ABSTRACT
HIV-specific T-cell responses play a key role in controlling HIV infection, and therapeutic vaccines for HIV that aim to improve viral control will likely need to improve on the T-cell responses induced by infection. However, in the setting of chronic infection, an effective therapeutic vaccine must overcome the enormous viral genetic diversity and the presence of pre-existing T-cell responses that are biased toward immunodominant T-cell epitopes that can readily mutate to evade host immunity and thus potentially provide inferior protection. To address these issues, we investigated a novel, epidermally administered DNA vaccine expressing SIV capsid (p27Gag) homologues of highly conserved elements (CE) of the HIV proteome in macaques experiencing chronic but controlled SHIV infection. We assessed the ability to boost or induce de novo T-cell responses against the conserved but immunologically subdominant CE epitopes. Two groups of animals were immunized with either the CE DNA vaccine or a full-length SIV p57gag DNA vaccine. Prior to vaccination, CE responses were similar in both groups. The full-length p57gag DNA vaccine, which contains the CE, increased overall Gag-specific responses but did not increase CE responses in any animals (0/4). In contrast, the CE DNA vaccine increased CE responses in all (4/4) vaccinated macaques. In SIV infected but unvaccinated macaques, those that developed stronger CE-specific responses during acute infection exhibited lower viral loads. We conclude that CE DNA vaccination can re-direct the immunodominance hierarchy towards CE in the setting of attenuated chronic infection and that induction of these responses by therapeutic vaccination may improve immune control of HIV.
Introduction
Antiretroviral therapy (ART) has a tremendous capacity to suppress HIV replication and prolong the life of HIV-infected persons. However, an effective ART regimen requires a combination of antivirals that are costly, cannot completely reverse immune dysfunction, can lead to the emergence of drug resistant viral variants, and cannot eradicate latently infected cells.Citation1-3 In addition, ART can have adverse side effects that can exacerbate non-compliance. Cessation of therapy typically results in viral rebound, thus requiring a lifetime of treatment.Citation4-8 Several additional approaches are being developed for administration in combination with ART, including gene therapies, latency reversal agents, and vaccines to boost antiviral immunity. These efforts seek to eradicate the residual population of cells harboring infectious proviruses, or achieve sustained viral remission, characterized by a protracted period of undetectable viremia after stopping ART.Citation9-15
Therapeutic vaccines offer a relatively inexpensive and widely applicable means to potentially contribute to or achieve these goals. A particular focus of therapeutic vaccines is to induce strong HIV-specific CD8+ cytotoxic T-lymphocyte (CTL) responses that can control viremia and eliminate residual cells expressing virus and viral antigens.Citation16-18 In support of this possibility, several therapeutic vaccines have been shown to augment virus-specific CTLs and in some cases, reduce viral loads in the plasma and tissues of rhesus macaques chronically infected with SIV.Citation19-22 We previously showed that an SIV therapeutic DNA vaccine delivered by particle mediated epidermal delivery (PMED) into the skin of SIV-infected macaques receiving suboptimal ART provided a substantial, 3–5 log reduction in viral load and induced durable control (> 1 year) of viremia in approximately half of vaccinated animals.Citation23
The vaccine employed in our previous therapeutic vaccine study consisted of full-length (FL) SIV immunogens, and although it broadened the mucosal CD8+ T-cell response, these responses were mostly directed against immunodominant epitopes rather than subdominant conserved regions.Citation24-26 A potential concern with the use of FL immunogens is that HIV diversity is so enormous and pathways to escape mutations are so varied that immunogens driving induction of immunodominant responses against variable epitopes, even if directed against multiple epitopes, are unlikely to provide a sustained block against the wide range of immune escape pathways.Citation27,Citation28
To address these concerns, we designed a conserved elements (CE) DNA vaccine,Citation29-32 that exclusively expresses CE sequences as a strategy to direct CD8+ T-cells to regions of the proteome that are highly conserved among viruses, where mutations are more likely to decrease viral fitness, and are associated with viremic control.Citation33 This approach was also developed based on findings showing that CD8+ T-cells targeting conserved regions, particularly within Gag, were associated with lower viral load, whereas responses to other, more variable regions were associated with high viral loadsCitation33-35; and that CD8+ T-cell responses to conserved epitopes were preferentially detected in elite controllers and long term non-progressors.Citation17,Citation18,Citation36-38,Citation39,Citation40 However, responses associated with viral control are typically subdominant in natural infections,Citation24,Citation33,Citation41,Citation42 with exceptions including individuals with “protective” HLA alleles, who tend to target epitopes whose escape results in a loss of viral fitness.Citation43 We hypothesize that a therapeutic vaccine that can direct immune responses to CE will be more effective at inducing durable viral remission by inhibiting a wider variety of viral variants, limiting escape mutations, and more often reducing viral fitness if escape occurs. In support of this concept, we previously demonstrated that prophylactic administration of DNA vaccines expressing CE from HIV and SIV Gag can induce stronger CE-specific responses in mice and uninfected rhesus macaques, compared to vaccinating with a FL HIV Gag immunogen.Citation29,Citation30,Citation32 Interestingly, in these studies, priming with the CE Gag DNA vaccine (hereafter referred to as the CE vaccine) and then boosting with CE plus FL Gag DNA was much more effective at generating CE-specific responses than priming and boosting with the CE vaccine alone.Citation29,Citation30,Citation32 However, we also found that priming T-cells with a FL Gag DNA substantially reduced recognition of CE epitopes when followed by a subsequent boost with CE,Citation30 suggesting that it may not be possible to redirect immunodominance by therapeutic vaccination during ongoing infection. To experimentally address this, we conducted a pilot study to determine if immunizing SIV-HIV hybrid (SHIV) virus infected rhesus macaques with a CE DNA vaccine could redirect a pre-existing immunodominant SIV-specific T-cell repertoire toward greater recognition of CE epitopes. The CE DNA vaccine significantly increased T-cell responses against subdominant CE epitopes in macaques with attenuated infection, demonstrating the feasibility of using therapeutic immunization to redirect the virus-primed T-cell repertoire toward greater targeting of CE. Furthermore, in a separate study, rhesus macaques that naturally developed CE responses during acute SIV infection were found to exhibit lower viral set points when compared to those that did not develop detectable CE responses, supporting the concept that increasing CE responses during therapy may improve control of HIV.
Materials and methods
Ethics statement and animal care
Sixteen Indian origin, adult rhesus macaques were used for these studies. The Washington National Primate Research Center (WaNPRC) is responsible for the humane care of laboratory animals. The WaNPRC is accredited by the American Association for the Accreditation of Laboratory Animal Care International (AAALAC). The University of Washington's Institutional Animal Care and Use Committee approved all experiments in these macaques. Animals were singly housed in comfortable, clean, adequately-sized cage. Cages, racks, and accessories were sanitized in mechanical cage washers at least once every two weeks and waste pans were cleaned daily. Temperatures in animal quarters were maintained between 72–82°F. Animals were fed a commercial monkey chow, supplemented daily with fruits and vegetables, and drinking water was available at all times provided by automatic watering devices. Throughout the study, animals were checked twice daily by the veterinary technicians to evaluate their physical and clinical condition. The macaques received environmental enrichment for the duration of the study including grooming contact, perches, toys, foraging experiences and access to additional environment enrichment devices. For the CE vaccination study, eight adult Indian origin rhesus macaques previously infected with SHIV89.6PCitation44 were reassigned to this project after completion of a separate, previous vaccine experiment consisting of a recombinant adenoviral vaccine prime (expressing either HIVIIIB tat,Citation45 SIV239 gag,Citation46 HIV89.6P gp140 and/or SIV239 nefCitation47) and a Tat, gp140, or Nef protein boost performed under the NIH/NIAID Simian Vaccine Evaluation Unit Program (Dr. Nancy Miller, Program Officer, NIH; Dr. Ruth Ruprecht, Principal Investigator). For the acute SIV infection study, eight adult Indian origin macaques were challenged with SIVΔB670 (kind gift, Dr. Michael Murphey-Corb, University of Pittsburgh) intravenously using a previously established protocol.Citation23 Cryopreserved SIVΔB670 was thawed and diluted in RPMI, and then 1 mL of RPMI containing 100 TCID50 was injected into the saphenous vein of each macaque. Macaques were MHC-I typed for Mamu alleles (A*01, A*02, A*08, A*11, B*01, B*08, B*17, and B*29) by PCR as previously described.Citation48-50
All biopsies were performed under ketamine (10 mg/kg) or Telazol (2.5-10 mg/kg) anesthesia and any continued discomfort or pain was alleviated at the discretion of veterinary staff. Euthanasia prior to necropsy was performed on the SHIV infected animals by administration of Euthasol® (Virbac Corp., Houston, TX) while the animal was under deep anesthesia in accordance with guidelines established by the 2007 American Veterinary Medical Association Guidelines on Euthanasia. None of the animals became severely ill during the course of the study and none required euthanasia prior to their experimental endpoint.
