ABSTRACT
Leptospirosis is one of the most important but neglected, infectious tropical diseases worldwide. Leptospira interrogans is now recognized as a leading cause of the disease. Little is known of the genetic diversity and phylogenetic characteristics of L. interrogans within China. To better understand the transmission and genetic diversity of L. interrogans populations, we characterized 271 isolates and seven vaccine strains from China during 1954–2014 using multilocus variable-number tandem repeat analysis (MLVA). 110 different L. interrogans MLVA profiles (MTs) were identified, of which five were predominant, reflecting a high level of genetic diversity in L. interrogans population in China. Different from that of circulating isolates, seven vaccine strains have different MT, of which some are phylogenetically away from the circulating isolates. The results showed that Icterohaemorrhagiae, Hebdomadis, and Canicola ranked as the top three serogroups among L. interrogans strains tested. The cluster analysis demonstrate the clonal links between rodent and human isolates, suggesting the rodent species played a key role in the transmission of leptospirosis to humans, and contributed to the circulation of the pathogen in humans. Taken together, these findings should provide insight into a better knowledge of the epidemiology and molecular evolution of L. interrogans in China. Furthermore, the results should facilitate the selection of candidate vaccine strains in the future.
Introduction
Leptospirosis is a zoonotic disease caused by pathogenic species of the Leptospira genus.Citation1 A wide variety of mammalian hosts can act as infection reservoirs.Citation2,Citation3 Humans usually get the disease following environmental exposure to Leptospira, which were released in the urine of infected animals.Citation3,Citation4 Infection occurs during occupational or recreational exposure to contaminated soil and/or water with organisms entering the accidental human host via abrasions or less commonly via the conjunctiva.Citation5 It is estimated that there are 1.03 million human leptospirosis cases and 58,900 deaths each year; suggesting that the disease has become as a leading zoonotic cause of morbidity and mortality worldwide.Citation6
In China, the earliest leptospirosis case could be traced to the 1930s and it becomes more frequently reported after 1955.Citation7 Ten outbreaks of leptospirosis with incidences of more than 10 cases per 100,000 populations and with > 2.5 million cases and > 20, 000 deaths reported over the past sixty years.Citation7 Improvements in sanitation, and vaccination of high-risk populations, have played crucial role in reducing the disease burden since 1980s in China, and the incidence of leptospirosis has recently been significantly decreased. However, small localized outbreaks still occur in some epidemic regions in China.Citation8–Citation10 Furthermore, due to the variety of geographic and climatic conditions in China, pathogenic leptospiral serovars are far more diverse than in other countries. Over 70 serovars in 18 serogroups of pathogenic Leptospira have been isolated and identified from different hosts,Citation11 of which some serogroups (i.e, Manhao) are only found in China.Citation10 With more recent genetic classification based on DNA relatedness, L. interrogans is now recognized as a leading cause of the disease, although other pathogenic species have also been found in China.Citation11,Citation12 However, little is known of the genetic diversity and phylogenetic characteristics of L. interrogans within China.
The current licensed Leptospira vaccine in China is a multivalent, inactivated whole cell vaccine.Citation11,Citation13 Three highly virulent (serogroups Icterohaemorrhagiae, Grippotyphosa, and Autumnalis) and four low-virulence strains (serogroups Canicola, Pomona, Australis, and Hebdomadis) are used as vaccine strains, which are the major L. interrogans serogroups and covered more than 80 % serogroup coverage of circulating strains in the country.Citation13 Considering the inherent side-reactions and serogroup-specific immunity from inactivated vaccines, it is suggested that universal recombinant vaccines should be developed in the future. Meanwhile, it is curial to select the appropriated vaccine strains for understand the genetic diversity of Leptospira isolates and vaccine strains.
In recent years, it is reported that multilocus variable-number tandem repeat (VNTR) analysis (MLVA) method can provide high discrimination for genotyping of endemic Leptospira prevalence and have favourable consistency with sequencing method.Citation14 Isolates of Leptospira interrogans and L. borgpetersenii in East Asia were analyzed by MLVA using 4 loci, demonstrating that there may be variability in the range of genetic diversity among different Leptospira serogroups, which may be attributed to maintenance host animals and environmental factors.Citation15 Pavan and his colleagues also investigated the genetic diversity and epidemiological relationships of L. interrogans isolates from Argentina using MLVA by the loci VNTR4, VNTR7, VNTR9, VNTR10, VNTR19, VNTR23 and VNTR31.Citation16 Therefore, to better understand the transmission and genetic diversity of L. interrogans populations, in this study, we explored the genetic diversity and phylodynamic transmission of L. interrogans circulating isolates and vaccine strains in China in the past sixty years using a MLVA method. Furthermore, the modes of transmission, implicated reservoir hosts, prevalent serogroups, and regional distribution of infection were also analyzed.
