ABSTRACT
The antigenic evolution of influenza is widely assumed to occur by antigenic drift, in which strains incrementally acquire mutations in highly variable epitopes under strong immune selective pressure, such as those in the major influenza antigen haemagglutinin. However, this is not easy to reconcile with epidemiological observations, which show that each influenza season is dominated by a limited number of strains. Here, we discuss this paradox in light of recent influenza epidemics that have been characterised by low vaccine effectiveness and dominated by strains of limited antigenic and genetic diversity.
Introduction
Influenza is a highly contagious viral respiratory illness.Citation1,Citation2 The WHO estimates that influenza causes 3 to 5 million cases of severe illness and 290,000–650,000 deaths worldwide annually.Citation3 Currently, the H1N1 and H3N2 subtypes of influenza A and Yamagata and Victoria lineages of influenza B circulate in the human population.Citation4 The burden of influenza disproportionately affects the elderly and young children, especially in developing countries.Citation5 Influenza seasons in the Northern and Southern hemisphere occur in the winter months and are dominated by a limited number of antigenically and genetically distinct strains.Citation6
The effectiveness of influenza vaccination in recent years
The first experimental influenza vaccine was produced in 1933.Citation7 Since 1973, the WHO has issued formal recommendations regarding the composition for the influenza vaccine, and since 1999, separate recommendations have been made for the Northern and Southern hemisphere influenza seasons. These recommendations are based on the circulating influenza strains collected by WHO surveillance centres and their antigenic similarity to the current influenza vaccine strains.Citation8
The influenza vaccine is administered either as trivalent vaccine (TIV) or a quadrivalent vaccine (QIV).Citation9 The TIV consists of influenza A H1N1 and H3N2 strains as well as a Victoria lineage influenza B strain. The QIV consists of an additional Yamagata lineage influenza B strain.Citation10 The vaccine is produced in an inactivated or live attenuated form. The circulating influenza strains change periodically, necessitating updates to the TIV and QIV every two to three years on average.Citation11
The effectiveness of the influenza vaccine varies between influenza seasons but is generally low in contrast to other vaccines.Citation12-Citation14 This has been apparent in recent seasons: the overall vaccine effectiveness (VE) was estimated to be 53% in 2013–2014,Citation15 19% in 2014–2015,Citation16 48% in 2015–2016,Citation17 43% in 2016–2017Citation18 and 36% in 2017–2018.Citation19 Moreover, the VE within each season varied between influenza strains and age groups. In fact, in 2015–2016 the vaccine had no statistically significant VE against the dominant circulating H3N2 strain.Citation16
Low VE is often attributed to vaccine mismatch: either incorrect formulation of the vaccines strains,Citation20,Citation21 or more recently, mutations due to the egg-based manufacture of the vaccine.Citation22
In the 2014–2015 season, mismatch was stated as a reason for the ineffectiveness of the vaccine. The 2014–2015 influenza vaccine contained a H3N2 strain from the 3C.1 clade (A/Victoria/361/2011) but antigenically distinct H3N2 strains from the 3C.2a and 3C.3a clades dominated that season.Citation21 Consequently, for the 2016–2017 season, the vaccine was updated to include a 3C.2a clade strain, A/Switzerland/9,715,293/2013.Citation23 It has been well documented since that the 3C.2a clade strain, A/Switzerland/9,715,293/2013, displayed attenuated growth in eggs. Consequently, the egg-based manufacture of the 2016–2017 vaccine facilitated the selection of a K160 reversion mutation due to its enhanced growth. This mutation disrupted the glycosylation site motif present in the vaccine strains antigenic site B, creating a mismatch between the vaccine strain and the strains circulating in the 2016–2017 season. Subsequent studies of sera taken prior to the 2016–2017 season from individuals aged 18–49 showed that the majority of antibodies bound to the non-glycosylated antigenic site B and were unable to bind the glycosylated form.Citation22 Consequently, it has been suggested that vaccination with the mismatched vaccine strain containing the non-glycosylated form of antigenic site B could have contributed to the reduced VE of the influenza vaccine during the 2016–2017 season. This reduced VE was found to be greater in younger adults and has been hypothesised to be due to exposure history.Citation22,Citation24
