ABSTRACT
Swine H1 influenza viruses were stable within pigs for nearly 70 years until in 1998 when a classical swine virus reassorted with avian and human influenza viruses to generate the novel triple reassortant H1N1 strain that eventually led to the 2009 influenza pandemic. Previously, our group demonstrated broad protection against a panel of human H1N1 viruses using HA antigens derived by the COBRA methodology. In this report, the effectiveness of COBRA HA antigens (SW1, SW2, SW3 and SW4), which were designed using only HA sequences from swine H1N1 and H1N2 isolates, were tested in BALB/c mice. The effectiveness of these vaccines were compared to HA sequences designed using both human and swine H1 HA sequences or human only sequences. SW2 and SW4 elicited antibodies that detected the pandemic-like virus, A/California/07/2009 (CA/09), had antibodies with HAI activity against almost all the classical swine influenza viruses isolated from 1973–2015 and all of the Eurasian viruses in our panel. However, sera collected from mice vaccinated with SW2 or SW4 had HAI activity against ~25% of the human seasonal-like influenza viruses isolated from 2009–2015. In contrast, the P1 COBRA HA vaccine (derived from both swine and human HA sequences) elicited antibodies that had HAI activity against both swine and human H1 viruses and protected against CA/09 challenge, but not a human seasonal-like swine H1N2 virus challenge. However, the SW1 vaccine protected against this challenge as well as the homologous vaccine. These results support the idea that a pan-swine-human H1 influenza virus vaccine is possible.
Introduction
The domestic pig (Sus scrofa domesticus), also referred to as swine or hog, can be infected by both avian and human influenza viruses through the expression of α2,3- and α2,6- sialic acid receptors in the respiratory tract. Annually, ~25% of the swine population in the United States is infected with a swine influenza virus.Citation1 Infected pigs show influenza-like symptoms including weight loss, fever, respiratory distress, coughing, and nasal discharge, but little mortality. Currently swine influenza viruses of the subtypes H1N1, H3N2, and H1N2 are circulating in North America.Citation2 In Asia, North America, and much of Europe, H1N1 swine influenza viruses are significantly prevalent in the populations and remain an important infection with zoonotic potential.Citation3,Citation4
The H1N1 influenza subtype was originally isolated from pigs in 1932, and these swine H1N1 influenza viruses remained genetically and antigenically stable in North American pigs until a series of reassortment events in 1998. The triple reassortant H3N2 virus originated from a classical swine lineage virus (Np, M, NS) reassorting with a human seasonal H3N2 virus (PB1, HA, NA) with an avian virus (PB2, PA). This H3N2 virus then went on to reassort with co-circulating classical lineage H1N1 swine viruses (HA and NA or HA). This was marked by the introduction of human and avian influenza virus gene segments with swine influenza virus genes and resulted in generation of novel strains with pandemic potential.Citation4 Based on phylogenetic analysis and ancestry studies, the swine H1 viruses belong to four distinctive clades: alpha, beta, gamma and delta.Citation5-Citation7 Alpha viruses are considered classical isolates and are related to viruses that circulated from 1930s to 1998 (cH1N1).Citation5,Citation8,Citation9 Viruses classified in the Beta clade developed due to reassortment events between cH1N1 isolates and H3N2 viruses in pigs that led to H1N1 viruses expressing cH1N1 HA and NA surface proteins with H3N2 internal genes. Gamma clade swine influenza viruses are referred to as H1N2-like isolates and are the result of a triple reassortant event between H3N2 and cH1N1 viruses. Swine influenza viruses of the Gamma clade include both H1N1 and H1N2 viruses.Citation5 Delta clade swine influenza viruses are highly divergent, containing both H1N1 and H1N2 swine influenza viruses, and contain H1 human seasonal influenza virus genes.Citation10 In 2009, the first pandemic of the 21st century occurred with the introduction of a swine-origin influenza virus of the cH1N1 subtype into the human population that transmitted easily between people.Citation11