Results
In total, nine strains were from five countries outside China and seven vaccines strains as well as 271 circulating isolates of L. interrogans from China used in the study were from the 11 provinces with the most widespread prevalence of leptospirosis in China (). All the seven vaccine strains (CMCC-1, CMCC-2, CMCC-3, 56,601, 56,606, 56,608, 56,609) were human originated. Among the 271 circulating isolates, 76 originated from humans (28%) while 195 were non-human originated (72%) (Supplementary Table 1). Serogroup of the isolates was determined using regular serological methods as previously described.Citation12 Of the 271 circulating isolates in China, serogroup Icterohaemorrhagiae (48%), Autumanlis (11%) and Hebdomadis (8%) ranked as the three most prevalent strains. Geographically, Sichuan (35%), Jiangxi (28%) and Anhui (18%) ranked as the three provinces which contributed most to isolate number in China (). Icterohaemorrhagiae was the most prevalent serogroup circulating in Anhui (80%), Sichuan (58%) and Jiangxi (44%). Serogroup Autumanlis (16%) and Hebdomadis (18%) were also prevalent in Sichuan province. Grippotyphosa (58%) was the most common serogroup circulating in Hunan province. Serogroups Icterohaemorrhagiae (22%), Hebdomadis (19%) and Canicola (8%) ranked as the three serogroups which contributed most to the pathogen’s circulation in humans. Furthermore, Icterohaemorrhagiae was the predominant serotype (53%) circulating in non-humans. Most of the non-human isolates were derived from murine sources. Details of species strain, serogroup, serovar, host, geographic source of isolation and year of isolation are listed in Supplementary Table 1.
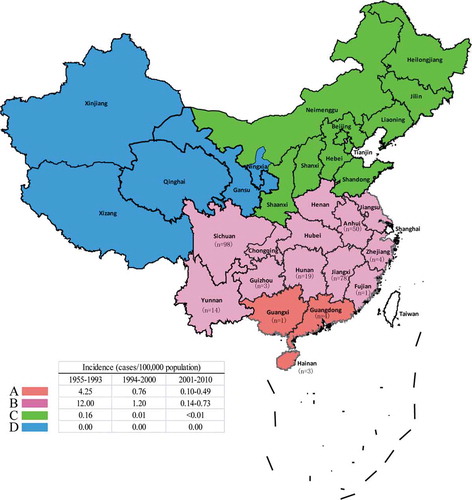
Figure 1. Geographic distribution of 271 L. interrogans isolates and seven vaccine strains from China. The number of isolates recovered from each province included in this study are listed. The provinces colored by pink and orange represent leptospirosis in epidemic areas B and A, respectively and the incidence data was also cited from previous studies. Citation7.
The MLVA results showed that the 271 circulating isolates and seven vaccine strains can be resolved into 110 MTs (designated MTs1-110), suggesting high genetic diversity among L. interrogans population in China (). 82 MTs were represented by single strain including six vaccine strains (CMCC-1, CMCC-2, CMCC-3, 56,606, 56,608, and 56,609) while 15 MTs were represented by two strains. 22 MTs were represented by at least three isolates, of which 5 contained at least 10 isolates (MT1, MT2, MT3, MT4, and MT5). Vaccine strains 56,601 and other two isolates belonged to MT20. Furthermore, different from that of Chinese isolates, nine international reference strains had unique MT types, respectively (Supplementary Table 1). All nine L. interrogans reference strains outside of China were located on the most peripheral positions of the MST tree (), and were distantly related to strains from China.
