Egg adaptation is usually required to improve virus growth in eggs during the development of CVVs, but egg-adaptive mutations sometimes confer altered antigenicity, as previous reported.Citation21–24
In this study, we constructed the reassortant H7N9 virus, RG-SF003, using a reverse genetics approach. HA was modified by the removal of the polybasic cleavage site (), reducing pathogenicity as previously described.Citation25 Wild-type SF003 virus harbored a mixture of R/Kat at position 292 in the NA 16and we chose to synthesize R instead of K at position 292 of the NA gene because the H7N9 R292K substitution decreases NA activity and impairs virus replication in previous study.Citation26 To obtain virus with the growth properties needed for vaccine production,Citation27 RG-SF003 virus was continuously passaged in embryonated eggs. Sequence analysis showed that the NA gene was stable and did not possess any changes during multiple passages in embryonated eggs. The HA gene sequence was a mixture at position 220 from V1E2 to V1E5. The percentage of R reduced with increasing of passage at position 220 in the HA and that of G was increased at same position. The G was gradually dominant during egg-adaptive passage as its percentage from 1% (V1E2) to 70% (V1E5) in RG-003 viruses. The amino acid I became to appear fromV1E3. We speculated that the amino acid at 220 position in HA was a hot-spot mutation during egg-adaptive passage.
To determine the effect of these HA variations, we compared the characteristics of the two reassortant viruses V1E1 and V1E5. Several experiments were conducted to verify the observed reduction in pathogenicity of the viruses including their replication ability in the presence or absence of TPCK-trypsin, their lethality towards embryonated eggs and their pathogenicity in chickens. The results showed that V1E5 and V1E1 had similar abilities for plaque formation on MDCK cells with or without TPCK-trypsin, as seen for the control virus PR8. Both V1E1 and V1E5 maintained low pathogenicity in embryonated eggs and chickens, indicating that the mutation at position 220 did not alter pathogenicity. Next, we evaluated the influence of the HA mutation on antigenicity. Antigenic analysis () revealed that V1E1 was antigenically identical to wild-type SF003 and exhibited much lower reactivity to V1E5 ferret sera with a 16-fold reduction in HI titer. This result might correlate with the amino acid variation 220 in HA, suggesting that genetic and antigenic alterations occurred in V1E5 after multiple passages in eggs. Our results are consistent with previous studiesCitation28 reporting that variations at residue 220 in HA, which is proximal to the receptor binding site, could alter viral antigenicity, but the mutation was R220S in their study. As shown in , V1E1 had lower immunogenicity to homologous virus than V1E5. It is worth noting that both antisera to V1E1 and V1E5 did cross-react with other CVVs. Finally, we investigated whether the mutation improved viral yield, as previous described.Citation23,Citation28 The replication ability was evaluated in MDCK cells at the same M.O.I and the results revealed that V1E5 exhibited significantly higher HA titers than those of V1E1 from 24 to 72 h p.i. The total viral protein of V1E5 was significantly higher than that of V1E1, which correlated with the growth characteristics of the respective viruses in MDCK cells. These results suggested that the 220 mutation was an egg-adaptive change that led to an improved viral yield but changed the antigenicity. Virus adaptation to propagation in eggs usually results in the introduction of amino acid substitutions in the HA glycoprotein.Citation21–24,Citation34 However, the majority high-yield CVVs and licensed influenza vaccines continue to be prepared in eggs.Citation21–24 Studies have shown that disappointing vaccine effectiveness for H3N2 component during the 2012–13 and 2016–2017 influenza season which was related to mutations in the egg-adapted vaccine strain.Citation35,Citation36
As the limited of animal experiment conditions, we did not do wild type HPAI A (H7N9) viruses challenge experiment and evaluate the protection.
Our results indicated that 220 was a key amino acid for the adaptation of RG-SF003 virus to eggs, significantly altering the antigenicity or immunogenicity of the virus. It is therefore important to closely monitor changes in viral sequences that may affect antigenicity during virus passage in eggs. Although V1E5 exhibited altered antigenicity, the antisera showed cross-reactivity to previous H7N9 CVVs.