Particle mediated epidermal delivery (PMED) of CE and FL DNA vaccines
We designed an SIV Gag CE DNA vaccineCitation32 corresponding to sequences homologous to the previously described HIV Gag CE DNA vaccineCitation29 to test the CE vaccine concept in rhesus macaques infected with SHIV89.6P. The SIV Gag CE DNA vaccine consists of two plasmids, each with a CMV promoter, a granulocyte macrophage-colony stimulating factor (GM-CSF) signal peptide to promote extracellular secretion, followed by a string of seven sequences encoding conserved elements of the viral proteome ranging from 12–24 amino acids in length separated by linker sequences designed for optimal immune-proteasome cleavage, and finally a bovine growth hormone polyadenylation signal ().Citation51,Citation52 To avoid a strongly hydrophobic amino terminus, which may interfere with intracellular trafficking, the CE1 sequence was placed at the C terminus.Citation29 To enhance coverage, the two plasmids corresponded to alternate versions of CE differing only by “toggled” amino acids. CE2, CE3, and CE5 contain a single toggle amino acid site. CE4 contains 2 additional toggled amino acid sites since those variants were always associated with the primary toggle site in the Los Alamos HIV database.Citation53 CE1, CE6, and CE7 do not contain a toggle site due to their high conservation among the available SIV sequences. The final SIV Gag CE DNA vaccine consisted of equal amounts of the toggled plasmids (p27CE1 and p27CE2). The FL DNA vaccine consisted of p57Gag from SIV17E-Fr, which is homologous to the SHIV89.P challenge virusCitation54,55 and contains each of the CE. Each SIV DNA vaccine (CE and FL) was co-delivered at a 10:1 ratio along with a 3rd plasmid expressing the heat-labile E. coli enterotoxin (LT) as a genetic adjuvant, previously shown to significantly increase systemic and mucosal immunogenicity of PMED-delivered DNA vaccines.Citation23,Citation56
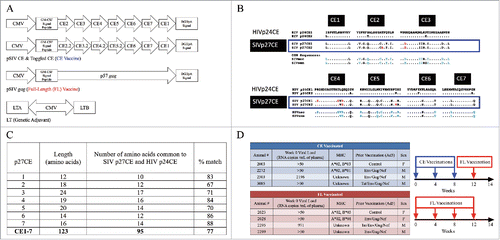
Figure 1. DNA vaccines and vaccine study design. (A) DNA vaccines. The CE DNA vaccine consists of two plasmids encoding seven different conserved sequences connected by optimized linker sequences. CE2, CE3, CE4, and CE5 differ by toggled amino acids analogous to the an HIV p24CE vaccine previously described (see Methods). The FL DNA vaccine expresses the p57 gag coding sequence from p57Gag from SIV17E-Fr. A genetic adjuvant plasmid expressing the heat-labile E. coli enterotoxin (LT) was co-delivered with both DNA vaccines (CE and FL) at each dose. (B) Comparison of p27Gag CE in HIV and SIV strains, adapted from Hu et al.Citation32 SIV p27Gag sequences from SIVmac (macaque origin) and SIVsmm (sooty mangabey), were compared to reported HIV Gag p24 CE sequences.Citation29 The SIV p27CE1 and p27CE2 toggled amino acids are indicated in red. Blue amino acids indicate SIV sequences that are dissimilar from HIV sequences. (C) The number of CE amino acids (AA) that are common in both SIV and the HIV-1 M group (see panel B), including toggle AA, for each CE is shown along with percent match between the HIV-1 p24CE and the SIV homologues. (D) Study Design. Macaques were stratified into either the CE (conserved elements) or FL (full length) DNA vaccine groups based on plasma viral load, MHC class I genotype, prior vaccination, and sex. The CE + LT DNA vaccinated animals (blue) received three doses of the CE + LT DNA vaccine administered into the epidermis of the skin by PMED at weeks 0, 4, and 8 followed by a single boost of the FL + LT DNA vaccine at week 12. The FL DNA vaccinated animals (red) received four doses of the FL SIV Gag DNA vaccine at weeks 0, 4, 8 and 12.
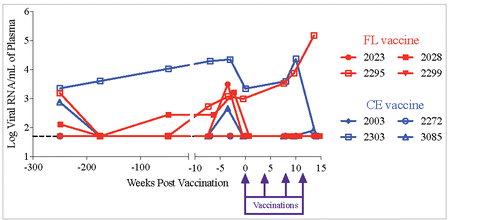
To administer the vaccines by PMED, the CE, FL Gag and LT plasmid DNAsCitation32,Citation56 were formulated separately onto gold particles as previously described.Citation23 Using the PowderJect® XR1 gene delivery device (PowderJect Vaccines, Inc.), each macaque received 32 μg of the CE or FL DNA vaccine along with 3.2 μg of the plasmid expressing the LT adjuvant administered over 16 skin sites (2 μg CE or FL DNA + 0.2 μg LT per site) along the abdomen and over the inguinal lymph nodes. Skin sites were clipped of fur and swabbed with alcohol prior to PMED administration. The two toggled SIV Gag DNA vaccine plasmids were previously shown to express similar levels of proteinCitation32 but since they were co-delivered, differences in the immunogenicity between the plasmids were not assessed in this study.
IFN-γ ELISpot assay
This assay was performed on PBMC or on Percoll® (GE Healthcare, Catalog #: 17-0891-01) purified mononuclear cells isolated from solid tissues as described below. Cells were stimulated with SIV Gag peptide pools containing 15-mer peptides overlapping by 11 amino acids and spanning the full length SIV Gag p57 viral protein (provided by the National Institutes of Health AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases). In addition, cells were stimulated with peptide pools spanning only the CE regions, including 15-mer peptides overlapping by 11 amino acids and 10-mer peptides overlapping by 9 amino acids. Peptides were resuspended in 80% DMSO and 20% water and then diluted in cell culture media so that the final concentration of DMSO in the culture media was <1%. Peptide stimulations were performed at a final concentration of 1μg/mL. Concanavalin A (5 μg/mL, Sigma-Aldrich, Catalog #: C2272) was used as a positive control, and DMSO at a concentration equal to peptide stimulations was used as a negative control. Antigen-specific cells secreting IFN-γ were detected using anti-macaque IFN-γ monoclonal antibodies (U-Cytech-BV) as previously described.Citation23 Spot forming cells (SFC) were enumerated using an Immunospot Analyzer with CTL Immunospot Profession Software (Cellular Technology Ltd.). Peptide-specific SFC were determined by subtracting background SFC measured in negative control wells. Responses were considered positive if they were at least twice that of background values and a minimum of 50 SFC per million cells.