Figure 2. Minimum spanning tree of multilocus variable number tandem-repeat analysis (MLVA) for L. interrogans isolates in China. The clustering used the categorical coefficient and eBURST priority rule of the highest number of single-locus changes. Each circle represents an MLVA type (MT). Thick lines, types differ by a single MLVA locus; thin lines, double-locus variants; dotted lines, 2 types differing by > 2 MLVA loci. The size of the circle reflects the number of isolates with a given MT. Only MT containing at least three isolates were labeled in the figure. The color codes for source of isolation, host and serogroups are respectively shown as Figure 2(a,b and c). Pie charts within a circle are used to indicate the proportion of isolates in a given MT. Vaccine strains are labeled with star and strain name respectively in Figure 2B.
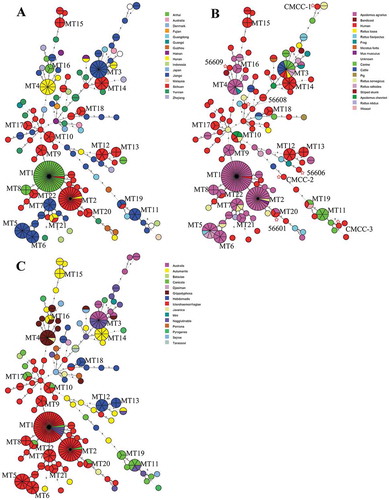
MT1 was the most predominant genotype among the isolates analyzed in this study, which was consisted of one strain from Sichuan province and the other 40 strains came from Anhui province. Interestingly, the strain from Sichuan province was separated from human and the other 40 strains from Apodemus agrarius, implicating that MT1 isolate may have spread from Anhui to Sichuan province (). MT2 consists of one strain from Hunan province with the other 23 strains coming from Sichuan province. All MT2 strains were separated from animal hosts, most (92%, 22/24) of which was Apodemus agrarius (). MT3 consisted of 13 strains from Jiangxi province and surprisingly four other animal groups except human (cattle, canine and in the rodents Rattus rattoides and Rattus losea) were found in this MT (). Similar to MT2, MT4 consisted of 10 strains from Hunan province along with two subspecies of murine in addition to an isolate of unknown origin. MT5 consisted of 10 isolates from Jiangxi province. Three murine subspecies existed in this MLVA subtype. Furthermore, there were 17 more MTs (MT6-MT22) scattered on the MST tree, whose number isolate ranged from 3 to 8(Supplementary Table 1).
Further analysis showed that isolates from human and animals located together in many MTs (), such as MT1 (human and murine), MT3 (human, cattle, canine, and murine), MT10 (human and murine) and MT19 (human, cattle, and weasel), suggesting that there were direct transmissions of L. interrogans pathogens from animals to humans. The numbers shown on the MST branch indicate MLVA distance between adjacent MTs based on the seven VNTR alleles. Strains in one cluster mean that there are no differences among these isolates. Number n means that the adjacent MTs differed from each other by n VNTR alleles. All MLVA distances on the MST tree were analyzed. If not more than 2 alleles difference were found between adjacent MTs, it is regarded that direct or indirect transmission events happened between the two MTs. Based on the MLVA distance data shown on the branches of the phylogenetic tree, we could demonstrate strain clonality between rodent and human isolates, suggesting rodent species played a key role in the transmission of leptospirosis to human in China. Canine and cattle are also important reservoirs for transmission of the pathogen to humans (). Transmission between different kinds of animal can also be found ().
Congruence of the phylogenetic MTs and serogroups was week for the strains used in this study (). One MT may contain several different serogroups (MT1, MT2, MT3, MT4, MT10, MT11, MT17, MT20, and MT22) and one serogroup may also exist in different gene clusters (e.g., Icterohaemorrhagiae, Grippotyphosa and Canicola). Strains of serogroup Icterohaemorrhagiae represent nearly half of the total stains. These strains are scattered on one side of the MST tree and not very far from each other, which seems like a result of a founder effect. The same situation also happened to serogroup Grippotyphosa and Australis.
Discussion
Leptospirosis has a broad geographical distribution due to wide range of serogroups and large spectrum of mammalian hosts.Citation4,Citation17,Citation18 The morbidity and mortality of the disease in different countries are not the same, which is depending on the location, climate, economic development degree.Citation19 In some developing countries with a tropical climate, the incidence of Leptospirosis are still high.Citation20,Citation21 The expansion of urban slums also provides conditions for the spread of the disease.Citation22–Citation25 These factors contribute to distinct epidemiology and genetic diversity of Leptospira in different countries and regions. To better control and prevent this disease, it is very important to understand the genetic diversity of L. interrogans circulating in China.