Ultimately, the expansive divergence of the swine hemagglutinin makes all the clades antigenically distinct; a wild-type H1 hemagglutinin vaccination would leave the host open to infection from a mismatched virus, of which, there are plenty. Therefore, to address the need for more broadly reactive influenza vaccines, our group has previously reported on the methodology of antigen design, termed computationally optimized broadly reactive antigen (COBRA), using multiple rounds of layered consensus building to generate influenza vaccine HA immunogens. COBRA HA vaccines elicit antibodies that target both the globular head and stem regions of HA. Recently, we reported the characterization of a COBRA-based vaccine for highly pathogenic H5N1Citation12-Citation16 and both human seasonal and pandemic H1N1 influenza virus isolates.Citation17 For H1N1, these COBRA HA candidates were designed to elicit antibodies with HAI activity against H1N1 viruses isolated within the last 30 years. In addition, several COBRA HA candidate designs were based on sequences of H1N1 viruses spanning the past 100 years, including modern pandemic H1N1 isolates. Three of the nine H1N1 COBRA HA proteins (X3, X6, and P1) had intense broad hemagglutination inhibition (HAI) activity against a panel of fifteen H1N1 viruses. These vaccines were derived using HA sequences from human isolates, except for P1, which was based on a combination of both human and H1N1 swine isolates. Mice vaccinated with a virus-like particle (VLP) expressing the P1 COBRA HA antigen had little or no detectable viral replication following challenge with the A/California/07/2009 (CA/09) pandemic-like H1N1 virus, which was comparable to a homologous matched CA/09 vaccine.
In this study, a set of H1 COBRA HA vaccines designed using only swine H1N1 and H1N2 viruses (SW1, SW2, SW3, SW4) were assessed for their ability to elicit protective antibodies with HAI activity against both human and swine H1 influenza viruses. Those results were then compared to the P1 swine-human COBRA HA, as well as, the human COBRA HA vaccines.Citation17 While swine COBRA HA vaccines elicited antibodies with HAI activity against a diverse panel of swine H1 viruses, these same elicited antibodies did not recognize human seasonal H1N1 viruses. The SW2 and SW4 vaccines elicited antibodies with HAI activity against classical swine H1 viruses, and the SW1 vaccine elicited protective immune responses against human seasonal-like H1 swine influenza viruses. Only the P1 COBRA vaccine was able to elicit antibodies that effectively recognized historical and contemporary human seasonal H1N1 viruses, in addition to, different lineages of H1N1 and H1N2 swine viruses.
Materials and methods
COBRA HA antigen construction and synthesis
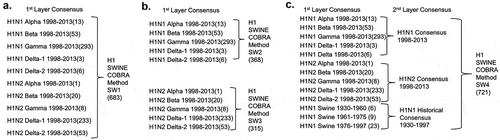
Nucleotide sequences (N = 721) for swine H1N1 and H1N2 influenza A hemagglutinin (HA) proteins, isolated from 1930–2013, were downloaded from the NCBI Influenza Virus Resource database.Citation13 The nucleotide sequences were translated into protein sequences using the standard genetic code. Full-length swine H1N1 and H1N2 sequences from viral infections covering the period from 1930 to 2013 were analyzed. Specifically, the SW1 COBRA was designed to include both H1N1 and H1N2 sequences from 1998 to 2013. SW2 incorporated only H1N1 sequences, and SW3 included sequences that included only H1N2 sequences from 1998 to 2013. SW4 included historical sequences from 1930 to 1997, in addition to, the 1998 to 2013 period (). The HA0 amino acid sequences were aligned, and the most common amino acid at each position was determined resulting in primary consensus sequences representing each genotypic group. The resulting primary sequences from each group were then realigned to generate a secondary consensus. This process was continued until a single final consensus was obtained. Finally, the ultimate amino acid sequence was reverse translated and optimized for expression in mammalian cells, including codon usage and RNA optimization (Genewiz, Washington, DC, USA). The resulting sequence was termed a computationally optimized broadly reactive antigen (COBRA). H1 HA genes were synthesized and inserted into the pTR600 expression vector, as previously described.Citation18
Viruses and HA antigens
H1 viruses were obtained through the Influenza Reagents Resource (IRR), BEI Resources, the Centers for Disease Control (CDC), or provided by Virapure, LLC (San Diego, CA, USA). Viruses were passaged once in the same growth conditions as they were received or as per the instructions provided by the World Health Organization, in either embryonated chicken eggs or Madin-Darby canine kidney (MDCK) cell culture.Citation19 Virus lots were aliquoted for single-use applications and stored at −80°C. Titer of the frozen aliquots was determined using turkey red blood cells (RBC).