Intracellular cytokine staining
Multiparameter flow cytometry was used to determine T-cell immune responses using peptide stimulated PBMC as previously described.Citation23 One million PBMC were stained for each condition (DMSO, PMA/Ionomycin, or peptide(s)) for 10–14 hours. PBMC were stained with the following antibodies: Live/Dead Yellow (Invitrogen®, Catalog #:L34959), CD3-APC (BD Biosciences, clone SP34-2, Catalog #: 551916), CD4-PerCP Cy5.5 (BD BioSciences, clone L200, Catalog #: 551980), CD8 APC-Cy7 (BD Biosciences, clone RPA-T8, Catalog #: 55760), IFNγ-BV650 (BioLegend, clone 4S.B3, Catalog #: 502537), IL-2-PE (BioLegend, clone MQ1-17H12, Catalog #: 500307), TNFα-PECy7 (BD Biosciences, clone Mab11, Catalog #: 557647), CD107a-FITC (BD Biosciences, clone H4A3, Catalog #: 555800). Cells were fixed in 1% paraformaldehyde and acquired using an LSR II flow cytometer (BD Biosciences) and the data analyzed using FlowJo software (Tree Star, Inc.). Samples were considered positive if peptide-specific responses were at least twice that of the negative control plus at least 0.01% after background subtraction.
Isolation of gut-associated mononuclear lymphocytes
15–20 cm of jejunum were removed surgically, then cut length-wise and mucous removed by gentle wiping with a paper towel. Next, fat tissue was excised and the remaining tissue was cut into small (1/3 × 1/3 cm) pieces and then enzymatically digested in serum free RPMI-1640 containing L-glutamine, penicillin/streptomycin, gentamycin, DNAse (4 μg/ml; Sigma-Aldrich, Catalog #: DN25) and Liberase® (40 μg/ml; Roche Inc., Catalog #: 5401119001) at 37°C and 5% CO2 for one hour. Tissue fragments were then crushed onto and passed through 70 μm filters and washed with R10 media. Cells were then counted on a Cellometer® Auto T4 (Nexcelom Bioscience) and cryopreserved. Jejunum and lymph nodes were collected at necropsy (Week 14). Due to limited numbers of cells isolated from the gut, only ELISpot was performed.
Plasma viral load quantification
SHIV89.6P plasma viral loads were determined by nucleic acid sequence-based amplicfication (NASBA) at Advanced BioScience Laboratories Inc. (ABL) as previously described.Citation44 The Virology Core at the WaNPRC quantified viral RNA in the plasma for the SIVΔB670 infected animals by RT-PCR as previously described.Citation23,Citation44
Statistical analyses
Statistical differences between the two groups were calculated using a two-sided Mann-Whitney test in GraphPad Prism (Version 6, GraphPad Software). Correlations between immune responses and viral loads were determined by a Spearman's rank correlation test. P ≤ 0.05 was considered significant for each test.
Results
SIV CE therapeutic DNA vaccine and study design
The CE DNA vaccine expresses seven highly conserved peptide sequences derived from p27Gag. The FL DNA vaccine expresses p57Gag from SIV17E-Fr. The LT genetic adjuvant plasmid expresses both the A and B subunits of the heat-labile enterotoxin of E. coli (LT) (). The SIV p27CE sequences were selected based on alignments with available SIV sequences and homology to HIV (). Comparison of SIV p27 Gag CE to the HIV p24 Gag CE sequences revealed a range of 75–100% amino acid similarity per CE (). As predicted from their strong conservation levels, the SIV p27 Gag CE are contained within both the FL SIV17E-Fr p57 Gag vaccine and the SHIV89.6P challenge stock that was previously used to infect the macaques in this study.Citation55,Citation57 To investigate the immunogenicity of the SIV CE DNA vaccines in the setting of attenuated chronic infection, we enrolled a group of eight rhesus macaques that had been previously immunized with either a recombinant adenovirus 5 (Ad5) vaccine expressing HIVIIIB tat, SIVmac239 gag, HIV89.6P gp140 and/or SIV239 nef or an empty Ad5 vector control.Citation44 All animals became infected following intravenous challenge with SHIV89.6P, were infected for nearly five years when enrolled in this study, but had attenuated infection, with viral loads ranging from undetectable to 104 copies/ml (). The eight macaques were stratified equally between the conserved elements (CE, N = 4) and full-length (FL, N = 4) DNA vaccination groups based on viral load, MHC, prior Ad5 vaccination, and sex (). The CE group received three priming doses of the CE DNA vaccine at weeks 0, 4, and 8 and one booster dose with the FL DNA vaccine at week 12 (), a regimen that was previously shown to induce strong CE responses in uninfected macaques.Citation30,Citation32 The FL vaccinated group (N = 4) received four doses at the same time points. The total amount of DNA per dose was equal in both groups and each dose was co-administered with the LT genetic adjuvant by PMED.
CE DNA vaccination increases CE-specific IFN-γ T-cells in both the blood and mucosal tissues
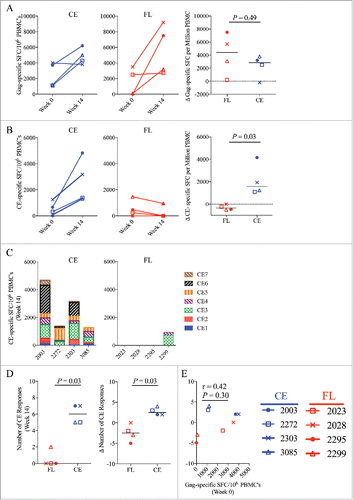
To determine the impact of the vaccines on T-cell responses in blood, the frequency of IFN-γ responses was determined by ELISpot at week 0 and week 14, the latter corresponding to two weeks following the last vaccination, using peptide pools representing either the full-length p57Gag or CE sequences alone. The magnitude of FL Gag-specific and CE-specific IFN-γ T-cell responses prior to vaccination (week 0) were similar between both groups (). Vaccinating with either the CE or FL DNA vaccine increased overall Gag-specific T–cell responses to similar levels (p = 0.49, ). In contrast, immunization with the CE vaccine followed by one dose of the FL vaccine significantly increased the magnitude of CE-specific responses in all four vaccinated animals in the CE group, whereas immunization with the FL vaccine resulted in no increase or a decline in CE responses. Overall, significantly higher CE responses were observed in the CE vaccine group (p = 0.03, ). Following vaccination, T-cell responses were detected against 5–7 CE in all four animals in the CE vaccine group versus only one animal in the FL group against 2 of the CE sequences (). Two CE vaccinated animals (2003 and 2303) developed T-cell responses against all 7 CE and all four animals responded to both CE3 and CE5. Overall, the CE vaccine induced greater magnitude, (, p = 0.03) and broader specificity (, p = 0.03) against otherwise subdominant CE-specific T-cell epitopes when compared to the FL only group. Notably, the magnitude of the overall Gag-specific T-cell response prior to vaccination (week 0) did not correlate with CE-breadth post-vaccination (week14), indicating that the levels of pre-existing Gag responses prior to vaccination did not influence the ability of the vaccine to broaden the CE-specific immune responses (p = 0.30, ). In agreement with prior vaccine studies in uninfected macaques,Citation30 we also did not observe a relationship between a particular MHC class I haplotype and the ability to develop CE responses (Supplemental Table 1).Citation30
Figure 2. CE-specific IFN-γ T-cell responses in the blood. T-cell responses in Spot Forming Cells (SFC) per on million PBMC were measured before and after vaccination by IFN-γ ELISpot following stimulation with SIV CE or p57 Gag peptide pools. Shown are the mean spot forming cells (SFC) per 106 PBMC corresponding to (A) Gag-specific and (B) CE-specific SFC measured at baseline (week 0) and 2 weeks after the final DNA vaccine dose (week 14) in the CE group (left panels) and FL group (middle panels). (C) The specificities of the CE-specific responses in each animal in the CE group (left panel) and the FL group (right panel) at week 14 were measured against each separate CE peptide pool. (D) Shown are the breadth of CE-specific responses (number of positive CE peptide pools) at week 14 (left panel) and the change of the CE breadth at week 14 versus week 0 (right panel). (E) Analysis of the correlation between the magnitude of Gag-specific SFC prior to vaccination (Week 0) versus the increase in CE breadth post-vaccination (Week 14) was determined by Spearman Rank correlation test. Median values are indicated. P-values were determined by non-parametric Mann-Whitney T tests. P values ≤ 0.05 were considered significant.