Based on the incidences of leptospirosis and latitude and longitude, four epidemic areas of leptospirosis (designated as A, B, C, and D) were divided in China.Citation7 Areas A and B, located in Southern China, are the areas with the most widespread prevalence of leptospirosis. With numerous rivers and lakes, a moist climate, and rice-planting tradition, these areas confer advantages for the spread and prevalence of L. interrogans. The incidences of leptospirosis in areas C (≤ 0.01) and D (0.00) was much lower than in areas A and B. All 278 L. interrogans strains (including 7 vaccine strains) tested in this study were collected from areas A and B. Furthermore, of 34 Chinese serogroup reference strains represented the diversity revealed by serogroup typing of > 300 isolates.Citation13 Thus, these strains used should provide a good representation of geographic and genetic diversity of L. interrogans population in China.
It is recently reported that 17 different STs were found in 120 serogroup icterohaemorrhagiae strains belonged to three pathogenic Leptospira species (L. interrogans, L. borgpetersenii, and L. kirschneri) in China, suggesting a high level of genetic diversity in the serogroup Icterohaemorrhagiae strains.Citation12 In this study, the MLVA analysis rendered 110 genotypes amongst L. interrogans strains circulating in China in the past sixty years, further providing the evidence to support high genetic diversity in L. interrogans population. Whole genomic analysis suggested that the same pathogenic Leptospira species may be undergo similar evolution from the most recent common ancestor.Citation26 The genetic heterogeneity among L. interrogans population also reflect that different mechanism may involved with intraspecies evolution.
The results showed that all nine L. interrogans reference strains outside of China were located on the most peripheral positions of the MST tree (), and were distantly related to strains from China. Thus, the emergence and re-emergence of L. interrogans in different regions of China was not caused by the introduction of exotic strains, but by the persistence of local reservoirs of infection. Basing the MST tree as a source of geographic information suggests that Jiangxi and Sichuan provinces played key roles in the circulation of L. interrogans in China. Though up to 50 isolates were also recovered from Anhui province. Almost all of these clustered together in MT1 with small links occurring between Anhui and other provinces. But another situation was found in Jiangxi and Sichuan province (). MT3, MT5, MT6, MT7, and MT11 were found in Jiangxi with up to 26 small size (1 or 2 isolates) MTs also found. Most of these MTs are phylogenetically far from each other and when scattered on the MST tree show a tight connection with other provinces, such as Sichuan, Hunan, Zhejiang, and Guangdong. The situation in Sichuan is the same as Jiangxi. MT2, MT9, MT10, MT12, MT13, MT15, MT17, MT18, and MT22 were found in Sichuan with up to 24 small size (1 or 2 isolates) MTs also found in this province. Most of these MTs are phylogenetically far from each other and when scattered on the MST tree show a tight connection with other provinces, such as Jiangxi, Yunnan, Hunan, Anhui, and Guangdong. Jiangxi and Sichuan are located on the Yangtze River Basin. Numerous rivers and lakes, a moist climate, a large agricultural population base and rice-planting are all advantageous for the spread and prevalence of L. interrogans within and out of these two provinces. A possible explanation is that circulating isolates derived from a single source, for example water contaminated by a strain of L. interrogans. Another explanation is that the pathogen may spread among these species over a short period.
The cluster analysis indicated that rodent species played a key role in the transmission of leptospirosis to humans in China. Their large numbers and extensive distribution in nature are important reasons.Citation1,Citation27 When water and soil are contaminated by murine urine or excrement, they can become a pathogen source. Leptospirosis case investigation reports and deaths recorded by China CDC investigators submitted from 2003–2017 were reviewed (earlier data were not available from the website). Most of the leptospirosis cases in China occurred from July to December with a peak in September(Supplementary Figure 1).Citation7 This is the time of rice planting and harvest in Southern China. Rats are also more active in these months.Citation28 Canine and cattle are also important reservoirs for the transmission of the pathogen to humans. Farmers may get infected from the domestic animals they feed or come into close contact with. That is why in rice-planting reasons in southern China, many cases of L. interrogans were reported in farmers.
