ABSTRACT
Inoculation with vaccine is the major intervention currently used to prevent influenza infections. However, it will be a challenge to produce and implement a new vaccine when a novel highly pathogenic influenza virus emerges in humans as significant infections. H7 subtype influenza viruses have similar epitopes on hemagglutinin, which can induce cross-reactive antibodies. In this study, a meta-analysis of the cross-reactivity of antibodies induced by one H7 subtype influenza vaccine against other H7 subtypes was performed. Database search was conducted in PubMed, Cochrane Library, EMBASE, MEDLINE, Chinese Biological Medicine Database (CBM), and Wanfang. A total of 9 articles comprising 811 human subjects were included in this meta-analysis. All assessed H7 influenza vaccines induced vaccine strain-specific protective antibodies [seroconversion rate (SCR) = 0.74, 95% CI (0.65, 0.82); seroprotection rate (SPR) = 0.81, 95% CI (0.78, 0.83)]. All H7 influenza virus monovalent vaccines exhibited cross-reactivity tested by hemagglutinin inhibition test (HI), microneutralization test (MN) and immunosorbent assay (ELISA) to other H7 subtype viruses. H7N1, H7N3, H7N7, and H7N9 vaccines elicited cross-reactive antibodies against other H7 subtype influenza viruses [SCR = 0.66, 95% CI (0.50, 0.82); SPR = 0.79, 95% CI (0.67, 0.91)]. The pooled SCR (95%CI) of cross-reactivity of H7N1 and H7N3 vaccines were 0.88 (0.85, 0.91) and 0.40 (0.26, 0.54), respectively. The consolidated SPR (95%CI) of H7N1 and H7N7 vaccines were 0.89 (0.86, 0.92) and 0.93 (0.81, 1.06). All H7 vaccines induced cross-reactive antibodies against H7N9 viruses [SCR = 0.69, 95% CI (0.52, 0.86); SPR = 0.85, 95% CI (0.76, 0.94)]. H7 vaccines can be used to limit influenza infection when a new highly pathogenic H7 virus appears.
Introduction
The first human case of H7N9 avian influenza virus was reported in China in March 2013.Citation1 The illness began with flu-like symptoms and progressed rapidly to acute pneumonia and acute respiratory distress syndrome.Citation2–Citation5 As of March 2018, a total of 1,567 laboratory-confirmed cases of human infection with H7N9 viruses, including at least 615 deaths, have been reported.Citation6 The novel H7N9 influenza virus was most likely generated by reassortment among wild bird H7N9, duck H7N3, and poultry H9N2 viruses.Citation1,Citation2,Citation7,Citation8 In addition to the recent emergence of the H7N9 virus in humans, patients infected with other H7 subtype influenza viruses, H7N7, H7N2, and H7N3, have been reported since 1959, with clinical symptoms of conjunctivitis, influenza-like manifestations,Citation9–Citation15 and acute respiratory distress syndrome.Citation16 The possibility of reassortment of new avian influenza viruses may be increased and influenza pandemics may happen, due to the migration of migratory birds and the variety of viruses that coexist in live poultry.
Subtypes of influenza A viruses are defined by the surface hemagglutinin (HA) and neuraminidase (NA), which have been classified into 18 (H1-H18) and 11 (N1-N11) subtypes, respectively, based on their amino acid sequences and structural features.Citation17–Citation19 The HA protein is composed of an immunodominant globular head domain and a stalk domain and it plays a major role in binding to host cell surface receptors.Citation20,Citation21 Most of the antibody responses induced by the influenza viruses or vaccine target the immunodominant HA head domain,Citation22,Citation23 thus the HA head represents the major influenza antigenic sites, and many of these have been defined, including epitopes Sa, Sb, Ca and Cb in H1, and epitopes A, B, C, D, and E in H3.Citation24–Citation28
Preventive vaccination is the major intervention currently used to prevent influenza infections.Citation29–Citation32 Several clinical trials have been performed analyzing the immunological responses to H7 influenza vaccines. Rudenko et al.Citation33 reported that adults vaccinated with H7N3 flu vaccine-induced protective antibodies against H7N3 and H7N9 viruses at rates of 44.8% and 34.8%, respectively. Madan et al.Citation34 found an increase in serum antibody titers in subjects vaccinated with the H7N9 flu vaccine supplemented with the AS03 adjuvant and identified seroprotection rates of 96.4% and 75% against H7N9 and H7N1 virus, respectively. After inoculation with H7N1 influenza vaccine supplemented with the AS03 adjuvant, the protection rates were 94.8% against H7N1 virus and 100% against H7N9 virus in the adult group, whereas the protection rates were 88.7% and 92% against the H7N1 and H7N9 viruses, respectively, in the elderly group.Citation35,Citation36
H7 subtype influenza vaccines include inactivated vaccines, live attenuated vaccines, subunit vaccines, and recombinant vaccines. All of them are in clinical phase I/II trials and have to date not been used on a large scale clinically. It has been reported that there is cross-protection between H7 subtypes, but the protection of cross-reactive antibodies still remains controversial because of the inconsistent results among studies. This report presents a meta-analysis of available data on the cross-reactivity of antibodies elicited by H7 influenza vaccine in order to provide a robust estimate of seroconversion and protection rates against non-vaccine incorporated H7 subtypes.