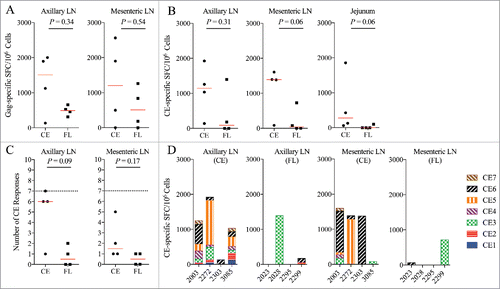
Lymph nodes and intestinal tissues are known viral reservoirs,58 and our previous results showed that the relative ability of a DNA vaccine to induce responses in these tissues correlated with a significant reduction in viral replication in these compartments.Citation23,Citation59 We therefore investigated the effects of the vaccines on immune responses in mucosal and lymphoid tissues. The CE and FL vaccinated animals had similar levels of ELISpot responses to the whole SIV p57 Gag in the axillary and mesenteric lymph nodes (LN) (). There was no significant difference between the FL and CE groups in the magnitude of the CE response in the axillary LN, but a strong trend (p = 0.06) for higher magnitude responses in the mesenteric LN and jejunum. In addition, all four animals in the CE group exhibited detectable responses to the CE portions of Gag in each tissue whereas only 1 or 2 animals in the FL group developed a detectable CE response in these tissues (). The CE group may have also had broader CE-specific ELISpot responses in the axillary (p = 0.09) and mesenteric LN (p = 0.17) with 4/4 animals developing responses against 1–7 CE, whereas only 2/4 animals in the FL vaccine group had CE responses, and only against 1–2 CE sequences (). CE breadth was not examined in the jejunum due to sample limitations. In the FL group, the strongest and most frequently targeted CE response in the blood () or lymph nodes () was biased towards CE3, whereas in the CE group, T cell responses were directed primarily to CE5 and CE6 ( & ), a result that is in agreement with previous CE DNA vaccine studies in uninfected rhesus macaques.Citation32 Taken together, these data demonstrate that therapeutic immunization with the CE vaccine was able to significantly increase the magnitude and breadth of CE-specific IFN-γ secreting cells in the blood as well as in lymphoid tissues in SHIV-infected macaques with attenuated infection.
Figure 3. CE-specific IFN-γ T-cell responses in tissues. (A) The total magnitude of Gag-specific IFN-γ T-cell responses as Spot Forming Cells (SFC) were determined by IFN-γ ELISpot at Week 14 following stimulation with pools of Gag peptides at Week 14 in the axillary (left panel) and mesenteric (right panel) lymph nodes (LN). (B) CE-specific T-cell responses are shown as cumulative number of SFC against each CE peptide pool and was determined by summing the responses against each individual CE (1-7) peptide pool in the axillary (left panel) and mesenteric (middle panel) lymph nodes and the jejunum (right panel). (C) The breadth of CE-specific IFN-γ T-cell responses was determined by measuring spot forming cells (SFC) against each individual CE peptide pool in the axillary (left panel) and mesenteric (right panel) lymph nodes. The dotted line at seven indicates the maximum number of CE that can be recognized. (D) The specificity of the CE responses was mapped in the axillary (left two panels) and mesenteric (right two panels) lymph nodes by ELISpot at week 14. Shown are responses against each peptide pool in animals immunized with either the CE or FL DNA vaccines. Median values are indicated in panels A-C with red horizontal lines and p-values were determined by non-parametric Mann-Whitney T test. P values ≤ 0.05 were considered significant.
CE vaccination induces multifunctional, cytolytic CE-specific CD8+ T-cell responses
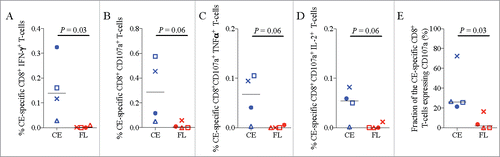
CD107a is a marker of degranulation, indicating cytolytic effector function, that has been previously shown to correlate with better control of viremia in SIV/SHIV-infected macaques and HIV infected humans.Citation60 Since both cytolytic and multifunctional CD8+ T-cell responses have been correlated with better viral control in HIV infected individuals,Citation61,Citation62 the ability of the CE vaccine to induce these responses may be important for enhanced viral clearance and control. Hence, CE-specific effector responses in the blood were characterized by intracellular cytokine staining (ICS) for IFN-γ, TNFα, IL-2, and for CD107a, at week 14. Similar to the IFN-γ ELISpot results, CE vaccinated animals had a greater fraction of CE-specific CD8 T-cells expressing IFN-γ detected by ICS (). There was also a strong trend for induction of higher magnitude CE-specific CD8+ T-cells expressing CD107a, CD107a+TNFα, or CD107a+IL-2 cells when compared to the FL vaccine (). Overall, the CE vaccine induced a higher proportion of CE-specific CD8+ T-cells expressing CD107a when compared to the FL DNA vaccine (p = 0.03, ). Cytolytic and multifunctional responses to the non-CE portions of Gag were not assessed in these animals so we were unable to determine if the FL or CE vaccinated animals had similar responses to epitopes outside of CE.
Figure 4. Multifunctional and cytolytic CE-specific CD8+ T-cell responses. The frequency of CE-specific CD8+ T-cells expressing the cytokines IFN-γ, IL-2, and/or TNF-⍺ and/or CD107a, a marker of cytolytic effector function, was determined two weeks after the final DNA vaccine dose (week 14) using cryopreserved PBMC stimulated overnight with CE peptide pools. (A-D) Shown are the frequencies of CE-specific CD8+ T-cells expressing the indicated cytokine or cytolytic functions. (E) The relative proportion of CE-specific CD8+ T-cells that are expressing the cytolytic degranulation marker, CD107a. The difference in mean response between the two groups was analyzed by a non-parametric Mann-Whitney T test. Median values are indicated with black horizontal lines and a p-value ≤ 0.05 was considered significant.
To determine if therapeutic DNA vaccination with either the CE or FL vaccines influenced viral load, plasma viral RNA levels were measured by RT-PCR. We found no significant difference in mean viral loads (VL) between the two groups at any timepoint, although only one animal per vaccination group exhibited significant viremia over the course of the study (). Even though VL in the CE vaccinated animal decreased while VL in the FL vaccinated animal increased, we cannot conclude that these results were due to either vaccine.
CE-specific T-cell responses are associated with lower viral set point in SIV-infected rhesus macaques
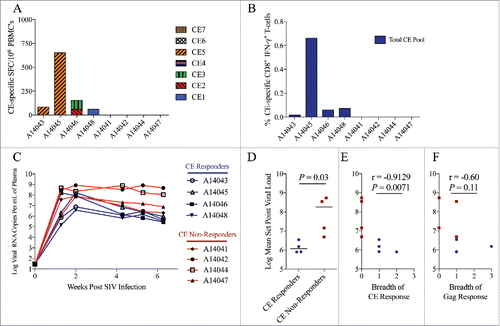
The SIV CE DNA vaccine was derived by homology to HIV CE.Citation32,Citation33 In acutely HIV infected ART naïve people, lower viral set point has been shown to correlate with the presence of stronger CE-specific responses, a finding that supports the hypothesis that increasing CE-specific responses during infection may provide a benefit in improving viral control.Citation33 However, it is unknown if CE-specific responses will have a similar correlation in the context of SIV or SHIV infections in macaques. Having demonstrated that it is possible to broaden or redirect the pre-existing immunodominant SIV Gag-specific T-cell repertoire toward increased targeting of SIV CE-specific epitopes in SHIV infected macaques, we sought to determine if, similar to humans, SIV-specific CE responses similarly correlated with lower viral set-point following acute infection in a second cohort of rhesus macaques. In this cohort, rhesus macaques were intravenously infected with SIVΔB670, a primary isolate that results in high viral loads during acute infection, reaches set point viremia by 6 weeks post-infection, and causes AIDS in the majority of animals within 11 months after infection.Citation23,Citation63 After infection, CE-specific T-cell responses, measured by both ELISpot () and ICS () were detected in 4 of the 8 macaques. Interestingly, CE-responsive animals exhibited significantly lower set point viremia than the 4 macaques that had no detectable CE responses (p = 0.03, & ). Furthermore, the breadth of CE-specific T-cell responses measured by ELISpot inversely correlated with set point viral load in these animals, as defined by the averaging the viral loads from weeks 4, 5, and 6 (p = 0.007, ), whereas the breadth of the response against the full length p57 Gag did not significantly correlate (p = 0.11, ). Similar to previous studies in macaques and humans,Citation30,Citation33 we observed no relationship between MHC class I haplotype and the development of CE responses in these animals (Supplemental Table 1). Furthermore, in agreement with our previous studies involving SIVΔB670,Citation23 we also found no association between MHC or TRIM5 genetics and viral load (Supplemental Table 1).