Consist with the previous findingCitation14, we also found week congruence between typing results of MLVA methods and serological tests for the 287 L. interrogans isolates. The two methods were associated with different types of genetic variation. MLVA methods can also show variation in repeated sequences, which are extensively located in the genome of pathogen.Citation14,Citation15 Serological tests were involved with the evolution of the membrane proteins. Molecular mutations on the membrane proteins can be a result of positive or negative selection.Citation1,Citation10 However, mutations detected by MLVA methods may be indicative of neutral selection.Citation14,Citation15,Citation29 Therefore, in order to understand the epidemiology and genetic evolution of pathogenic Leptospira, it is suggested that a combined molecular (MLVA or multilocus sequence typing) and serological analysis may be a better approach for the typing of this pathogen.
Vaccination of at-risk populations remains the most viable strategy to control leptospirosis. An inactivated whole-cell vaccine including seven major L. interrogans serogroups is still used in the Chinese Expanded Program on Immunization.Citation11,Citation13 Not surprising, seven vaccine strains have different MT, which were different from other circulating isolates. It is possibly associated with these vaccine strains belonged to old circulating isolates and replaced by new type isolates. Vaccine strains, such as CMCC-1, 56,606 and 56,609, are phylogenetically away from the circulating isolates from human ()). According to this study, candidate vaccines should be selected around MT17, MT12, MT18 or MT14, which was the gathering location of circulating isolates in human on the MST tree. Furthermore, considering the short-lived serogroup-specific immunity acquired from current inactivated vaccines, it is necessary to develop a universal recombinant leptospirosis vaccine.Citation13 The data of genotyping for circulating isolates and vaccine strains will contribute to the choices of vaccine strain and efficacy evaluation of leptospirosis vaccine product.
Conclusions
Systematic large-scale studies were carried out that provided detailed information on the epidemiologic and phylogenetic characteristics of L. interrogans prevalence in China. This study described the genetic diversity and phylodynamic transmission of L. interrogans in China, with special focus on modes of transmission. Our results implicated: reservoir hosts; prevalent serovars; and regional distributions of the pathogen. The results of phylogenetic analysis indicated that 278 isolates circulating in China were resolved into 110 MTs. Five major MTs were found. Cluster analysis suggested that rodent species played a key role in the transmission of leptospirosis to humans. Our results should facilitate the selection of candidate vaccine strains, which contribute to the development of new Leptospiravaccine .
Materials and methods
Bacterial strains and culture
Two hundred and seventy-one L. interrogans representative of isolates from eleven provinces during 1954–2014 and seven vaccine strains were used in this study (). The related epidemic information for each isolate including: host, source of isolation and year of isolation were collected. Furthermore, nine L. interrogans reference strains from other countries were also included during this study (Supplementary Table 1).
All isolates were grown in Leptospira Medium Base Ellinghausen McCullough Johnson Harris (EMJH) supplemented with 10% rabbit serum to late log phase and harvested by centrifugationCitation27. Genomic DNA was extracted using a Wizard Genomic DNA Purification Kit (Promega, Southampton, UK) according to the manufacturer’s instructions. Serogroup of the isolates were identified using a microscopic agglutination test as previously described. Citation12
A map of the distribution of isolates showing the geographic origin and number of isolates per province in China was developed (). We also reviewed leptospirosis case investigation reports and deaths by Chinese CDC investigators that were submitted from 2003–2017 (previous data are not available from the website). These data can be downloaded from website of China CDC (www.chinacdc.cn).
MLVA analysis
Seven VNTR loci, including VNTR4, VNTR7, V27, V29, V30, V36, and V50, were selected, and the primers for each VNTR loci were used as previous described.Citation29,Citation30 PCR were performed using a 50 μl volume, containing: 1× PCR buffer, 1.5 mM MgCl2, 200 uM dNTPs, 0.2 umM forward and reverse primer, 1.25 U AmpliTaq Gold DNA polymerase (Applied Biosystems), 2.5 ng DNA template, and double-distilled water to make up the final volume. Thermocycling parameters included an initial denaturation at 94°C for 10 min followed by 25 cycles of 94°C for 30 s, a specific annealing temperature for each VNTR loci for 30 s and 72°C for 1 min, followed by a final extension at 72°C for 10 min. The PCR product was then sequenced by using an ABI 3730XL system (Applied Biosystems) to identify the size of each fragment.