Materials and methods
Generation of H7N9 reassortant viruses by reverse genetics
The HA and NA genes were synthesized (GenScript) based on the sequence of the A/Guangdong/SF003/2016 virus, removing the multibasic cleavage site for HA and position 292 of the NA gene. The synthesis genes were cloned into a reverse genetics vector pHW2000 and designated pHW2000-SF003HA and pHW2000-SF003NA, respectively. (6+2) H7N9 reassortant influenza viruses (RG-SF003) were generated by a reverse genetics approach. Plasmids containing the six internal genes from PR8 along with pHW2000–003HA and pHW2000-SF003NA were co-transfected into Vero cells using TurboFect transfection reagent (Thermo Fisher Scientific, R0531) according to the manufacturer's instructions. Three days after transfection, the supernatant in the co-culture was inoculated into 9-day-old SPF eggs for virus propagation. Then, allantoic fluids with positive HA titers were collected at 48–72 h p.i. and were designated V1E1. V1E2–V1E5 virus stocks were prepared by inoculation into the allantoic cavity of 9-day-old SPF eggs using limiting dilution. Then, allantoic fluids with positive HA titers were collected at 48–72 h p.i. and stored at ˗80°C. All viruses were sequenced in our laboratory using Next Generation Sequencing as previously described.Citation29 We used BioEdit (Version 7.1.3.0) to analyze the viruses genetically. The 50% tissue culture infective dose (TCID50) was determined in MDCK cells and the 50% egg infectious dose (EID50) was determined in SPF embryonated eggs. TCID50 and EID50 were calculated by the Reed–Muench method.Citation30
Plaque formation in MDCK cells with or without trypsin
The viral plaque characteristics were determined in MDCK cells. Briefly, MDCK cells were grown in six-well culture plates in DMEM supplemented with 10% FBS and antibiotics (100 mg/ml penicillin and 100 mg/ml streptomycin). After inoculation with serial dilutions of the virus stock, cell monolayers were overlaid with Avicell in 2 × MEM without serum, with or without 2 µg/ml TPCK-trypsin. Cells were fixed with ice-cold 4% paraformaldehyde complemented with Triton X-100 and detected by staining of the nucleoprotein. Plates were incubated at 37°C for 24 h, then mouse-anti-influenza A monoclonal antibody (Merck Millipore, MAB8257 and MAB8258) was added to the cells and incubated at 4°C for 1 h, followed by incubation with goat-anti-mouse antibody (KPL, 074–1806) at 4°C for 1 h. Finally, true blue was added to visualize the plaques using Elispot Reader (AID, version4.0).
Chicken embryo lethality test
Nine-day-old SPF chicken embryos were inoculated in the allantoic cavity with 0.1 ml of a 10-fold dilution of each virus preparation with a known infectious titer.Citation31 Embryo viability was recorded at 48 h post-inoculation. The virus dose that caused death in 50% of embryos was calculated by the Reed–Muench method and reported as the median chicken embryo lethal dose (CELD50).Citation31
Pathogenicity in chickens
Fresh, infective, allantoic fluid with a HA titer >512 was diluted 1:10 in sterile isotonic saline. Ten 6-week-old SPF chickens were injected intravenously with 0.1 ml of the 1/10 dilution of viruses. Chickens were examined at 24-h intervals for 10 days. The IVPI was determined as described for chicken pathogenicity tests in the Manual of Diagnostic Tests and Vaccines for Terrestrial Animals.Citation17 At each observation, each chicken was scored 0 if normal, 1 if sick, 2 if severely sick, and 3 if deceased, as described in the OIE Guidelines.Citation17
Growth kinetics of rescued viruses in MDCK cells
Reassortant viruses were inoculated into a monolayer of MDCK cells at a M.O.I of 0.001. The supernatants from the infected cells were collected at specific time points (24, 48, 72 and 96 h p.i.) and stored at ˗80°C prior to processing. Viruses were titrated by a hemagglutination assay with 1% turkey erythrocytes. The data were analyzed by two-way ANOVA using the GraphPad Prism 5 software package (version 5.0).