Figure 6. Higher CE-specific T-cells responses correlate with lower viral set point during acute SIV infection. CE-specific T-cell responses in rhesus macaques acutely infected with SIVΔB670 were measured six weeks following intravenous infection using (A) IFN-γ ELISpot against individual CE peptides and (B) IFN-γ ICS against a pool of all seven CE peptides. CE responders were defined as having a detectable CE response measured by ELISpot and/or ICS. (C) Viral loads were measured in the plasma by quantitative RT-PCR. (D) Mean viral set point was determined by averaging the viral loads measured at weeks 4, 5, and 6 and compared between CE responders (animals with detectable CE responses) and CE non-responders (animals with undetectable CE responses) by non-parametric Mann-Whitney T test. Horizontal lines indicate median values. Correlation between the breadth of (E) CE and (F) Gag-specific T-cell responses and mean set point plasma viral load was determined by a Spearman Rank correlation. A p value ≤ 0.05 was considered significant.
Discussion
The results reported here demonstrate that therapeutic immunization with an SIV CE DNA vaccine can broaden T-cell responses primed by SHIV infection toward increased targeting of CE sequences. Our results also provide evidence that CE-specific T-cell responses induced by DNA vaccination have stronger cytolytic effector function when compared to T-cell responses induced by a standard FL DNA vaccine and as such, may be more effective in controlling viral replication. Consistent with this possibility, we found that unvaccinated rhesus macaques that developed stronger CE-specific responses during the acute stage of a highly pathogenic SIV infection also developed lower viral loads. These results are consistent with data from HIV-infected humans where it has been shown that CE responses that appear early in infection correlate with lower viral set pointCitation33 supporting the concept that CE-specific CD8+ T-cell responses may limit SIV replication and suggests that therapeutic immunization with CE vaccines may be able to improve treatment of HIV infection by increasing subdominant CE responses primed by the virus.
Previous studies have suggested that T-cell responses against subdominant Gag epitopes are preferentially lost during chronic HIV and SIV infection, perhaps due to T-cell exhaustion.Citation64-66 Therefore, vaccination with full-length antigens may mimic, or worsen, the effects of natural infection and fail to induce or even decrease CE-specific T-cell responses, as found here. Our data indicates that immunizing with a CE DNA vaccine can circumvent this problem, likely by excluding immunodominant decoy elements that divert T-cells responses, thereby boosting or inducing de novo T-cells with greater TCR affinity to the subdominant conserved regions of the viral proteome. The superior ability of the CE vaccine to induce broader CE responses and greater functional responses against subdominant epitopes when compared to a FL vaccine is in agreement with our prophylactic DNA vaccine studies in uninfected macaques.Citation30,Citation32 In contrast, Stephenson et al reported that prophylactic immunization with full-length mosaic immunogens via recombinant adenovirus vectors in rhesus macaques (Ad35 and Ad26) was superior at inducing responses against conserved regions when compared to Adenoviruses expressing a conserved-region-only HIV immunogen.Citation67 This could be due to the different composition and configuration of the conserved immunogens, differences between consensus and mosaic immunogens and the single FL immunogen used here, and/or the use of adenoviral vectors versus DNA vaccine delivery, since Ad vectors contain immunodominant epitopes that may compete with subdominant conserved HIV epitopes.Citation68 Previously, we demonstrated that in uninfected rhesus macaques, CE DNA priming followed by FL DNA boosting was more effective at increasing the magnitude of CE responses, compared to priming with CE without a heterologous boost or priming with a FL DNA vaccine and then boosting with a CE DNA vaccine. The latter result suggested a limited ability to improve CE responses in animals that were previously primed with immunodominant responses to variable regions of the viral proteome.Citation30 Unexpectedly then, we found that therapeutic immunization increased CE-specific T-cell epitopes in SHIV-infected macaques primed with full-length antigens by the viral challenge, albeit in animals with attenuated infection. In this setting, therapeutic DNA vaccination may have augmented subdominant CE-specific cellular responses that were primed by the virus. This suggests that therapeutic vaccination, in a setting were viral replication is suppressed by antiretroviral drugs may be able to overcome the “original antigenic sin” associated with SIV/HIV infection that typically favors induction of immunodominant responses to more variable sequences.
The enhanced cytotoxic function of the CE-specific responses we observed following CE vaccination is consistent with previous studies in uninfected rhesus macaques immunized with an HIV Gag CE DNA vaccine.Citation30,Citation32 However, due to the small numbers of animals in this study and low initial viremia in both groups, we cannot determine if CE therapeutic vaccination provided a benefit in the control of viremia.
CE vaccinated animals also exhibited a trend toward greater CE-specific T-cell responses in the gut mucosa and lymphoid tissues. Since the gut and lymphoid tissues contain virally infected cellsCitation23,Citation26,Citation58,Citation59,Citation69,Citation70 that can contribute to viral rebound after cessation of ART,Citation71 inducing CE-specific responses in these tissues may be important to decrease the size of the reservoir and provide more durable control of viremia in the absence of drug.
In the SHIV study, we found no relationship between the magnitude of Gag-specific responses measured prior to therapeutic vaccination and the development of de novo CE-specific responses induced post-vaccination. This indicates that the presence of immunodominant HIV or SIV CD8+ T-cell responses against Gag prior to vaccination did not significantly impair the immunogenicity of the CE DNA vaccine. However, we did not measure responses against other viral antigens that were not included in the vaccine, such as Env, Pol, or Nef that are also rich in immunodominant CD8+ T-cell epitopes, so we cannot exclude the possibility that immunodominant responses against other viral antigens could influence immunogenicity of the CE vaccine.
HIV CE sequences were selected based on sequence conservation and fitness constraints as previously described and are generally >98% conserved among all HIV-1 group M isolates evaluated to date. This is the key advantage of using this vaccine strategy over other vaccines in that we can expect a high degree of similarity between the vaccine and any virus that would theoretically be found in humans. The SIV Gag CE DNA vaccine used for these studies was not based on sequences in the challenge virus but rather, they were designed based on homology to the HIV CE sequences as previously described.Citation32 The 75–100% amino acid similarity between SIV p27CE and HIV p24CE further highlights the high degree of conservation of the CE sequences across not only HIV strains but also SIV strains. The full length (FL) Gag DNA vaccine used in this study was derived from SIV/17-Fr sequence, a macrophage tropic, congenic isolate of SIVmac239.Citation72 These two strains are about 15% divergent in the Env geneCitation23 but as is the case for most SIV strains, the Gag sequences for the two strains are 100% identical,Citation57 and each contain CE sequences. The challenge virus SHIV89.6P is derived from SIVmac239,Citation55 so it too was at least substantially homologous to the FL vaccine, although we did not sequence virus in the SHIV89.6P-infected macaques in this study, and the sequences may have diverged from the challenge stock prior to therapeutic vaccination.
Taken together, these results show that CE DNA vaccination can significantly increase the magnitude, function and breadth of CE-specific responses even in the presence of a strong immunodominant memory T-cell response induced by chronic SHIV infection in rhesus macaques, although we are unable to draw conclusions about the antiviral efficacy of the CE vaccine in this study. As the overall goal of an effective therapeutic vaccine is to induce viral remission or cure of the infection, future studies will investigate the ability of this vaccine, in combination with antiretroviral drug therapy and potentially reservoir activating agents and immune modulators, to reduce viral replication or block viral rebound after analytic treatment interruption in SIV or SHIV infected macaques. If successful, these studies would support further development of a therapeutic CE vaccine for people living with HIV.
Supplemental Table 1. MHC and TRIM5 genetics.