A similar situation arose in the 2013–2014 season, during which H1N1 influenza was highly prevalent, and adults aged 30 to 59 exhibited increased morbidity and mortality.Citation20 Sera from individuals within this age range disproportionately targeted an epitope in the H1 haemagglutinin protein (HA) containing a K180 residue (linear numbering) within the Sa antigenic site. A K180Q mutation arose during the 2013–2014 season, and subsequently, this mutation has been suggested as a possible reason explanation for the increased morbidity and mortality in 30 to 59 year olds.Citation20,Citation25 The 2013–2014 influenza vaccine was shown to boost responses to an epitope containing K180 and so the K180 mutation is likely to have also contributed to its low VE.Citation20
Original antigenic sin and its impact on vaccination
The recent epidemiological findings relating to low VE appear at odds with typical experimental measures of antigenic drift. The classical serological tests used by surveillance laboratories involve determining the antigenic similarity of new circulating viruses to historical viruses using a panel of ferret anti-sera in haemagglutinin inhibition and virus neutralisation assays.Citation21,Citation26,Citation27
In contrast, recent publications demonstrate that human sera is dominated by antibodies which bind to regions of the HA which are similar to those of previously encountered viruses.Citation20,Citation22,Citation25,Citation27,Citation28 This idea was first proposed in 1960 by Robert Francis and coined ‘original antigenic sin’ and more recently as ‘immune focusing’.Citation29 Other manifestations of this phenomenon have been demonstrated for influenza: the antibodies of individuals infected for the first time with the 2009 pandemic H1N1 virus tended to target epitopes that were similar to those they had encountered during childhood.Citation25 Furthermore, studies have shown that if an individuals first infection was with a H2N2 influenza strain, which circulated between 1957 and 1968, they were at greater risk of mortality during the 2009 H1N1 pandemic.Citation30 Moreover, Gostic et al 2016 recently demonstrated an association between the subtype of an individual’s first infection and the risk of mortality from certain avian influenza subtypes.Citation31
An understanding is now emerging based on recent studies: if the HA of a circulating influenza strain has similar regions to those that certain demographics of the population have previously encountered, then the immune response of individuals within those demographics will focus on these regions and mutation within them will lead to viral escape.Citation24 In fact, it is likely that regions focused by the immune system will be the first to change, due to the greater immune selective pressure exerted on them.Citation25 To combat this, several groups have suggested that sera from humans should be used to determine vaccine escape instead of, or in addition to, ferret anti-sera.Citation20,Citation24
The ‘diversity paradox’
The paradox at the centre of influenza research is that whilst we intrinsically think of influenza as highly variable, we are able to vaccinate against influenza precisely because influenza seasons tend to be dominated by a small number of antigenically and genetically distinct strains.
Theoretical works have been trying to explain this paradox and elucidate its relevance to vaccine development.Citation32-Citation34 A prominent example is the ‘antigenic thrift’ model that postulates that this paradox can be resolved by proposing the existence of multiple immunogenic epiotopes of limited variability, which cycle in an asynchronous manner as the influenza virus evolves.Citation32,Citation35,Citation36
Our group has recently identified such an epitope of limited variability in the head domain of the H1 HA. We have shown that it cycles over time between a limited number of conformations as host population immunity changesCitation37 – in line with the antigenic thrift theory. It is likely that such epitopes are the same ones targeted by the human immune system due to their capacity to mutate more slowly. Furthermore, if these epitopes are targeted preferentially, they could explain age-dependent VE (see for illustration of this process).Citation22
Figure 1. Illustration of Influenza Immunity Over Time.
The diagram shows how the immunity of three individuals changes (horizontal black lines) over a period of years. Three epitopes are represented on the haemagglutinin (HA) structure (protein structures overlaid with purple, yellow and green shapes), which vary through time (different shapes). After an encounter with a specific strain, individuals acquire immunity to the encountered epitopes, which is represented by the presence of different shapes for each individual. Immunity is commutative through time and can increased by multiple encounters (stacked shapes). Similar epitopes are encountered periodically. This leads to a different immunity profile of individuals based on their year of birth.