Methods
Search strategy
Two reviewers (Xiaoqin Gou and Xiaoxue Wu) independently searched articles in Chinese and English databases using the search strategy (H7N1 OR H7N2 OR H7N3 OR H7N4 OR H7N5 OR H7N6 OR H7N7 OR H7N8 OR H7N9 OR H7N10 OR H7N11 OR H7 subtype) and the search strategy (vaccine). Published studies were retrieved in PubMed, Cochrane Library, EMBASE, MEDLINE, CBM, and Wanfang database. All retrievals were implemented by using the Mesh and free word (established to December 1, 2018).
Selection criteria
All included studies met the following criteria. (1) Healthy subjects with a description of cross-reactivity induced by an H7 influenza vaccine, regardless of nationality, race, age and follow-up time. (2) Inoculation with an H7 influenza vaccine. (3) One of the main outcomes: influenza infection rate, incidence of influenza-like symptoms, serological changes of cross-reactive antibodies after vaccination [SCR, SPR, geometric mean titers (GMTs), and mean geometric increase (MGI) measured by HI, MN, or ELISA]. (4) Cohort study, randomized controlled trial, controlled before-after vaccination study or clinical trial.
Exclusion criteria were: (1) not H7 subtype influenza vaccine; (2) reporting vaccine types, vaccine management and production, not cross-reactivity induced by a H7 vaccine; (3) animal research; (4) subjects were also vaccinated with other vaccines, such as H1N1, H3N2 or H5N1 vaccines, which may affect the cross-reactivity induced by H7 vaccine; (5) no cross-reactivity data.
Data extraction
Two independent reviewers extracted data from all selected studies, including participant characteristics (total number, age group), vaccine dose, adjuvant, inoculation times, vaccine strain, heterologous strain, and outcomes. Any disagreements or discrepancies were resolved by discussion or by the third reviewer. If there was some missing data, the original authors were contacted for additional information on unreported data. For the results of antibody titersCitation37,Citation38 and fold changes of antibody titersCitation39 displayed as images without digital data, images were imported into digital software (Engauge Digitizer 4.1)Citation40 to convert the outlines into x and y coordinates. Consequently, the values of the antibody titers or fold changes were displayed and then exported into Excel files.
Assessment of study quality
Study quality, risk of bias, was assessed by the Cochrane Handbook (5.1.0).Citation41 The levels of risk of bias in the random sequence generation, the allocation concealment, the blinding of participants and personnel, the blinding of outcome assessment, incomplete outcome data, selective reporting, and other bias (whether the baseline is comparable) were judged as “low risk”, “high risk” and “uncertainty”.
Statistical methods
Estimates of SCR and SPR were pooled using Stata12.0 software.Citation42 Comparison of sub-groups (risk ratio, RR) was performed using Review Manager 5.3.Citation43 The figure with scatter plots was made to display the SCR and SPR of individual study using Origin 2017 software. To assess heterogeneity between studies, I2 values were calculated. The fixed-effects model (FEM) was used when I2 < 50% indicating no statistical heterogeneity between studies, otherwise the random-effects model (REM) was used after excluding significant clinical heterogeneity effects if I2 ≥ 50%. Egger’s test was performed to evaluate the publication bias of specific antibody responses using Stata12.0 software.Citation42 Fail-Safe Number was calculated to evaluate cross-reactivity using formula (∑Z/1.64)2-k (k represents the number of studies included. Obtain Z by checking the standard normal distribution table according to P of each independent study).
Results
Selection of studies
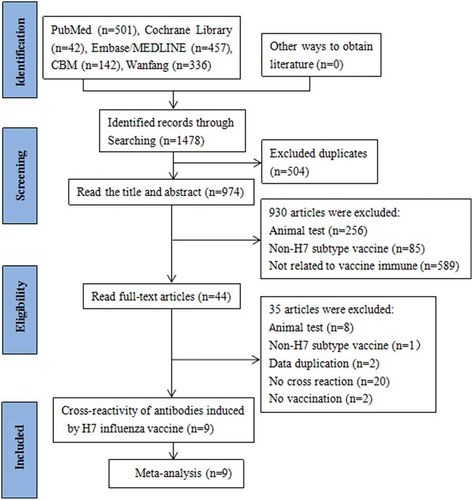
A total of 1478 articles were obtained from the databases. Five hundred and four duplicates were removed. By two selection rounds on titles and abstracts, 44 studies were potentially eligible. After carefully reading full text and discussion, 9 studies identified to meet the inclusion criteria and focus on the H7 subtype vaccine cross-reactivity were retained for systematic review and meta-analysis, totaling 811 participantsCitation33–Citation39,Citation44,Citation45 ().