Shown for each animal are TRIM5 haplotype, MHC type (A*01, A*02, A*08, A*11, B*01, B*08, B*17, and B*29), SIV or SHIV infecting strain, vaccination regimen, and the viral load at the end of study (2 weeks post 4th vaccination in SHIV89.6P infected animals and 6 weeks post-infection in SIVΔB670 infected animals), as well as the specificity of CE-specific cellular immune responses. Animals in blue are CE responders whereas animals indicated in red are non-responders.
Abbreviations
| ART | = | antiretroviral therapy |
| CE | = | conserved elements |
| FL | = | full-length |
| LT | = | heat labile E. coli enterotoxin |
Disclosure of potential conflicts of interest
The authors claim no conflict of interest.
Previous presentation of data
Portions of this manuscript have not been previously published.
KHVI_A_1448328_Supplemental.pdf
Download PDF (61.7 KB)Acknowledgments
The authors would like to thank all veterinary and research support staff of the Washington National Primate Research Center with special thanks to Drew May, Solomon Wangari, Dr. Jennifer Lane, Dr. Cassie Moats, Dr. Jeremy Smedley, and Dr. Robert Murnane. We also wish to thank Dr. Nancy Miller of the Division of AIDS, National Institutes of Health. Gag peptides were kindly provided by the NIH AIDS Research and Reference Reagent Program. Additionally, we acknowledge the MHC Genotyping Service at the University of Miami, which is supported by the NIH grant 5R24RR016038 awarded to Dr. David Watkins, for providing the MHC data in this manuscript. This manuscript is dedicated to the late Dr. Michael Agy who was the PI of the Simian Vaccine Evaluation Unit grant that, in part, funded this project.
Funding
This work was supported by National Cancer Institute (NCI) and the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under award numbers [T32CA080416 to Paul Munson]; [N01AI60006-7 (M Agy, PI)] and [R01 AI104679 (Fuller/Mullins, MPI)].
References
- Schackman BR, Fleishman JA, Su AE, Berkowitz BK, Moore RD, Walensky RP, Becker JE, Voss C, Paltiel AD, Weinstein MC, et al. The Lifetime Medical Cost Savings from Preventing HIV in the United States. Med Care. 2015;53:293-301. PMID:25710311.
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365-71. doi:10.1038/nm1511. PMID:17115046.
- Gupta RK, Jordan MR, Sultan BJ, Hill A, Davis DH, Gregson J, Sawyer AW, Hamers RL, Ndembi N, Pillay D, et al. Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. The Lancet. 2012;380:1250-8. doi:10.1016/S0140-6736(12)61038-1.
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183-8. doi:10.1038/387183a0. PMID:9144289.
- Butler KM, Gavin P, Coughlan S, Rochford A, Donagh SM, Cunningham O, Poulsom H, Watters S, Klein N. Rapid Viral Rebound after 4 Years of Suppressive Therapy in a Seronegative HIV-1 Infected Infant Treated from Birth. Pediatr Infect Dis J. 2014; PMID:25144802.
- Calza L. Renal toxicity associated with antiretroviral therapy. HIV Clin Trials. 2012;13:189-211. doi:10.1310/hct1304-189. PMID:22849961.
- Chun T-W, Nickle DC, Justement JS, Meyers JH, Roby G, Hallahan CW, Kottilil S, Moir S, Mican JM, Mullins JI, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197:714-20. doi:10.1086/527324. PMID:18260759.
- Crooks AM, Bateson R, Cope AB, Dahl NP, Griggs MK, Kuruc JD, Gay CL, Eron JJ, Margolis DM, Bosch RJ, et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J Infect Dis. 2015;212:1361-5. doi:10.1093/infdis/jiv218. PMID:25877550.
- Mylvaganam GH, Silvestri G, Amara RR. HIV therapeutic vaccines: moving towards a functional cure. Curr Opin Immunol. 2015;35:1-8. doi:10.1016/j.coi.2015.05.001. PMID:25996629.
- de Mendoza C, Barreiro P, Benitez L, Soriano V. Gene therapy for HIV infection. Expert Opin Biol Ther. 2015;15:319-27. doi:10.1517/14712598.2015.967208. PMID:25323559.
- García F, Plana M, Climent N, León A, Gatell JM, Gallart T. Dendritic cell based vaccines for HIV infection: the way ahead. Hum Vaccines Immunother. 2013;9:2445-52. doi:10.4161/hv.25876.
- Rasmussen TA, Tolstrup M, Søgaard OS. Reversal of Latency as Part of a Cure for HIV-1. Trends Microbiol. 2016;24:90-7. doi:10.1016/j.tim.2015.11.003. PMID:26690612.
- von Gegerfelt AS, Rosati M, Alicea C, Valentin A, Roth P, Bear J, Franchini G, Albert PS, Bischofberger N, Boyer JD, et al. Long-lasting decrease in viremia in macaques chronically infected with simian immunodeficiency virus SIVmac251 after therapeutic DNA immunization. J Virol. 2007;81:1972-9. doi:10.1128/JVI.01990-06. PMID:17135321.
- Halwani R, Boyer JD, Yassine-Diab B, Haddad EK, Robinson TM, Kumar S, Parkinson R, Wu L, Sidhu MK, Phillipson-Weiner R, et al. Therapeutic vaccination with simian immunodeficiency virus (SIV)-DNA + IL-12 or IL-15 induces distinct CD8 memory subsets in SIV-infected macaques. J Immunol Baltim Md 1950. 2008;180:7969-79.
- Valentin A, von Gegerfelt A, Rosati M, Miteloudis G, Alicea C, Bergamaschi C, Jalah R, Patel V, Khan AS, Draghia-Akli R, et al. Repeated DNA therapeutic vaccination of chronically SIV-infected macaques provides additional virological benefit. Vaccine. 2010;28:1962-74. doi:10.1016/j.vaccine.2009.10.099. PMID:20188252.
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857-60. doi:10.1126/science.283.5403.857. PMID:9933172.
- Mothe B, Llano A, Ibarrondo J, Zamarreño J, Schiaulini M, Miranda C, Ruiz-Riol M, Berger CT, Herrero MJ, Palou E, et al. CTL Responses of High Functional Avidity and Broad Variant Cross-Reactivity Are Associated with HIV Control. PLoS ONE. 2012;7:e29717. doi:10.1371/journal.pone.0029717. PMID:22238642.
- Rolland M, Heckerman D, Deng W, Rousseau CM, Coovadia H, Bishop K, Goulder PJR, Walker BD, Brander C, Mullins JI. Broad and Gag-Biased HIV-1 Epitope Repertoires Are Associated with Lower Viral Loads. PLoS ONE. 2008;3:e1424. doi:10.1371/journal.pone.0001424. PMID:18183304.
- Tubiana R, Carcelain G, Vray M, Gourlain K, Dalban C, Chermak A, Rabian C, Vittecoq D, Simon A, Bouvet E, et al. Therapeutic immunization with a human immunodeficiency virus (HIV) type 1-recombinant canarypox vaccine in chronically HIV-infected patients: The Vacciter Study (ANRS 094). Vaccine. 2005;23:4292-301. doi:10.1016/j.vaccine.2005.04.013. PMID:15927325.
- De Rose R, Fernandez CS, Smith MZ, Batten CJ, Alcântara S, Peut V, Rollman E, Loh L, Mason RD, Wilson K, et al. Control of viremia and prevention of AIDS following immunotherapy of SIV-infected macaques with peptide-pulsed blood. PLoS Pathog. 2008;4:e1000055. doi:10.1371/journal.ppat.1000055. PMID:18451982.
- Hel Z, Venzon D, Poudyal M, Tsai WP, Giuliani L, Woodward R, Chougnet C, Shearer G, Altman JD, Watkins D, et al. Viremia control following antiretroviral treatment and therapeutic immunization during primary SIV251 infection of macaques. Nat Med. 2000;6:1140-6. doi:10.1038/80481. PMID:11017146.