The classification of the swine HA was determined using the Swine H1 Clade Classification Tool (http://www.fludb.org),Citation9 which analyzes the HA nucleotide sequence and infers both the globalCitation9 and USCitation20,Citation21 swine H1 clade classification. The protein accession numbers for the HA amino acid sequences used in this study are provided. If no accession number was available, the amino acid sequences are provided (Supplementary Table 1). wild-type H1N1 strains that matched the challenge strains included A/California/07/2009 (CA/09; YP_009118626.1) and A/Swine/North Carolina/152702/2015 (SW/NC/152702/15). Additional H1N1 swine influenza viruses used in this study included: A/Swine/Wisconsin/125/1997 (SW/WI/97; AAF87274.1), A/Swine/Indiana/P12439/2000 (SW/IN/00; AAL87870.1), A/Swine/Spain/50047/2003 (SW/Spain/03; ABD78104.1), A/Swine/Korea/Asan04/2006 (SW/Korea/06; ACE77933.1), A/Swine/Zhejiang/1/2007 (SW/Zhejiang/07; ACJ06667.1), A/Swine/North Carolina/02744/2009 (SW/NC/02744/09), A/Swine/North Carolina/34543/2009 (SW/NC/34543/09; AEX25796.1), and A/Swine/Minnesota/A01489606/2015 (SW/MN/15; AKD00877.1), A/Swine/Iowa/1973 (SW/IA/73; ABV25637.1), A/Swine/North Carolina/93523/2001 (SW/NC/01; AAL87867.1), A/Swine/Ohio/511445/2007 (SW/OH/07; ACH69547.1), A/Swine/Colorado/SG1322/2009 (SW/CO/09; AHB21556.1), A/Swine/North Carolina/5043–1/2009 (SW/NC/5043–1/09; ADV69084.1), A/Swine/Missouri/A01203163/2012 (SW/MO/12; AFM47013.1), A/Swine/Nebraska/A10444614/2013 (SW/NE/13; AGF68975.1) and A/Swine/North Carolina/A01377454/2014 (SW/NC/14; AJM70701.1).
Lastly, some human H1N1 strains were used including A/Chile/1/1983 (Chile/83; AFM72054.1), A/Singapore/6/1986 (Sing/86; ABO38395.1), A/New Caledonia/20/1999 (New Cal/99; AGQ47728.1), A/Texas/36/1991 (TX/91; ABF21276.1), A/Beijing/262/1995 (Beijing/95; ACF41867.1), and A/Solomon Islands/3/2006 (SI/06; ABU99109.1), and A/Brisbane/59/2007 (Bris/07; ACA28846.1). This 25-member panel of viruses used in the HAI assay represented human viruses from 1980 to the introduction of CA/09 and swine viruses from different phylogenetic lineages and clades. Wild-type HA vaccine antigens were codon optimized for expression in mammalian cells. The additional eight swine and three human strains were included in the HAI panel to expand the breadth of the human and swine phylogenetic coverage from 1930 to 2015.