Characteristics of studies included
The characteristics of the included studies were described in . All nine articles were conducted in American and Canada from 2014 to 2017. Of nine, fourCitation33-Citation36 were randomized controlled trials containing saline or placebo groups, and fiveCitation37–Citation39,Citation44,Citation45 were self-controlled clinical trials. H7N1, H7N3, H7N7, and H7N9 viruses were used as vaccine strains. None of the studies included influenza infection rates, vaccine protection rates, or described influenza-like symptoms, but serological antibody levels were included. The cross-reactivity of antibodies was assessed using SCR and SPR in meta-analysis. SCR was defined as the percentage of the pre-vaccination serum antibody titer <1:10 and post-vaccination antibody titer ≥1:40, or pre-vaccination serum antibody titer ≥1:10 and at least a four-fold increase in post-vaccination antibody titer. SPR was defined as the percentage of the serum antibody titer ≥1:40 after vaccination.
Table 1. Main characteristics of the included studies.
Methodological bias risk
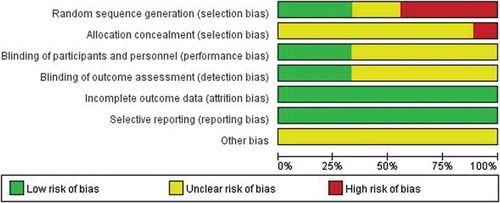
In the nine articles included for meta-analysis, one systematic review was at low or moderate risk of bias across most domains, with high risk in allocation in one study (). Three studiesCitation33,Citation35,Citation36 give specific methods for generating random sequences, two articlesCitation34,Citation39 only mention random sequences without specific methods, whereas four articlesCitation37,Citation38,Citation44,Citation45 do not mention random sequences. One literatureCitation33 did not implement allocation concealment. Six articlesCitation34,Citation37–Citation39,Citation44,Citation45 did not explain blindness. All literature data is complete and there are no selective reports.
Vaccine-specific antibody responses
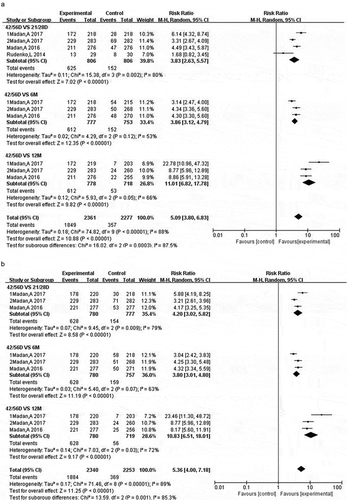
Antibodies induced by certain antigen can bind to other antigen containing the same or similar epitopes, which is called cross-reaction.Citation46 The cross-reactivity depends on specific antibody level and affinity. In this systematic review, all H7 subtype vaccines, including H7N1, H7N3, H7N7, and H7N9, elicited protective vaccine-specific antibodies.Citation33-Citation39,Citation41–Citation45 All nine studies were used to assess the pooled cross-reactivity by meta-analysis. All subjects were inoculated with H7 vaccines on day 0 (first dose) and day 21/28 (second dose). Since antibody responses peak in 3 or 4 weeks after vaccination, the titers of antibodies at day 21/28 and day 42/56 were used to represent the antibody response of one dose and two doses, respectively. We first analyzed the specific antibody responses at days 21/28 (3 or 4 weeks after the first inoculation), 42/56 (3 or 4 weeks after the second inoculation), 6 months and 12 months to determine the time point for the highest SCR and SPR. Our results showed that SCR [0.74 (0.65, 0.82)] and SPR [0.81 (0.78, 0.83)] of vaccine-specific antibodies were the highest at day 42/56 (two doses), meeting vaccine production licensing criteria CBER and CHMP,Citation46–Citation48 whereas one dose exhibited lower antibody responses (). We then analyzed the RR values of SCR, SPR between day 42/56 and other time points using the formula RR = the SCR or SPR at day 42/56: the SCR or SPR at day 21/28, 6-month or 12-month. The RRs were greater than 1, and P < .05 when comparing the responses at day 42/56 to those at day 21/28, suggesting that antibody responses of two doses were higher than one dose. The RRs were greater than 1 when comparing the SCR and SPR of 42/56d to 6-month and 12-month, suggesting that antibody titers declined from the peak of response (42/56d) after the second dose (). These results indicated that the vaccine-specific antibody levels were the highest at day 42/56 and the time point is best for analysis of the cross-reaction.
Table 2. Vaccine-specific antibody responses.