Immunization of ferrets and the preparation of antisera
Antisera to reassortant viruses were produced in ferrets pre-screened for absence of antibodies to seasonal influenza viruses. Ferrets were inoculated intranasally (500ul per nostril) with reassortant viruses and blood samples collected on day 17 post-inoculation were tested in a HI assay to determine the HI titer as previously described.Citation24
Antigenic characterization by HI assay
Antigenic characterization of relevant H7N9 viruses, recommended as previous H7N9 CVVs by the WHO,Citation10 was performed using ferret antisera in a HI assay with 1% turkey erythrocytes according to standard protocols.Citation32 The HI titers are presented as the reciprocal value of the highest serum dilution that inhibited hemagglutination.
Viral protein and determination of the HA protein proportion
Viruses were purified by density gradient ultracentrifugation and the total protein content of purified viral protein was determined by a BCA protein assay kit (Solarbio, PC0020–500) using the protein BCA unit of a NanoDrop spectrophotometer (Thermo Fisher Scientific 2000/2000c). Viral protein of the same concentration was deglycosylated using PNGase F (NEB, P0704S) and a 1/50 dilution of PNGaseF enzyme was added to the sample as described previously.Citation33 The proteins were separated on a 12% SDS-PAGE precast gel (Bio-Rad Laboratories Mini-proteaN TGX), and gels were stained with Coomassie brilliant blue as previously described.Citation33 The proportion of HA protein in the observed band was determined using Image lab software 3.0. At least two independent virus concentrates were generated for each virus.
Ethical statement
All animal experiments were conducted in accordance with the Guidelines for Animal Experiments described and approved by the Animal Care Welfare Committee (No. 20170609021).
Abbreviations
| CVV | = | candidate vaccine virus |
| EID50 | = | 50% egg infectious dose |
| HI | = | hemagglutination inhibition |
| HPAI | = | highly pathogenic avian influenza |
| HA | = | hemagglutinin |
| IVPI | = | intravenous pathogenicity index |
| M.O.I. | = | multiplicity of infection |
| NA | = | neuraminidase |
| PR8 | = | A/Puerto Rico/8/34 |
| p.i. | = | post-infection |
| RG | = | reverse genetics |
| SPF | = | specific pathogen free |
| SF003 | = | A/Guangdong/SF003/2016 |
| TPCK | = | N-p-tosyl-L-phenylalanine chloromethyl ketone-treated |
| WHO | = | World Health Organization |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888–97. doi:10.1056/NEJMoa1304459.
- Kreijtz JH, Kroeze EJ, Stittelaar KJ, de Waal L, van Amerongen G, van Trierum S, van Run P, Bestebroer T, Kuiken T, Fouchier RA, et al. Low pathogenic avian influenza A(H7N9) virus causes high mortality in ferrets upon intratracheal challenge: A model to study intervention strategies. Vaccine. 2013;31(43):4995–9. doi:10.1016/j.vaccine.2013.06.071.
- Wang D, Yang L, Zhu W, Zhang Y, Zou S, Bo H, Gao R, Dong J, Huang W, Guo J, et al. Two outbreak sources of influenza A (H7N9) viruses have been established in China. J Virol. 2016;90(12):5561–73. doi:10.1128/JVI.03173-15.
- Ip DK, Liao Q, Wu P, Gao Z, Cao B, Feng L, Xu X, Jiang H, Li M, Bao J, et al. Detection of mild to moderate influenza A/H7N9 infection by China's national sentinel surveillance system for influenza-like illness: Case series. BMJ. 2013;346:f3693. doi:10.1136/bmj.f3693.
- Peiris JS, Cowling BJ, Wu JT, Feng L, Guan Y, Yu H, Leung GM. Interventions to reduce zoonotic and pandemic risks from avian influenza in Asia. Lancet Infect Dis. 2016;16(2):252–8. doi:10.1016/S1473-3099(15)00502-2.
- World Health Organization. Monthly risk assessment summary. [accessed 2017 Sep 2]. http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/.
- World Health Organization. Human infection with avian influenza A(H7N9) virus –China. Disease outbreak news. 27 February 2017. [accessed 2017 Sep 2]. http://www.who.int/csr/don/27-february-2017-ah7n9-china/en/.