Phylogenetic comparison of swine, human, and COBRA HA sequences
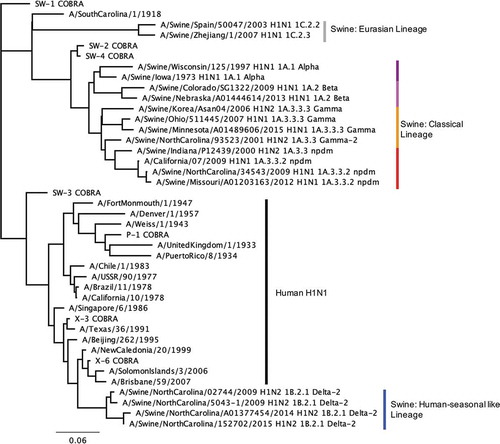
Viruses used for the vaccination panel and HAI panel were visualized on a phylogenetic tree to confirm full coverage of swine and human influenza clades (). Briefly, the HA0 was aligned utilizing Geneious alignment with global alignment with free end gaps and a cost matrix Blosum62 with open gap penalty 12, and gap extension penalty 3, with refinement iterations of 2 (Geneious v11.1.5). The HA1 portions (1-327AA) were extracted from the alignment. Using only sequences with a full HA1 amino acid sequence, Geneious Tree Builder, which observed the same alignment characteristics, was used to obtain a neighbor-joining Jukes-Cantor phylogenic tree with no indicated outgroup. The scale bar represents 0.06 amino acid substitutions per site, of a total of 327 amino acid sites.Citation22
Figure 2. The unrooted human and swine H1 HA phylogenetic tree was inferred from database HA amino acid sequences with the inclusion of COBRA HA sequences. Neighbor-joining tree built on the alignment of the amino acid sequences of the HA1 (1–327) region. The swine isolates include the influenza subtype, H1 swine global classification, and swine North American classification. Scale bar length represents the number of amino acid substitutions per amino acid site.
Predicted HA structures by Pymol
The known crystal structure were used for A/California/07/2009 (PDB accession: 3lzgCitation23) and A/Swine/Indiana/P12439/2000 (PDB accession: 4f3zCitation24). For all other HA vaccines the predicted protein structural models were constructed using SWISS-MODEL workspace (https://swiss-model.expasy.org).Citation25,Citation26 The template for the target sequence was identified through parallel searches of the SWISS-MODEL template library (SMTL) using BLASTCitation27 and HHBlits.Citation28 For all the target HA sequences two templates were chosen for model building.Citation29 The crystal structures of A/Jiangsu/ALSI/2011 (H1N1) HA (PDB accession: 6d8w) was selected for the Eurasian strains, and A/Netherlands/002P1/1951 (H1N1) HA (PDB accession: 6n41) was selected for all remaining HAs. The template’s quality was predicted from features of the target-template alignment. The templates with the highest quality were then selected for model building. Models were built based on the target-template alignment using ProMod3. Conserved coordinates between the target and the template were copied from the template to the model. Insertions and deletions were remodeled using a fragment library. Side chains were then rebuilt. Finally, by using a force field the geometry of the resulting model was regularized. Incase loop modeling with ProMod3 failed, an alternative model was built with PROMOD-II.Citation30 The global model quality estimation (GMQE) determines the expected accuracy of the model based upon the alignment of the target to the template as well as the coverage of the target. The global and per-residue model quality was assessed using the QMEAN scoring function based upon geometrical properties.Citation31 QMEAN Z-scores close to zero indicate high quality, whereas less than −4.0 indicate low quality models.Citation30,Citation32 The quaternary structure prediction score (QSQE) indicates the quality of fit for the quaternary structure of the protein given that HA is naturally isolated as a homotrimer. Putative glycosylation sites were predicted using the online tool NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/).Citation33 The antigenic regions were defined as: Sa (124–125, 153–157, 159–164); Sb (184–195); Ca1 (166–170, 203–205, 235–237); Ca2 (137–142, 221–222); and Cb (70–75).
Calculation of P-epitope values
Due to the inclusion of the pandemic HAs, the A/California/04/2009 numbering scheme was used.Citation34 The amino acid numbering begins following the seventeen amino acids in the signal peptide.Citation35-Citation37 The epitope sites include the amino acids predicted to be important to vaccine efficacy: neutralizing-antibody binding residues, structure/sequence homologues of known H3 epitopes, and protein surface residues with high information entropy (Supplementary Table 2).Citation34 The p-epitope provides a quantitative value for the antigenic distance between two hemagglutinin amino acid sequences based upon the comparison of the five epitopes. For any two given sequences, the p-value was calculated for each epitope region to be the proportion of differing amino acids. The largest p-value of the five was then considered the p-dominant epitope, and the corresponding epitope was named the dominant epitope region (). A p-epitope of less than 0.442 has been correlated with a vaccine effectiveness greater than zero in humans.Citation38 Therefore, this cutoff was observed for the study.