Cross-reactivity between H7 subtype vaccines
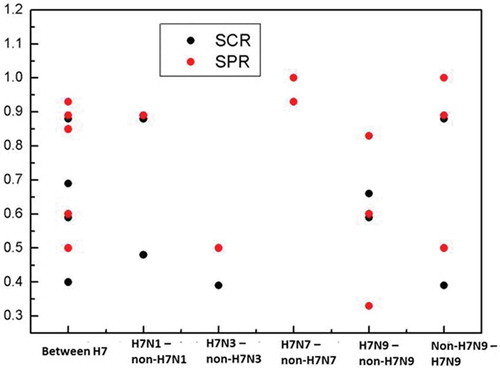
Systematic analysis of the nine articles, cross-reactivity of vaccines was detected in subjects vaccinated with H7N9 against H7N1, H7N2, H7N3, H7N7, and H7N8; H7N7 against H7N3 and H7N9; H7N3 against H7N1 and H7N9; H7N1 against H7N3 and H7N9. Three articlesCitation35,Citation36,Citation39 descripted the cross-reactivity induced by an H7N1 vaccine against H7N9 and H7N3 viruses. Two articlesCitation33,Citation45 described cross-reactivity induced by an H7N3 vaccine against H7N9 and H7N1 viruses. Only one article represented cross-reactivity induced by an H7N9 vaccine against H7N1,Citation34 H7N2,Citation44 H7N8,Citation44 H7N7,Citation37 H7N3,Citation37 by an H7N3 against H7N1,Citation45 or by an H7N7 against H7N3,Citation38 H7N9,Citation38 respectively. Four articlesCitation34–Citation36,Citation45 have both SCR and SPR, whereas five studiesCitation33,Citation37-Citation39,Citation44 only descripted SCR or SPR. Based on the results of the specific antibody responses, we analyzed the cross-reactivity of H7 vaccines at day 42/56, which was detected by HI and MN assays. Our results showed that antibodies elicited by H7N1, H7N3, H7N7, and H7N9 vaccines have cross-protection against other H7 influenza virus, with SCR of 0.66 (0.50, 0.82) and SPR of 0.79 (0.67, 0.91), meeting vaccine production license criteria CBER and CHMP. H7N1 vaccines displayed cross-reactivity to non-H7N1 viruses (H7N9) [SCR: 0.88 (0.85, 0.91), SPR: 0.89 (0.86, 0.92)]. However, H7N3 vaccines displayed low cross-reactivity to non-H7N3 viruses (H7N9 and H7N1) [SCR: 0.40 (0.26, 0.54), SPR: 0.50 (0.27, 0.73)]. This may due to the unadjuvanted H7N3 vaccines used in the subjects. High pooled SPR was found in the cross-reactivity of H7N7 vaccines against non-H7N7 viruses (H7N9 and H7N3) [0.93 (0.81, 1.06)] (, ).
Table 3. The cross-reactivity induced by H7 vaccines.
Cross-reactivity of H7 subtype vaccines against H7N9
H7N9 avian influenza virus is one of the H7 subtype viruses, which has caused severe human infections with high mortality in the past several years.Citation1,Citation49 Of the nine studies, five showed that H7N1, H7N3, and H7N7 elicited cross-reactive antibodies against H7N9, which were measured by HI. To investigate the cross-protection of other subtype H7 vaccines against H7N9 virus, data from five articles were analyzed, involving H7N1 (both inactivated and live attenuated), H7N7 (inactivated) and H7N3 (live attenuated) vaccines. The SCR and SPR of non-H7N9 (H7N1, H7N3, and H7N7) vaccines against H7N9 viruses were 0.69 (0.52, 0.86) and 0.85 (0.76, 0.94), respectively (, ), meeting CBER and CHMP criteria. Study conducted by KrammerCitation45 showed that three of six vaccinees who did not induce H7N3-specific antibodies displayed no cross-reactivity of the vaccine, whereas the other three did. These results indicate that H7 subtype influenza vaccines, H7N1, H7N3 and H7N7 exhibit cross-protection from H7N9 viruses.
Publication bias
Due to the limited literature, the funnel plots cannot be wisely evaluated for publication bias. The publication bias of specific antibody responses (RR of SCR at 24/28d vs 48/56d) was evaluated using Stata12.0 for Egger’s test. No publication bias was found (p = .841). The Fail-Safe Number was 1071 (>5k+10) calculated when assessing the publication bias of cross-reaction (SPR between H7, baseline as the control group), indicating no publication bias in all included articles.
Discussion
Inoculation with seasonal influenza vaccine is the major intervention currently used to prevent influenza infections. However, some studies reported that there was no cross-reaction between seasonal influenza vaccine and H7N9 influenza virus.Citation50–Citation53 It takes 4–6 months to manufacture a new vaccineCitation54-Citation57 and this manufacturing delay is not helpful for the prevention of infection when a novel influenza virus emerges in humans as significant infections will likely occur before a vaccine is made available. Generation of cross-reactivity takes advantage of the fact that the same HA subtype influenza viruses share similar epitopes.Citation7 Antibodies elicited by these strains can bind to other viruses having similar epitopes.Citation34-Citation36 Phylogenetic analysis has shown a high degree of homology in the HA gene sequence of various H7 viruses.Citation7 Stadlbauer et al.Citation44 showed that an increase in the mean geometric increase of the cross-reaction of H7N9 influenza vaccine with H7 subtypes was 14.2, while H1, H3, H4, H14, H10, and H15 viruses were 1.3, 1.9, 3.5, 2.9, 3.5 and 5.0, respectively. As such, it is likely that vaccination against one H7 subtype may be sufficient to elicit cross-reaction to several of H7 subtype viruses. However, a comprehensive analysis of H7 induced cross-reactivity is lacking.