- Zhou L, Tan Y, Kang M, Liu F, Ren R, Wang Y, Chen T, Yang Y, Li C, Wu J, et al. Preliminary epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus, China, 2017. Emerg Infect Dis. 2017;23:1355–9. https://doi.org/10.3201/eid2308.170640. doi:10.3201/eid2308.170640.
- World Health Organization. Monthly risk assessment summary; 30 Oct 2017. [accessed 2017 Nov 23]. http://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_10_30_2017.pdf?ua=1.
- World Health Organization. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Wkly Epidemiol Rec. 2017; 24;92(12):129–44.
- De Groot AS, Ardito M, Terry F, Levitz L, Ross T, Moise L, Martin W. Low immunogenicity predicted for emerging avian-origin H7N9: Implication for influenza vaccine design. Hum Vaccin Immunother. 2013;9(5):950–6. doi:10.4161/hv.24939.
- Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, et al. Epidemiology of the avian influenza a (h7n9) outbreak in china. N Engl J Med. 2013;370(6):520–32. doi:10.1056/NEJMoa1304617.
- Kile JC, Ren R, Liu L, Greene CM, Roguski K, Iuliano AD, Jang Y, Jones J, ThorS, SongY, et al. Update: Increase in human infections with novel asian lineage avian influenza a(h7n9) viruses during the fifth epidemic — china, october 1, 2016–august 7, 2017. MMWR Morb Mortal Wkly Rep. 2017;66(35):928–932. doi:10.15585/mmwr.mm6635a2.
- World Health Organization. Update of WHO biosafety risk assessment andguidelines for the production and quality control of human influenza vaccines against avian influenza A (H7N9) virus; 10 May 2013. [accessed 2017 Sep 2]. http://www.who.int/biological/areas/vaccines/influenza/biosafetyrisk assessment_10may2013.pdf.
- World Health Organization. A description of the process of seasonal and h5n1 influenza vaccine virus selection and development; 19 Nov 2007. [accessed 2017 Sep 2]. http://apps.who.int/gb/pip/pdf_files/Fluvaccvirusselection.pdf?ua=1.
- Zhu W, Zhou J, Li Z, Yang L, Li X, Huang W, Zou S, Chen W, Wei H, Tang J, et al. Biological characterisation of the emerged highly pathogenic avian influenza (HPAI) A(H7N9) viruses in humans, in mainland China, 2016 to 2017. Euro Surveill. 2017;22(19):pii: 30533. doi:10.2807/1560-7917.ES.2017.22.19.30533.
- World Organization for Animal Health(OIE). Avian influenza (infection with avian influenza viruses). In: Manual of diagnostic tests and vaccines for terrestrial animals. Paris: OIE; 2016. p.1–23.
- Iuliano AD, Jang Y, Jones J, Davis CT, Wentworth DE, Uyeki TM, Roguski K, Thompson MG, Gubareva L, Fry AM, et al. Increase in human infections with avian influenza a(h7n9) virus during the fifth epidemic — china, October 2016–February 2017. MMWR Morb Mortal Wkly Rep. 2017;66(9):254–255. doi:10.15585/mmwr.mm6609e2.
- Kageyama T, Fujisaki S, Takashita E, Xu H, YamadaS, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013; 11;18(15):20453.
- Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: A modelling study. Lancet Infect Dis. 2012;12(9):687–95. doi:10.1016/S1473-3099(12)70121-4.
- Wang W, Lu J, Cotter CR, Wen K, Jin H, Chen Z. Identification of critical residues in the hemagglutinin and neuraminidase of influenza virus h1n1pdm for vaccine virus replication in embryonated chicken eggs. J Virol. 2013;87(8):4642–9. doi:10.1128/JVI.03271-12.
- Nakamura K, Shirakura M, Suzuki Y, Naito T, Fujisaki S, Tashiro M, Nobusawa E. Development of a high–yield reassortant influenza vaccine virus derived from the A/Anhui /1/2013(H7N9) strain. Vaccine. 2016;34(3):328–33. doi:10.1016/j.vaccine.2015.11.050.
- Barman S, Franks J, Turner JC, Yoon SW, Webster RG, Webby RJ. Egg-adaptive mutations in H3N2v vaccine virus enhance egg-based production without loss of antigenicity or immunogenicity. Vaccine. 2015;33(28):3186–92. doi:10.1016/j.vaccine.2015.05.011.