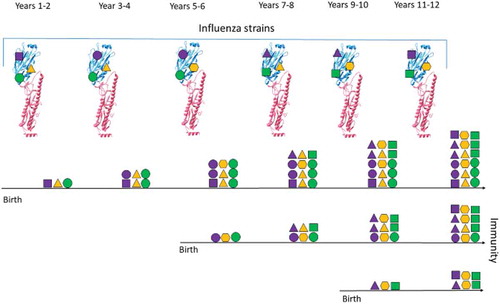
The existence of other epitopes of limited variability is evident from analysis of the literature: Raymond et al 2018 and Huang et al 2015 have purified monoclonal antibodies that bind to an epitope that includes position 180 (linear numbering). Collectively, these antibodies neutralise viruses that last circulated in 2009, 1977 and 1918 but not viruses that circulated in 2006, 1999 and 1934. As mentioned previously, population immunity directed at this epitope was further characterised by Linderman et al 2014.Citation20 Other broadly neutralising antibodies could indicate the location of further epitopes of limited variability. More recently, an monoclonal antibody named ‘KPF1ʹ was identified, which binds to a region inclusive of position 129 (linear numbering, Nogales et al. 2018). This antibody appears to bind to influenza viruses that circulated in 2009, 1999, 1991 and 1918 but not 1977, and only weakly to the A/PR/8/1934 strain, which was collected in 1934. The 5j8 and CH65 broadly neutralising antibodies also bind to distinct conformations of the epitope of limited variability that we have identified.Citation39,Citation40
Influenza vaccines in development
To circumvent the problems associated with the TIV and QIV vaccines outlined here and the intrinsic variation of the influenza virus, a number of groups are developing ‘universal’ influenza vaccines. These vaccines target conserved regions within the influenza virus and can induce a humeral and/or cellular immune response.
One such method that has proven popular is targeting the stem of HA, as it is more conserved than the head domain of HA. Doud et al 2018 have also shown that the stem is more recalcitrant to mutation and antibody escape than the head.Citation41,Citation42 Several approaches which enable the stem to be targeted by stabilising it or targeting it through a prime and boost regimen using chimeric HAs are currently moving through pre-clinical and Phase I clinical trials.Citation43-Citation45 Recent studies have shown that many of the antibodies targeting the stem act via antibody-dependent cellular cytotoxicity (ADCC).Citation46
T-cell vaccines have been developed as another innovative methods of targeting conserved influenza epitopes.Citation47,Citation48 One of the most advanced T-cell vaccines uses a Adenovirus/MVA prime-boost strategy to target the immune response against conserved epitopes of in the viral internal proteins NP and M1.Citation47 Individuals vaccinated with vaccine show increased levels of IFNγ-producing CD8+ T cells and a corresponding increase in protection against influenza virus infection.Citation47-Citation49 This vaccine is currently undergoing Phase II clinical trials in individuals over the age of 65.
One further vaccine at an advanced stage, which targets conserved epitopes, is developed by BiondVax Pharmaceuticals. It consists of a synthetic peptide, named M-001, containing conserved epitopes from HA, NP and M1. Consequently, it is reported to stimulate both humeral and cellular immunity.Citation50 In 2018, M-001 will begin Phase III clinical trials.
Importantly, most vaccines seek to capture the intrinsic variation of influenza by targeting conserved regions but bypass the way in which it evolves. Whilst initial results are promising placing previously conserved regions of the virus under greater selective pressure may yet produce escape mutants. A potential alternative was recently suggested by our groupCitation37: not using inactivated or attenuated viruses to target highly variable epitopes, or targeting conserved epitopes, but instead targeting those highly immunogenic epitopes which are limited in variability and likely to be the natural focus of humeral immunity.