In this study, we report the meta-analysis of cross-reactive antibodies induced by H7 subtype avian influenza vaccine for the first time. To investigate the best cross-reactivity between H7 subtype vaccines, the vaccine-specific antibody responses were assessed by random effect model. The highest SCR and SPR of protective antibodies were 74% and 81%, respectively, 3–4 weeks after the second inoculation, meeting the international vaccine licensing standards, while one does not. The antibody responses at day 42/56 were higher than those at day 21/28, 6-month and 12-month, indicating that two doses induced stronger antibody responses than one dose and day 42/56 is the best time for analysis of the cross-reaction. Interestingly, in the Madan 2017 study, the only study in this meta-analysis that included aged individual (>65), the RR was 22.8 and 23.45 for SCR and SPR, respectively, when comparing peak of the response (42/56d) to 12 months, which was much higher than those in younger adults, suggesting a big decline in antibody titers in older adults. Immune protection in older adults may last a shorter time than in younger adults. The cross-reactivity of H7 vaccines at day42/56 was further pooled to assess the cross-protection against other H7 viruses. The results showed that all H7 influenza virus vaccines, including H7N1, H7N3, H7N7 and H7N9, induced cross-reactive antibodies against other H7 subtype viruses (H7N9, H7N1, and H7N3). Both consolidated SCR (66%) and SPR (79%) of the antibodies met vaccine production license standards, CBER and CHMP. The H7N3 vaccines had low cross-reactivity because of insufficient vaccine-specific antibody responses elicited by the unadjuvanted vaccine. In addition, these H7N9 vaccines also exhibited cross-reaction to H7N2, H7N7, and H7N8, as described early in this systematic review. These results suggest that all H7 subtype vaccines that induced effective vaccine-specific antibody responses can prevent H7 influenza infection.
H7N9 is a novel H7 subtype influenza virus with high human mortality. Since 2013, China has experienced five epidemics of human infection with H7N9 viruses, a significant mortality rate of 39%.Citation1,Citation49,Citation58,Citation59 Although other H7 influenza viruses, such as H7N1, H7N2, and H7N3, have been reported to infect humans, the patients usually showed mild to moderate clinical symptoms.Citation9–Citation15 We further analyzed the cross-reactivity induced by H7 subtype influenza vaccines against H7N9 viruses. The results showed that the H7 vaccine, H7N1, H7N3, and H7N7, displayed cross-protection against H7N9, with pooled SCR of 69% and SPR of 85%. Moreover, H7N9 vaccines elicited protective antibodies against H7N1, H7N2, H7N3, H7N7, and H7N8. In general, H7 subtype influenza vaccines can produce cross-protection against other H7 subtype influenza virus (where enough specific antibodies can be produced).
Several limitations in our meta-analysis are worthy of mentioning. First, this paper only analyzed the cross-reaction between H7 subtype vaccines, or against H7N9, or H7N1 and H7N3 against H7N9, due to the limited literature. Future large-scale studies are required to carry out cross-reactivity analysis between specific subtypes. Second, heterogeneities were detected in the research. Among the nine articles included, Rudenko et al.Citation33 showed that the specific response and cross-reaction of H7N3 vaccine after two doses of vaccination were low, possibly due to the unadjuvanted vaccine. Previous studies have shown that the immune effect of H7 subtype vaccine after adding adjuvant is significantly higher than that without adjuvant,Citation60–Citation62 which may be the main reason for the heterogeneity of the study. Moreover, most studies had unclear bias risk, which might weaken the result’s reliability. Therefore, high-quality, large-scale clinical trials are needed for further validation.
In conclusion, the recent human infections with a new avian influenza A/H7N9 indicate an increased risk of pandemic of unpredictable reassortant viruses capable of transmitting from animal to humans. Our study confirmed the cross-reactivity between the H7 subtype vaccines, indicating that the H7 vaccines, regardless of their specific subtype, induce cross-protection against other subtypes of the H7 viruses. However, the composition of vaccine can influence antibody responses. The effect of vaccine types, such as adjuvanted vs. non-adjuvanted, inactivated whole virus vs. recombinant proteins or live-attenuated vaccines, etc., on the cross-reactivity of H7 subtype vaccines is needed further clarification by large trails.
Author contributions
XG and JH designed the study. XW, XG, and YS developed the search strategy. XW and XG searched the databases, screened the eligibility of the retrieved studies and extracted the information from the studies. XW, XG, and KZ performed the data analysis. XG and JH wrote the original draft, which was reviewed and edited by JH. All authors then approved the final written manuscript.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dr. Shannon P. Hilchey, University of Rochester Medical Center, Rochester, NY, USA, for the critical review of this paper.
Additional information
Funding
References
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–97. doi:10.1056/NEJMoa1304459.
- Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–25. doi:10.1016/S0140-6736(13)60903-4.