Data analysis
Allelic sizes were then converted into repeat copy numbers using the formula: repeat number = [fragment size (bp) – flanking regions (bp)]/repeat unit size (bp). The raw repeat copy numbers were then converted into integers. The assignment of an MLVA type was based on the combination of repeat counts for VNTR4, VNTR7, V27, V29, V30, V36 and V50. MLVA data, expressed as allelic profiles for isolates, were analyzed using Bionumerics software v7.5 (Applied-Maths). A minimum-spanning tree based upon multilocus, variable number of tandem repeat analysis, was constructed.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Supplemental Material
Download Zip (73.5 KB)Supplementary material
supplementary material data can be accessed here.
Additional information
Funding
References
- Adler B., de la Peña Moctezuma A. dlPM. leptospira and leptospirosis. Vet Microbiol. 2010.;140:287–296. doi:10.1016/j.vetmic.2009.03.012.
- Obiegala A, Woll D, Karnath C, Silaghi C, Schex S, Eßbauer S, Pfeffer. M, Picardeau M. Prevalence and genotype allocation of pathogenic leptospira species in small mammals from various habitat types in Germany. PLoS Negl Trop Dis. 2016.;10:e0004501. doi:10.1371/journal.pntd.0004501.
- Evangelista KV, Coburn J. Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiol. 2010.;5:1413–1425. doi:10.2217/fmb.10.102.
- Levett PN. Leptospirosis. Clin Microbiol Rev. 2001.;14:296–326. doi:10.1128/CMR.14.2.296-326.2001.
- Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–771.
- Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, Stein C, Abela-Ridder B, Ko AI, Small PLC. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis. 2015;9:e0003898. doi:10.1371/journal.pntd.0003898.
- Zhang C, Wang H, Yan J. Leptospirosis prevalence in Chinese populations in the last two decades. Microbes Infect. 2012.;14:317–323. doi:10.1016/j.micinf.2011.11.007.
- Liu Y, Li SJ, Yao GH, Huang H, Ma Q, Zhou JZ, Tang GP, Wang DM. Epidemiological investigation of two leptospirosis death cases in Guizhou Province. Zhonghua Yu Fang Yi Xue Za Zhi. 2017;51:243–247.
- Chung WS, Chu YH, Lin CL, Kao Ch. Increased risk of acute coronary syndrome among leptospirosis patients: A nationwide cohort analysis. Int J Cardiol. 2015;184:576–580. doi:10.1016/j.ijcard.2015.03.021.
- Xu Y, Zheng H, Zhang Y, Wang Y, Zhang J, Li Z, Cui S, Xin X, Ye Q, Chang YF, et al. Genomic analysis of a new serovar of leptospira weilii serogroup manhao. Front Microbiol. 2017;8:149.
- Hu W, Lin X, Yan J. Leptospira and leptospirosis in China. Curr Opin Infect Dis. 2014.;27:432–436. doi:10.1097/QCO.0000000000000097.
- Zhang C, Yang H, Li X, Cao Z, Zhou H, Zeng L, Xu J, Xu Y, Chang YF, Guo X, et al. Molecular typing of pathogenic leptospira serogroup icterohaemorrhagiae strains circulating in China during the past 50 years. PLoS Negl Trop Dis. 2015.;9:e0003762. doi:10.1371/journal.pntd.0003762.
- Xu Y, Ye Q. Human leptospirosis vaccines in China. Hum Vaccin Immunother. 2018.;14:984–993. doi:10.1080/21645515.2017.1405884.
- Hamond C, Pinna M, Medeiros MA, Bourhy P, Lilenbaum W, Picardeau M. A multilocus variable number tandem repeat analysis assay provides high discrimination for genotyping Leptospira santarosai strains. J Med Microbiol. 2015.;64:507–512. doi:10.1099/jmm.0.000045.
- Koizumi N, Izumiya H, Mu JJ3, Arent Z, Okano S, Nakajima C, Suzuki Y6, Mizutani Muto M, Tanikawa T, Taylor KR, et al. Multiple-locus variable-number tandem repeat analysis of Leptospira interrogans and Leptospira borgpetersenii isolated from small feral and wild mammals in East Asia. Infect Genet Evol. 2015;36:434–440. doi:10.1016/j.meegid.2015.08.013.