- Pollard RB, Rockstroh JK, Pantaleo G, Asmuth DM, Peters B, Lazzarin A, Garcia F, Ellefsen K, Podzamczer D, van Lunzen J, et al. Safety and efficacy of the peptide-based therapeutic vaccine for HIV-1, Vacc-4×: a phase 2 randomised, double-blind, placebo-controlled trial. Lancet Infect Dis. 2014;14:291-300. doi:10.1016/S1473-3099(13)70343-8. PMID:24525316.
- Fuller DH, Rajakumar P, Che JW, Narendran A, Nyaundi J, Michael H, Yager EJ, Stagnar C, Wahlberg B, Taber R, et al. Therapeutic DNA Vaccine Induces Broad T Cell Responses in the Gut and Sustained Protection from Viral Rebound and AIDS in SIV-Infected Rhesus Macaques. PLoS ONE. 2012; 7:e33715. doi:10.1371/journal.pone.0033715. PMID:22442716.
- Liu Y, McNevin J, Rolland M, Zhao H, Deng W, Maenza J, Stevens CE, Collier AC, McElrath MJ, Mullins JI. Conserved HIV-1 epitopes continuously elicit subdominant cytotoxic T-lymphocyte responses. J Infect Dis. 2009;200:1825-33. doi:10.1086/648401. PMID:19909083.
- Li F, Finnefrock AC, Dubey SA, Korber BTM, Szinger J, Cole S, McElrath MJ, Shiver JW, Casimiro DR, Corey L, et al. Mapping HIV-1 vaccine induced T-cell responses: bias towards less-conserved regions and potential impact on vaccine efficacy in the Step study. PloS One. 2011;6:e20479. doi:10.1371/journal.pone.0020479. PMID:21695251.
- Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, Lai J, McHugh HL, Hao H, Zhang H, et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381-5. doi:10.1038/nature14053. PMID:25561180.
- Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, Sanders-Buell E, Heath L, Magaret CA, Bose M, Bradfield A, et al. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med. 2011;17:366-71. doi:10.1038/nm.2316. PMID:21358627.
- Herbeck JT, Rolland M, Liu Y, McLaughlin S, McNevin J, Zhao H, Wong K, Stoddard JN, Raugi D, Sorensen S, et al. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J Virol. 2011;85:7523-34. doi:10.1128/JVI.02697-10. PMID:21593162.
- Kulkarni V, Rosati M, Valentin A, Ganneru B, Singh AK, Yan J, Rolland M, Alicea C, Beach RK, Zhang G-M, et al. HIV-1 p24(gag) derived conserved element DNA vaccine increases the breadth of immune response in mice. PloS One. 2013;8:e60245. doi:10.1371/journal.pone.0060245. PMID:23555935.
- Kulkarni V, Valentin A, Rosati M, Alicea C, Singh AK, Jalah R, Broderick KE, Sardesai NY, Le Gall S, Mothe B, et al. Altered Response Hierarchy and Increased T-Cell Breadth upon HIV-1 Conserved Element DNA Vaccination in Macaques. PloS One. 2014;9:e86254. doi:10.1371/journal.pone.0086254. PMID:24465991.
- Kulkarni V, Valentin A, Rosati M, Rolland M, Mullins JI, Pavlakis GN, Felber BK. HIV-1 Conserved Elements p24CE DNA Vaccine Induces Humoral Immune Responses with Broad Epitope Recognition in Macaques. PLOS ONE. 2014;9:e111085. doi:10.1371/journal.pone.0111085. PMID:25338098.
- Hu X, Valentin A, Dayton F, Kulkarni V, Alicea C, Rosati M, Chowdhury B, Gautam R, Broderick KE, Sardesai NY, et al. DNA Prime-Boost Vaccine Regimen To Increase Breadth, Magnitude, and Cytotoxicity of the Cellular Immune Responses to Subdominant Gag Epitopes of Simian Immunodeficiency Virus and HIV. J Immunol. 2016;1600697.
- Kunwar P, Hawkins N, Dinges WL, Liu Y, Gabriel EE, Swan DA, Stevens CE, Maenza J, Collier AC, Mullins JI, et al. Superior control of HIV-1 replication by CD8+ T cells targeting conserved epitopes: implications for HIV vaccine design. PloS One. 2013;8:e64405. doi:10.1371/journal.pone.0064405. PMID:23741326.
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007; 13:46-53. doi:10.1038/nm1520. PMID:17173051.
- Zuñiga R, Lucchetti A, Galvan P, Sanchez S, Sanchez C, Hernandez A, Sanchez H, Frahm N, Linde CH, Hewitt HS, et al. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol. 2006;80:3122-5. doi:10.1128/JVI.80.6.3122-3125.2006. PMID:16501126.
- Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97:2709-14. doi:10.1073/pnas.050567397. PMID:10694578.
- Streeck H, Lichterfeld M, Alter G, Meier A, Teigen N, Yassine-Diab B, Sidhu HK, Little S, Kelleher A, Routy J-P, et al. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J Virol. 2007;81:7725-31. doi:10.1128/JVI.00708-07. PMID:17494064.
- Schneidewind A, Brockman MA, Sidney J, Wang YE, Chen H, Suscovich TJ, Li B, Adam RI, Allgaier RL, Mothé BR, et al. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J Virol. 2008;82:5594-605. doi:10.1128/JVI.02356-07. PMID:18385228.
- Dinges WL, Richardt J, Friedrich D, Jalbert E, Liu Y, Stevens CE, Maenza J, Collier AC, Geraghty DE, Smith J, et al. Virus-Specific CD8+ T-Cell Responses Better Define HIV Disease Progression than HLA Genotype. J Virol. 2010;84:4461-8. doi:10.1128/JVI.02438-09. PMID:20147397.
- Leitman EM, Willberg CB, De Burgh-Thomas A, Streeck H, Goulder PJR, Matthews PC. Subdominant Gag-specific anti-HIV efficacy in an HLA-B∗57-positive elite controller. AIDS Lond Engl. 2016;30:972-4. doi:10.1097/QAD.0000000000001022.
- Friedrich TC, Valentine LE, Yant LJ, Rakasz EG, Piaskowski SM, Furlott JR, Weisgrau KL, Burwitz B, May GE, León EJ, et al. Subdominant CD8+ T-Cell Responses Are Involved in Durable Control of AIDS Virus Replication. J Virol. 2007;81:3465-76. doi:10.1128/JVI.02392-06. PMID:17251286.
- Frahm N, Kiepiela P, Adams S, Linde CH, Hewitt HS, Sango K, Feeney ME, Addo MM, Lichterfeld M, Lahaie MP, et al. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat Immunol. 2006;7:173-8. doi:10.1038/ni1281. PMID:16369537.
- Brockman MA, Brumme ZL, Brumme CJ, Miura T, Sela J, Rosato PC, Kadie CM, Carlson JM, Markle TJ, Streeck H, et al. Early Selection in Gag by Protective HLA Alleles Contributes to Reduced HIV-1 Replication Capacity That May Be Largely Compensated for in Chronic Infection. J Virol. 2010;84:11937-49. doi:10.1128/JVI.01086-10. PMID:20810731.
- Demberg T, Florese RH, Heath MJ, Larsen K, Kalisz I, Kalyanaraman VS, Lee EM, Pal R, Venzon D, Grant R, et al. A replication-competent adenovirus-human immunodeficiency virus (Ad-HIV) tat and Ad-HIV env priming/Tat and envelope protein boosting regimen elicits enhanced protective efficacy against simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J Virol. 2007;81:3414-27. doi:10.1128/JVI.02453-06. PMID:17229693.
- Zhao J, Voltan R, Peng B, Davis-Warren A, Kalyanaraman VS, Alvord WG, Aldrich K, Bernasconi D, Buttò S, Cafaro A, et al. Enhanced cellular immunity to SIV Gag following co-administration of adenoviruses encoding wild-type or mutant HIV Tat and SIV Gag. Virology. 2005;342:1-12. doi:10.1016/j.virol.2005.07.016. PMID:16109434.
- Zhao J, Lou Y, Pinczewski J, Malkevitch N, Aldrich K, Kalyanaraman VS, Venzon D, Peng B, Patterson LJ, Edghill-Smith Y, et al. Boosting of SIV-specific immune responses in rhesus macaques by repeated administration of Ad5hr-SIVenv/rev and Ad5hr-SIVgag recombinants. Vaccine. 2003;21:4022-35. doi:10.1016/S0264-410X(03)00266-4. PMID:12922139.