- Lu S, Xi X, Zheng Y, Cao Y, Liu X, Lu H. Analysis of the clinical characteristics and treatment of two patients with avian influenza virus (H7N9). Biosci Trends. 2013;7:109–12.
- Yu H, Cowling BJ, Feng L, Lau EH, Liao Q, Tsang TK, Peng Z, Wu P, Liu F, Fang VJ, et al. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet. 2013;382:138–45. doi:10.1016/S0140-6736(13)61207-6.
- Chen E, Chen Y, Fu L, Chen Z, Gong Z, Mao H, Wang D, M Y N, Wu P, Yu Z, et al. Human infection with avian influenza A (H7N9) virus re-emerges in China in winter 2013. Euro Surveill. 2013;18:20616. doi:10.2807/1560-7917.ES2013.18.43.20616.
- World Health Organization. Influenza: influenza at the human-animal interface. 2018. http://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_02_03_2018.pdf?ua=1.
- Shi JZ, Deng GH, Liu PH. Isolation and characterization of H7N9 viruses from live poultry markets—implication of the source of current H7N9 infection in humans. Chin Sci Bull. 2013;58:1857–63. doi:10.1007/s11434-013-5873-4.
- Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–32. doi:10.1016/S0140-6736(13)60938-1.
- DeLay PD, Casey HL, Tubiash HS. Comparative study of fowl plague virus and a virus isolated from man. Public Health Rep. 1967;82:615. doi:10.2307/4593084.
- Campbell CH, Webster RG, Breese SS. Fowl plague virus from man. J Infect Dis. 1970;122:513–16. doi:10.1093/infdis/122.6.513.
- Kurtz J, Manvell RJ, Banks J. Avian influenza virus isolated from a woman with conjunctivitis. Lancet. 1996;348:901–02. doi:10.1016/S0140-6736(05)64783-6.
- Banks J, Speidel E, Alexander DJ. Characterisation of an avian influenza A virus isolated from a human–is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch. Virol. 1998;143:781–87.
- Eurosurveillance Editorial Team. Avian influenza A/(H7N2) outbreak in the United Kingdom. Euro Surveill. 2007;12:E070531.
- Puzelli S, Trani L, Fabiani C, Campitelli L, De Marco MA, Capua I, Aguilera JF, Zambon M, Donatelli I. Serological analysis of serum samples from humans exposed to avian H7 influenza viruses in Italy between 1999 and 2003. J Infect Dis. 2005;192:1318–22. doi:10.1086/444390.
- Nguyen-Van-Tam JS, Nair P, Acheson P, Baker A, Barker M, Bracebridge S, Croft J, Ellis J, Gelletlie R, Gent N, et al. Outbreak of low pathogenicity H7N3 avian influenza in UK, including associated case of human conjunctivitis. Euro Surveill. 2006;11:E060504. 2.
- Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–61. doi:10.1073/pnas.0308352100.
- Andrews SF, Joyce MG, Chambers MJ, Gillespie RA, Kanekiyo M, Leung K, Yang ES, Tsybovsky Y, Wheatley AK, Crank MC, et al. Preferential induction of cross-group influenza A hemagglutinin stem–specific memory B cells after H7N9 immunization in humans. Sci Immunol. 2017;2:2676. doi:10.1126/sciimmunol.aan2676.
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109:4269–74. doi:10.1073/pnas.1116200109.
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi:10.1371/journal.ppat.1003657.
- Jang YH, Seong BL. Options and obstacles for designing a universal influenza vaccine. Viruses. 2014;6:3159–80. doi:10.3390/v6083159.
- Klenk HD, Rott R, Orlich M, Blodorn J. Activation of influenza A viruses by trypsin treatment. Virology. 1975;68:426–39. doi:10.1016/0042-6822(75)90284-6.
- Knossow M, Gaudier M, Douglas A, Barrere B, Bizebard T, Barbey C, Gigant B, Skehel JJ. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology. 2002;302:294–98. doi:10.1006/viro.2002.1625.
- Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A. 2013;110:264–69. doi:10.1073/pnas.1218256109.
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. doi:10.1146/annurev.biochem.69.1.531.
- Han T, Marasco WA. Structural basis of influenza virus neutralization. Ann N Y Acad Sci. 2011;1217:178–90. doi:10.1111/j.1749-6632.2010.05829.x.
- Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–60. doi:10.1126/science.1186430.
- Laver WG, Air GM, Webster RG, Gerhard W, Ward CW, Dopheide TA. Antigenic drift in type A influenza virus: sequence differences in the hemagglutinin of Hong Kong (H3N2) variants selected with monoclonal hybridoma antibodies. Virology. 1979;98:226–37. doi:10.1016/0042-6822(79)90540-3.
- Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373. doi:10.1038/289806a0.
- Haaheim LR, Madhun AS, Cox R. Pandemic influenza vaccines–the challenges. Viruses. 2009;1:1089–109. doi:10.3390/v1031089.
- Luke CJ, Subbarao K. Vaccines for pandemic influenza. Emerg Infect Dis. 2006;12:66.