Summary
Despite decades of research, many aspects of the biology of influenza remain elusive. However, the way in which original antigenic sin impacts the evolution of influenza and vaccine design is becoming more apparent. As a consequence, a coherent narrative regarding how influenza history affects the focus of human immunity and how regions targeted by the immune system change over time is becoming established. Hopefully, this route will eventually lead to more efficient and broad influenza vaccines.
Disclosure of potential conflicts of interest
Craig Thompson has submitted a vaccine patent through the University of Oxford relating to an influenza vaccine targeting epitopes of limited variability.
Additional information
Funding
References
- Miller MA, Viboud C, Balinska M, Simonsen L. 2009. The signature features of influenza pandemics — implications for policy. N Engl J Med. 360:2595–2598. https://www.nejm.org/doi/full/10.1056/NEJMp0903906
- Kilbourne ED. 2006. Influenza pandemics of the 20th century. Emerg Infect Dis. 12:9–14. doi:10.3201/eid1201.051254.
- WHO. Influenza factsheet. 2018. Geneva, Switzerland: World Health Organisation
- WHO. Recommended composition of influenza virus vaccines for use in the 2016 southern hemisphere influenza season. WHO Weeky Epidemiol Rec. 2015;90:545–560.
- Paules C, Subbarao K. 2017. Influenza. Lancet. 390:697–708. doi:10.1016/S0140-6736(17)30129-0.
- Rambaut, A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi:10.1038/nature06945.
- Francis T, Magill TP. The antibody response of human subjects vaccinated with the virus of human influenza. J Exp Med. 65;1937:251–259.
- WHO. WHO report on global surveillance of epidemic-prone infectious diseases. Who (2000). <http://www.who.int/en/>
- Schotsaert M, García-Sastre A. 2017. Inactivated influenza virus vaccines: the future of TIV and QIV. Curr Opin Virol. 23:102–106. doi:10.1016/j.coviro.2017.04.005.
- Moa AM, Chughtai AA, Muscatello DJ, Turner RM, MacIntyre CR. Immunogenicity and safety of inactivated quadrivalent influenza vaccine in adults: A systematic review and meta-analysis of randomised controlled trials. Vaccine. 34;2016:4092–4102.
- Plant EP, Fredell LJ, Hatcher BA, Li X, Chiang MJ, Kosikova M, Xie H, Zoueva O, Cost AA, Ye Z, Cooper MJ. Different repeat annual influenza vaccinations improve the antibody response to drifted influenza strains. Sci Rep. 2017;7. doi:10.1038/s41598-017-05579-4.
- Snape MD, Pollard AJ. 2013. The beginning of the end for serogroup B meningococcus? Lancet. 381:785–787. doi:10.1016/S0140-6736(12)62194-1.
- Christensen H, Trotter CL, Hickman M, Edmunds WJ. Re-evaluating cost effectiveness of universal meningitis vaccination (Bexsero) in England: modelling study. BMJ. 2014;349. doi:10.1136/bmj.g5725.
- Harper DM, DeMars LR. 2017. HPV vaccines – A review of the first decade. Gynecol Oncol. 146:196–204. doi:10.1016/j.ygyno.2017.04.004.
- Gaglani M, Pruszynski J, Murthy K, Clipper L, Robertson A, Reis M, Chung JR, Piedra PA, Avadhanula V, Nowalk MP, et al. Influenza vaccine effectiveness against 2009 pandemic influenza A(H1N1) virus differed by vaccine type during 2013-2014 in the United States. J Infect Dis. 2016;213:1546–1556. doi:10.1093/infdis/jiv577.
- Zimmerman RK, Nowalk MP, Chung J, Jackson ML, Jackson LA, Petrie JG, Monto AS, McLean HQ, Belongia EA, Gaglani M, et al. 2014–2015 influenza vaccine effectiveness in the United States by vaccine type. Clin Infect Dis. 2016;63:1564–1573. doi:10.1093/cid/ciw635.
- Jackson ML, Chung JR, Jackson LA, Phillips CH, Benoit J, Monto AS, Martin ET, Belongia EA, McLean HQ, Gaglani M, et al. Influenza vaccine effectiveness in the United States during the 2015–2016 season. N Engl J Med. 2017;377:534–543. doi:10.1056/NEJMoa1700153.