- Patterson LJ, Malkevitch N, Pinczewski J, Venzon D, Lou Y, Peng B, Munch C, Leonard M, Richardson E, Aldrich K, et al. Potent, Persistent Induction and Modulation of Cellular Immune Responses in Rhesus Macaques Primed with Ad5hr-Simian Immunodeficiency Virus (SIV) env/rev, gag, and/or nef Vaccines and Boosted with SIV gp120. J Virol. 2003;77:8607-20. doi:10.1128/JVI.77.16.8607-8620.2003. PMID:12885879.
- Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'Connor DH, Carrington M, Watkins DI. The high-frequency major histocompatibility complex class I Allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074-7. doi:10.1128/JVI.80.10.5074-5077.2006. PMID:16641299.
- Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, et al. Mamu-B*08-Positive Macaques Control Simian Immunodeficiency Virus Replication. J Virol. 2007;81:8827-32. doi:10.1128/JVI.00895-07. PMID:17537848.
- Giraldo-Vela JP, Rudersdorf R, Chung C, Qi Y, Wallace LT, Bimber B, Borchardt GJ, Fisk DL, Glidden CE, Loffredo JT, et al. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J Virol. 2008;82:859-70. doi:10.1128/JVI.01816-07. PMID:17989178.
- Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. J Clin Invest. 2007;117:3563-75. doi:10.1172/JCI32047. PMID:17975674.
- Zhang SC, Martin E, Shimada M, Godfrey SB, Fricke J, Locastro S, Lai NY, Liebesny P, Carlson JM, Brumme CJ, et al. Aminopeptidase substrate preference affects HIV epitope presentation and predicts immune escape patterns in HIV-infected individuals. J Immunol Baltim Md 1950. 2012;188:5924-34.
- Rolland M, Nickle DC, Mullins JI. HIV-1 group M conserved elements vaccine. PLoS Pathog. 2007;3:e157. doi:10.1371/journal.ppat.0030157. PMID:18052528.
- Mankowski JL, Flaherty MT, Spelman JP, Hauer DA, Didier PJ, Amedee AM, Murphey-Corb M, Kirstein LM, Muñoz A, Clements JE, et al. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J Virol. 1997;71:6055-60. PMID:9223498.
- Reimann KA, Li JT, Veazey R, Halloran M, Park IW, Karlsson GB, Sodroski J, Letvin NL. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922-8. PMID:8794335.
- Arrington J, Braun RP, Dong L, Fuller DH, Macklin MD, Umlauf SW, Wagner SJ, Wu MS, Payne LG, Haynes JR. Plasmid Vectors Encoding Cholera Toxin or the Heat-Labile Enterotoxin from Escherichia coli Are Strong Adjuvants for DNA Vaccines. J Virol. 2002;76:4536-46. doi:10.1128/JVI.76.9.4536-4546.2002. PMID:11932419.
- Flaherty MT, Hauer DA, Mankowski JL, Zink MC, Clements JE. Molecular and biological characterization of a neurovirulent molecular clone of simian immunodeficiency virus. J Virol. 1997;71:5790-8. PMID:9223467.
- Ling B, Mohan M, Lackner AA, Green LC, Marx PA, Doyle LA, Veazey RS. The Large Intestine as a Major Reservoir for Simian Immunodeficiency Virus in Macaques with Long-Term, Nonprogressing Infection. J Infect Dis. 2010; 202:1846-54. doi:10.1086/657413. PMID:21050120.
- Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, Mahyari E, Hagen SI, Bae JY, Reyes MD 3rd, et al. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat Med. 2012;18:1673-81. doi:10.1038/nm.2934. PMID:22961108.
- Freel SA, Lamoreaux L, Chattopadhyay PK, Saunders K, Zarkowsky D, Overman RG, Ochsenbauer C, Edmonds TG, Kappes JC, Cunningham CK, et al. Phenotypic and functional profile of HIV-inhibitory CD8 T cells elicited by natural infection and heterologous prime/boost vaccination. J Virol. 2010;84:4998-5006. doi:10.1128/JVI.00138-10. PMID:20200250.
- Betts MR, Nason MC, West SM, Rosa SCD, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781-9. doi:10.1182/blood-2005-12-4818. PMID:16467198.
- Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, Kovacs CM, Rodriguez B, Sieg SF, Teixeira-Johnson L, et al. Perforin expression directly ex vivo by HIV-specific CD8+ T-cells is a correlate of HIV elite control. PLoS Pathog. 2010;6:e1000917. doi:10.1371/journal.ppat.1000917. PMID:20523897.
- Trichel AM, Rajakumar PA, Murphey-Corb M. Species-specific variation in SIV disease progression between Chinese and Indian subspecies of rhesus macaque. J Med Primatol. 2002;31:171-8. doi:10.1034/j.1600-0684.2002.02003.x. PMID:12390539.
- Geldmacher C, Gray C, Nason M, Currier JR, Haule A, Njovu L, Geis S, Hoffmann O, Maboko L, Meyerhans A, et al. A high viral burden predicts the loss of CD8 T-cell responses specific for subdominant gag epitopes during chronic human immunodeficiency virus infection. J Virol. 2007;81:13809-15. doi:10.1128/JVI.01566-07. PMID:17898052.
- Osuna CE, Gonzalez AM, Chang H-H, Hung AS, Ehlinger E, Anasti K, Alam SM, Letvin NL. TCR affinity associated with functional differences between dominant and subdominant SIV epitope-specific CD8+ T cells in mamu-A*01+ rhesus monkeys. PLOS Pathog. 2014;10:e1004069. doi:10.1371/journal.ppat.1004069. PMID:24743648.
- Conrad JA, Ramalingam RK, Smith RM, Barnett L, Lorey SL, Wei J, Simons BC, Sadagopal S, Meyer-Olson D, Kalams SA. Dominant clonotypes within HIV-specific T cell responses are programmed death-1high and CD127low and display reduced variant cross-reactivity. J Immunol Baltim Md 1950. 2011;186:6871-85.
- Stephenson KE, SanMiguel A, Simmons NL, Smith K, Lewis MG, Szinger JJ, Korber B, Barouch DH. Full-Length HIV-1 Immunogens Induce Greater Magnitude and Comparable Breadth of T Lymphocyte Responses to Conserved HIV-1 Regions Compared with Conserved-Region-Only HIV-1 Immunogens in Rhesus Monkeys. J Virol. 2012;86:11434-40. doi:10.1128/JVI.01779-12. PMID:22896617.
- Schirmbeck R, Reimann J, Kochanek S, Kreppel F. The immunogenicity of adenovirus vectors limits the multispecificity of CD8 T-cell responses to vector-encoded transgenic antigens. Mol Ther J Am Soc Gene Ther. 2008;16:1609-16. doi:10.1038/mt.2008.141.
- Horiike M, Iwami S, Kodama M, Sato A, Watanabe Y, Yasui M, Ishida Y, Kobayashi T, Miura T, Igarashi T. Lymph nodes harbor viral reservoirs that cause rebound of plasma viremia in SIV-infected macaques upon cessation of combined antiretroviral therapy. Virology. 2012;423:107-18. doi:10.1016/j.virol.2011.11.024. PMID:22196013.
- Belyakov IM, Hel Z, Kelsall B, Kuznetsov VA, Ahlers JD, Nacsa J, Watkins DI, Allen TM, Sette A, Altman J, et al. Mucosal AIDS vaccine reduces disease and viral load in gut reservoir and blood after mucosal infection of macaques. Nat Med. 2001;7:1320-6. doi:10.1038/nm1201-1320. PMID:11726972.
- Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A. 2015;112:E1126-1134. doi:10.1073/pnas.1414926112. PMID:25713386.
- Mankowski JL, Flaherty MT, Spelman JP, Hauer DA, Didier PJ, Amedee AM, Murphey-Corb M, Kirstein LM, Muñoz A, Clements JE, et al. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J Virol. 1997;71:6055-60. PMID:9223498.