- Rolfes MA, Flannery B, Chung J, O’Halloran A, Garg S, Belongia EA, Gaglani M, Zimmerman R, Jackson ML, Monto AS, et al. Effects of influenza vaccination in the United States during the 2017–2018 influenza season. Clin Infect Dis. 2019. doi:10.1093/cid/ciz075.
- Havers FP, Chung JR, Belongia EA, McLean HQ, Gaglani M, Murthy K, Zimmerman RK, Nowalk MP, Jackson ML, Jackson LA, et al. Influenza vaccine effectiveness and statin use among adults in the United States, 2011-2017. Clin Infect Dis. 2018;68:1616–22. dio:10.1093/cid/ciy780.
- Rudenko L, Kiseleva I, Naykhin AN, Erofeeva M, Stukova M, Donina S, Petukhova G, Pisareva M, Krivitskaya V, Grudinin M, et al. Assessment of human immune responses to H7 avian influenza virus of pandemic potential: results from a placebo-controlled, randomized double–blind phase I study of live attenuated H7N3 influenza vaccine. PLoS One. 2014;9:e87962. doi:10.1371/journal.pone.0087962.
- Madan A, Segall N, Ferguson M, Frenette L, Kroll R, Friel D, Soni J, Li P, Innis BL, Schuind A. Immunogenicity and safety of anAS03-adjuvanted H7N9 pandemic influenza vaccine in a randomized trial in healthy adults. J Infect Dis. 2016;214:1717–27. doi:10.1093/infdis/jiw414.
- Madan A, Ferguson M, Sheldon E, Segall N, Chu L, Toma A, Rheault P, Friel D, Soni J, Li P. Immunogenicity and safety of an AS03- ajuvanted H7N1 vaccine in healthy adults: A phase I/II, observer-blind, randomized,controlled trial. Vaccine. 2017;35:1431–39. doi:10.1016/j.vaccine.2017.01.054.
- Madan A, Ferguson M, Rheault P, Seiden D, Toma A, Friel D, Soni J, Li P, B L I, Schuind A. Immunogenicity and safety of an AS03-ajuvanted H7N1 vaccine in adults 65 years of age and older: A phase II, observer-blind,randomized, controlled rial. Vaccine. 2017;35:1865–72. doi:10.1016/j.vaccine.2017.02.057.
- Sobhanie M, Matsuoka Y, Jegaskanda S, Fitzgerald T, Mallory R, Chen Z, Luke C, Treanor J, Subbarao K. Evaluation of the safety and immunogenicity of a candidate pandemic live attenuated influenza vaccine (pLAIV) against influenza A (H7N9). J Infect Dis. 2015;213:922–29. doi:10.1093/infdis/jiv526.
- Babu TM, Levine M, Fitzgerald T, Luke C, Sangster MY, Jin H, Topham D, Katz J, Treanor J, Subbarao K. Live attenuated H7N7 influenza vaccine primes for a vigorous antibody response to inactivated H7N7 influenza vaccine. Vaccine. 2014;32:6798–804. doi:10.1016/j.vaccine.2014.09.070.
- Krammer F, Jul-Larsen A, Margine I, Hirsh A, Sjursen H, Zambon M, Cox RJ. An H7N1 influenza virus vaccine induces broadly reactive antibody responses against H7N9 in humans. Clin Vaccine Immunol. 2014;21:1153–63. doi:10.1128/CVI.00272-14.
- Wang L, Xin FL, Lin NP, Wang YC, Liu XL, Liu JF. Metallothioneins may be a potential prognostic biomarker for tumors: A Prisma-compliant meta-analysis. Medicine (Baltimore). 2018;97:e13786. doi:10.1097/MD.0000000000013786.
- Higgins J, Green S. Cochrane handbook for systematic reviews of interventions Version 5.1. 0. The Cochrane Collaboration. Confidence intervals; 2011.
- Review Manager (RevMan) [Computer program]. 5.3 ed. Copenhagen: The Nordic Cochrane Centre: The Cochrane Collaboration, Oxford, UK; 2014.
- Sterne JA, Egger M, Smith GD. Systematic reviews in health care: investigating and dealing with publication and other biases in meta-analysis. BMJ. 2001;323:101. doi:10.1136/bmj.323.7304.101.
- Stadlbauer D, Rajabhathor A, Amanat F, Kaplan D, Masud A, Treanor JJ, Izikson R, Cox MM, Nachbagauer R, Krammer F. Vaccination with a recombinant H7 hemagglutinin-based influenza virus vaccine induces broadly reactive antibodies in humans. mSphere. 2017;2:e00502–17. doi:10.1128/mSphere.00502-17.
- Krammer F, Albrecht RA, Tan GS, Margine I, Hai R, Schmolke M, Runstadler J, Andrews SF, Wilson PC, Cox RJ, et al. Divergent H7 immunogens offer protection from H7N9 challenge. J Virol. 2014;21:1153–63.