- Ridenour C, Johnson A, Winne E, Hossain J, Mateu-Petit G, Balish A, Santana W, Kim T, Davis C, Cox NJ, et al. Development of influenza A(H7N9) candidate vaccine viruses with improved hemagglutinin antigen yield in eggs. Influenza and Other Respiratory Viruses. 2015;9(5):263–70. doi:10.1111/irv.12322.
- Liu Q, Lu L, Sun Z, Chen GW, Wen Y, Jiang S. Genomic signature and protein sequence analysis of a novel influenza A (H7N9) virus that causes an outbreak in humans in China. Microbes Infect. 2013; 15(6–7):432–9. doi:10.1016/j.micinf.2013.04.004.
- Wu Y, Bi Y, Vavricka CJ, Sun X, Zhang Y, Gao F, Zhao M, Xiao H, Qin C, He J, et al. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res. 2013;23(12):1347–55. doi:10.1038/cr.2013.144.
- Fulvini AA, Ramanunninair M, Le J, Pokorny BA, Arroyo JM, Silverman J, Devis R, Bucher D. Gene constellation of influenza a virus reassortants with high growth phenotype prepared as seed candidates for vaccine production. PLoS ONE. 2011;6(6):e20823. doi:10.1371/journal.pone.0020823.
- Chen Z, Baz M, Lu J, Paskel M, Santos C, Subbarao K, Jin H, Matsuoka Y. Development of a high-yield live attenuated h7n9 influenza virus vaccine that provides protection against homologous and heterologous h7 wild-type viruses in ferrets. J Virol. 2014;88(12):7016–23. doi:10.1128/JVI.00100-14.
- Zhang H, Liu M, Zeng X, Zhao X, Deng Z, Yang L, Chen W, Li Z, Jiao M, Xia W, et al. Identification of a novel reassortant A (H9N6) virus in live poultry markets in Poyang Lake region, China. Arch Virol. 2017;162(12):3681–3690. doi:10.1007/s00705-017-3507-x.
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–7.
- Zhou F, Zhou J, Ma L, Song S, Zhang X, Li W, Jiang S, Wang Y, Liao G. High-yield production of a stable Vero cell-based vaccine candidate against the highly pathogenic avian influenza virus H5N1. Biochem Biophys Res Commun. 2012;421(4):850–4. doi:10.1016/j.bbrc.2012.04.101.
- World Health Organization Global Influenza Surveillance Network. Manual for the laboratory diagnosis and virological surveillance of influenza. Geneva (Switzerland):WHO Press; 2011.
- Li C, Shao M, Cui X, Song Y, Li J, Yuan L, Fang H, Liang Z, Cyr TD, Li F, et al. Application of deglycosylation and electrophoresis to the quantification of influenza viral hemagglutinins facilitating the production of 2009 pandemic influenza (H1N1) vaccines at multiple manufacturing sites in China. Biologicals. 2010; 38(2):284–9. doi:10.1016/j.biologicals.2009.12.004.
- Parker L, Wharton SA, Martin SR, Cross K, Lin Y, Liu Y, Feizi T, Daniels RS, McCauley JW. Effects of egg-adaptation on receptor-binding and antigenic properties of recent influenza A (H3N2) vaccine viruses. J GenVirol. 2016;97(6):1333–44.
- Skowronski DM, Janjua NZ, De Serres G, Sabaiduc S, Eshaghi A, Dickinson JA, Fonseca K, Winter AL, Gubbay JB, Krajden M, et al. Low 2012–13 influenza vaccine effectiveness associated with mutation in the egg-adapted h3n2 vaccine strain not antigenic drift in circulating viruses. PLoS ONE. 2014;9(3):e92153. doi:10.1371/journal.pone.0092153.
- Zost SJ, Parkhouse K, Gumina ME, Kim K, Diaz Perez S, Wilson PC, Treanor JJ, Sant AJ, Cobey S, Hensley SE. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodieselicited by egg-adapted vaccine strains. Proc Natl Acad Sci. 2017; pii: 201712377.