- Flannery B, Chung JR, Belongia EA, McLean HQ, Gaglani M, Murthy K, Zimmerman RK, Nowalk MP, Jackson ML, Jackson LA, et al. Interim estimates of 2016–17 seasonal influenza vaccine effectiveness — United States, February 2017. MMWR Morb Mortal Wkly Rep. 2017;66:167–171. doi:10.15585/mmwr.mm6606a3.
- Flannery B, Chung JR, Thaker SN, Monto AS, Martin ET, Belongia EA, McLean HQ, Gaglani M, Murthy K, Zimmerman RK, et al. Interim estimates of 2017–18 seasonal influenza vaccine effectiveness — United States, February 2018. Am J Transpl. 2018;18:1020–1025. doi:10.1111/ajt.14730.
- Linderman SL, Chambers BS, Zost SJ, Parkhouse K, Li Y, Herrmann C, Ellebedy AH, Carter DM, Andrews SF, Zheng NY, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc Natl Acad Sci. 2014;111:15798–15803. doi:10.1073/pnas.1409171111.
- Xie H, Wan XF, Ye Z, Plant EP, Zhao Y, Xu Y, Li X, Finch C, Zhao N, Kawano T, et al. H3N2 mismatch of 2014-15 northern hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Sci Rep. 2015;5. doi:10.1038/srep15279.
- Zost SJ, Parkhouse K, Gumina ME, Kim K, Diaz Perez S, Wilson PC, Treanor JJ, Sant AJ, Cobey S, Hensley SE, et al.Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci. 2017;114: 201712377. doi:10.1073/pnas.1712377114.
- WHO. Recommended composition of influenza virus vaccines for use in the 2016–2017 northern hemisphere influenza season. Wkly Epidemiol Rec. 2016;91:121–132.
- Cobey S, Hensley SE. 2017. Immune history and influenza virus susceptibility. Curr Opin Virol. 22:105–111. doi:10.1016/j.coviro.2016.12.004.
- Huang KY, Rijal P, Schimanski L, Powell TJ, Lin TY, McCauley JW, Daniels RS, Townsend AR. Focused antibody response to influenza linked to antigenic drift. J Clin Invest. 2015;125:2631–2645. doi:10.1172/JCI80323.
- Liu M, Zhao X, Hua S, Du X, Peng Y, Li X, Lan Y, Wang D, Wu A, Shu Y, et al. Antigenic patterns and evolution of the human influenza A (H1N1) virus. Sci Rep. 2015;5:14171. doi:10.1038/srep14171.
- Carter DM, Bloom CE, Nascimento EJ, Marques ET, Craigo JK, Cherry JL, Lipman DJ, Ross TM. Sequential seasonal H1N1 influenza virus infections protect ferrets against novel 2009 H1N1 influenza virus. J Virol. 2013;87:1400–1410. doi:10.1128/JVI.02257-12.
- Li Y, Myers JL, Bostick DL, Sullivan CB, Madara J, Linderman SL, Liu Q, Carter DM, Wrammert J, Esposito S, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med. 2013;210:1493–1500. doi:10.1084/jem.20130166.
- Francis T. On the doctrine of original antigenic sin ON THE DOCTRINE OF ORIGINAL ANTIGENIC SIN *. Source Proc Am Philos Soc. 104;1960:572–578.
- Gagnon A, Acosta E, Hallman S, Bourbeau R, Dillon LY, Ouellette N, Earn DD, Herring DA, Inwood K, Madrenas J, et al. Pandemic paradox: early life H2N2 pandemic influenza infection enhanced susceptibility to death during the 2009 H1N1 pandemic. MBio. 2018;9. doi:10.1128/mBio.02091-17.
- Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO. 2016. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 354:722–726. doi:10.1126/science.aag1322.
- Recker M, Pybus OG, Nee S, Gupta S. 2007. The generation of influenza outbreaks by a network of host immune responses against a limited set of antigenic types. Proc Natl Acad Sci. 104:7711–7716. doi:10.1073/pnas.0702154104.