- Pavan ME, Cairó F, Pettinari MJ, Samartino L, Brihuega B. Genotyping of Leptospira interrogans strains from argentina by multiple-locus variable-number tandem repeat analysis (MLVA). Comp Immunol Microbiol Infect Dis. 2011.;34:135–141. doi:10.1016/j.cimid.2010.06.002.
- Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009.;7:736–747. doi:10.1038/nrmicro2208.
- Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis. 2008.;12:351–357. doi:10.1016/j.ijid.2007.09.011.
- Lau CL, Smythe LD, Craig SB, Weinstein P. Climate change, flooding, urbanisation and leptospirosis: fuelling the fire? Trans R Soc Trop Med Hyg. 2010.;104:631–638. doi:10.1016/j.trstmh.2010.07.002.
- Wuthiekanun V, Sirisukkarn N, Daengsupa P, Sakaraserane P, Sangkakam A, Chierakul W, Smythe LD, Symonds ML, Dohnt MF, Slack AT, et al. Clinical diagnosis and geographic distribution of leptospirosis, Thailand. Emerg Infect Dis. 2007;13:124–126. doi:10.3201/eid1301.060718.
- Agampodi S, Peacock SJ, Thevanesam V. The potential emergence of leptospirosis in Sri Lanka. Lancet Infect Dis. 2009.;9:524–526. doi:10.1016/S1473-3099(09)70211-7.
- Maciel EA, de Carvalho AL, Nascimento SF, de Matos RB, Gouveia EL, Reis MG, Ko AL. Household transmission of leptospira infection in urban slum communities. PLoS Negl Trop Dis. 2008.;2:e154. doi:10.1371/journal.pntd.0000326.
- Ganoza CA, Matthias MA, Collins-Richards D, Brouwer KC, Cunningham CB, Segura ER, Gilman RH, Gotuzzo E, Vinetz JM, White N. Determining risk for severe leptospirosis by molecular analysis of environmental surface waters for pathogenic Leptospira. PLoS Med. 2006.;3:e308. doi:10.1371/journal.pmed.0030308.
- Felzemburgh RD, Ribeiro GS, Costa F, Reis RB, Hagan JE, Melendez AX, Fraga D, Santana FS, Mohr S, Dos Santos BL, et al. Prospective study of leptospirosis transmission in an urban slum community: role of poor environment in repeated exposures to the Leptospira agent. PLoS Negl Trop Dis. 2014.;8:e2927. doi:10.1371/journal.pntd.0002927.
- Costa F, Ribeiro GS, Felzemburgh RD, Santos N, Reis RB, Santos AC, Fraga DB, Araujo WN, Santana C, Childs JE, et al. Influence of household rat infestation on leptospira transmission in the urban slum environment. PLoS Negl Trop Dis. 2014.;8:e3338. doi:10.1371/journal.pntd.0003338.
- Xu Y, Zhu Y, Wang Y, Chang YF, Zhang Y, Jiang X, Zhuang X, Zhang J, Zeng L, Yang M, et al. Whole genome sequencing revealed host adaptation-focused genomic plasticity of pathogenic Leptospira. Sci Rep. 2016.;6:20020. doi:10.1038/srep20020.
- Faine S, Adler B, Bolin C. Leptospira and Leptospirosis. 2nd. Melbourne (Australia): MediSci; 1999.
- Li S, Wang D, Zhang C, Wei X, Tian K, Li X, Nie Y, Liu Y, Yao G, Zhou J, et al. Source tracking of human leptospirosis: serotyping and genotyping of Leptospira isolated from rodents in the epidemic area of Guizhou province, China. BMC Microbiol. 2013;13:75. doi:10.1186/1471-2180-13-75.
- Slack A, Symonds M, Dohnt M, Smythe L. An improved multiple-locus variable number of tandem repeats analysis for Leptospira interrogans serovar Australis: a comparison with fluorescent amplified fragment length polymorphism analysis and its use to redefine the molecular epidemiology of this serovar in Queensland, Australia. J Med Microbiol. 2006.;55:1549–1557. doi:10.1099/jmm.0.46779-0.
- Majed Z, Bellenger E, Postic D, Pourcel C, Baranton G, Picardeau M. Identification of variable-number tandem-repeat loci in Leptospira interrogans sensu stricto. J Clin Microbiol. 2005.;43:539–545. doi:10.1128/JCM.43.2.539-545.2005.