- Guideline on influenza vaccines prepared from viruses with the potential to cause a pandemic and intended for use outside of the core dossier context. Committee for Guidance for Human Medicinal Products (CHMP). EMEA/CHMP/VWP/263499/2006; 2007. p. 2.
- Guideline on dossier structure and content for pandemic influenza vaccine marketing authorisation application (revision). Committee for guidance for human medicinal products (CHMP). EMEA/CPMP/VEG/4717/2003- Rev. 1; 2008.
- Guidance for industry: clinical data needed to support the licensure of pandemic influenza vaccines. Center for Biologics Evaluation and Research (CBER); 2007. https://www.fda.gov/media/73691/download.
- Su W, Cheng KL, Chu DKW, Zhou J, Mao X, Zhong Z, Song Y, Peiris M, Wu J, Yen HL. Genetic analysis of H7N9 highly pathogenic avian influenza virus in Guangdong, China, 2016–2017. J Infect. 2018;76:93–96. doi:10.1016/j.jinf.2017.09.001.
- Guo W, Xu J, Wu J, Zhao S, He H, Shi W, Yu D, Li J, Gao H, Chen J. Safety and immunogenicity of seasonal inactivated influenza vaccine (split virion) and cross-reactive antibody responses to the H7N9 avian. Zhonghua Liu Xing Bing Xue Za Zhi. 2014;35:949–52.
- Shen Y, Hu Y, Meng F, Du W, Li W, Song Y, Ji X, Huo L, Fu Z, Yin W. Safety, immunogenicity and cross-reactivity of a Northern hemisphere 2013–2014 seasonal trivalent inactivated split influenza virus vaccine, Anflu®. Hum Vaccin Immunother. 2016;12:1229–34. doi:10.1080/21645515.2015.1123357.
- Wang W, Alvarado FE, Chen Q, Anderson CM, Scott D, Vassell R, Weiss CD. Serum samples from middle-aged adults vaccinated annually with seasonal influenza vaccines cross-neutralize some potential pandemic influenza viruses. J Infect Dis. 2015;213:403–06. doi:10.1093/infdis/jiv407.
- Chu DH, Sakoda Y, Nishi T, Hiono T, Shichinohe S, Okamatsu M, Kida H. Potency of an inactivated influenza vaccine prepared from A/duck/Mongolia/119/2008 (H7N9) against the challenge with A/Anhui/1/2013 (H7N9). Vaccine. 2014;32:3473–79. doi:10.1016/j.vaccine.2014.04.060.
- Khurana S, Verma S, Verma N, Crevar CJ, Carter DM, Manischewitz J, King LR, Ross TM, Golding H. Bacterial HA1 vaccine against pandemic H5N1 influenza virus: evidence of oligomerization, hemagglutination, and cross-protective immunity in ferrets. J Virol. 2011;85:1246–56. doi:10.1128/JVI.02107-10.
- Jennings LC, Monto AS, Chan PK, Szucs TD, Nicholson KG. Stockpiling prepandemic influenza vaccines: a new cornerstone of pandemic preparedness plans. Lancet Infect Dis. 2008;8:650–58. doi:10.1016/S1473-3099(08)70232-9.
- Sambhara S, Poland GA. H5N1 Avian influenza: preventive and therapeutic strategies against a pandemic. Annu Rev Med. 2010;61:187–98. doi:10.1146/annurev.med.050908.132031.
- Leroux-Roels I, Borkowski A, Vanwolleghem T, Drame M, Clement F, Hons E, Devaster JM, Leroux-Roels G. Antigen sparing and cross-reactive immunity with an adjuvanted rH5N1 prototype pandemic influenza vaccine: a randomised controlled trial. Lancet. 2007;370:580–89. doi:10.1016/S0140-6736(07)61297-5.
- Su S, Gu M, Liu D, Cui J, Gao GF, Zhou J, Liu X. Epidemiology, evolution, and pathogenesis of H7N9 influenza viruses in five epidemic waves since 2013 in China. Trends Microbiol. 2017;25:713–28. doi:10.1016/j.tim.2017.06.008.
- Shen Y, Lu H. Global concern regarding the fifth epidemic of human infection with avian influenza A (H7N9) virus in China. Biosci Trends. 2017;11:120–21. doi:10.5582/bst.2017.01040.
- Cox RJ, Madhun AS, Hauge S, Sjursen H, Major D, Kuhne M, Hoschler K, Saville M, Vogel FR, Barclay W, et al. A phase I clinical trial of a PER. C6® cell grown influenza H7 virus vaccine. Vaccine. 2009;27:1889–97. doi:10.1016/j.vaccine.2009.01.116.
- Couch RB, Patel SM, Wade-Bowers CL, Nino DA. randomized clinical trial of an inactivated avian influenza A (H7N7) vaccine. PLoS One. 2012;7:e49704. doi:10.1371/journal.pone.0049704.
- Fries LF, Smith GE, Glenn GM. A recombinant virus-like particle influenza A (H7N9) vaccine. N Engl J Med. 2013;369:2564–66. doi:10.1056/NEJMc1313186.