- Koelle K, Cobey S, Grenfell B, Pascual M. 2006. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science. 314:1898–1903. doi:10.1126/science.1132745.
- Ferguson NM, Galvani AP, Bush RM. 2003. Ecological and immunological determinants of influenza evolution. Nature. 422:428–433. doi:10.1038/nature01509.
- Zinder D, Bedford T, Gupta S, Pascual M. The roles of competition and mutation in shaping antigenic and genetic diversity in influenza. PLoS Pathog. 2013;9. doi:10.1371/journal.ppat.1003104.
- Gupta S, Ferguson N, Anderson R. 1998. Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science. 280:912–915. doi:10.1126/science.280.5365.912.
- Thompson CP, Lourenço J, Walters AA, Obolski U, Edmans M, Palmer DS, Kooblall K, Carnell GW, O’Connor D, Bowden TA, et al. A naturally protective epitope of limited variability as an influenza vaccine target. Nat Commun. 2018. doi:10.1038/s41467-018-06228-8. [Epub ahead of print]
- Nogales A, Piepenbrink MS, Wang J, Ortega S, Basu M, Fucile CF, Treanor JJ, Rosenberg AF, Zand MS, Keefer MC, et al. A highly potent and broadly neutralizing H1 influenza-specific human monoclonal antibody. Sci Rep. 2018;8. doi:10.1038/s41598-018-22307-8.
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE Jr. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85:10905–10908. doi:10.1128/JVI.00700-11.
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci. 2011;108:14216–14221. doi:10.1073/pnas.1111497108.
- Doud MB, Lee JM, Bloom JD. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat Commun. 2018;9. doi:10.1038/s41467-018-03665-3.
- Doud MB, Hensley SE, Bloom JD. Complete mapping of viral escape from neutralizing antibodies. PLoS Pathog. 2017;13. doi:10.1371/journal.ppat.1006271.
- Krammer F, Pica N, Hai R, Margine I, Palese P. 2013. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. 87:6542–6550. doi:10.1128/JVI.00641-13.
- Byrd-Leotis L, Galloway SE, Agbogu E, Steinhauer DA. 2015. Influenza hemagglutinin (HA) stem region mutations that stabilize or destabilize the structure of multiple HA subtypes. J Virol. 89:4504–4516. doi:10.1128/JVI.00057-15.
- Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi:10.1126/science.aac7263.
- Dilillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcR interactions for protection against influenza virus in vivo. Nat Med. 20:143–151. doi:10.1038/nm.3443.
- Coughlan L, Sridhar S, Payne R, Edmans M, Milicic A, Venkatraman N, Lugonja B, Clifton L, Qi C, Folegatti PM, et al. Heterologous two-dose vaccination with simian adenovirus and poxvirus vectors elicits long-lasting cellular immunity to influenza virus a in healthy adults [EBioMedicine 29 (2018) 146–154] (S2352396418300653) (10.1016/j.ebiom.2018.02.011)). EBioMedicine. 2018;31:321. doi:10.1016/j.ebiom.2018.05.001.
- Powell TJ, Silk JD, Sharps J, Fodor E, Townsend ARM. 2012. Pseudotyped influenza a virus as a vaccine for the induction of heterotypic immunity. J Virol. 86:13397–13406. doi:10.1128/JVI.01820-12.
- Antrobus RD, Lillie PJ, Berthoud TK, Spencer AJ, McLaren JE, Ladell K, Lambe T, Milicic A, Price DA, Hill AV, et al. A T cell-inducing influenza vaccine for the elderly: safety and immunogenicity of MVA-NP+M1 in adults aged over 50 years. PLoS One. 2012;7. doi:10.1371/journal.pone.0048322.
- van Doorn E, Liu H, Ben-Yedidia T, Hassin S, Visontai I, Norley S, Frijlink HW, Hak E. Evaluating the immunogenicity and safety of a BiondVax-developed universal influenza vaccine (Multimeric-001) either as a standalone vaccine or as a primer to H5N1 influenza vaccine. Med (United States). 2017;96:e6339.