Table 1. Computationally optimized broadly reactive antigens used in the study.
Vaccine preparation
Human embryonic kidney 293T (HEK-293T) cells (1 x 106) were transiently transfected with 1 μg DNA of each of the three pTR600 mammalian expression vectorsCitation39 expressing the influenza neuraminidase (A/Mallard/Alberta/24/2001; H7N3), the HIV p55 Gag sequence, and one of the various H1 wild-type or COBRA HAs. Following 72 h of incubation at 37°C, supernatants from transiently transfected cells were collected, centrifuged to remove cellular debris, and filtered through a 0.22 μm pore membrane. Mammalian virus-like particles (VLPs) were purified and sedimented by ultracentrifugation on a 20% glycerol cushion at 135,000 x g for 4 h at 4°C. VLPs were resuspended in phosphate buffered saline (PBS), and total protein concentration was determined with the Micro BCA Protein Assay Reagent kit (Pierce Biotechnology, Rockford, IL, USA). Hemagglutination activity of each preparation of VLP was determined by serially diluting volumes of VLPs and adding equal volume 0.8% turkey red blood cells (RBCs) (Lampire Biologicals, Pipersville, PA, USA) suspended in PBS to a V-bottom 96-well plate with a 30 min incubation at room temperature (RT). Prepared RBCs were stored at 4°C and used within 72 h. The highest dilution of VLP with full agglutination of RBCs was considered the endpoint HA titer.
Virus-like particles (VLP) vaccines were generated with one of several wild-type or COBRA HA antigens. Swine COBRA H1 HA antigens developed in this study were designated SW-1 COBRA (SW1), SW-2 COBRA (SW2), SW-3 COBRA (SW3), SW-4 COBRA (SW4). In addition, previously characterized COBRA H1 antigens, P-1 COBRA (P1), X-3 COBRA (X3), X-6 COBRA (X6) were used as comparator controls.
Determination of HA content
Purified VLPs were mixed with 6X reducing Laemmli’s SDS-sample buffer and incubated at 100°C for 5 min (Boston BioProducts, Ashland, MA, USA). Reduced VLPs were then electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAG) and transferred to a polyvinylidene difluoride (PVDF) membrane. The blot was probed 1:1000 with commercially sourced mouse anti-human HA clade 1 (15B7; Immune Technology Corporation, Lexington Ave, NY, USA). HA-antibody complexes were then detected using 1:4000 goat anti-mouse IgG labeled with horseradish peroxidase (HRP) (Southern Biotech, Birmingham, AL, USA). HRP activity was detected using ClarityTM Western ECL substrate (Bio-Rad Laboratories, Hercules, CA, USA) and digitally imaged using a cooled charged-coupled device camera (myECL Imager, ThermoFisher Scientific, Waltham, MA, USA). Linear regression standard curve analysis was performed using the known concentrations of the standard recombinant antigen (H1N1 A/California/07/2009) to estimate HA content in VLP lots. Quantifications were performed in duplicate.
Mouse vaccination and challenge studies
BALB/c mice (Mus musculus, females, 6 to 8 weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME, USA) and housed in microisolator units and allowed free access to food and water. Mice (9 and 11 mice per group for CA/09 and SW/NC/152702/15, respectively) were vaccinated with a 1:1 mixture of purified VLP (1.0 μg HA/mouse) and AFO3, an emulsified MF59-like squalene-based oil-in-water adjuvant (Sanofi Pasteur, Lyon, France). Mice were vaccinated via intramuscular injection at week 0 and boosted with the same vaccine formulation at the same dose at weeks 4 and 8. PBS mixed 1:1 with adjuvant served as a mock vaccination. Blood samples were collected from mice via cheek bleeds twenty-eight days after each vaccination in 1.5 ml microcentrifuge tubes. The samples were incubated at RT for 30 min and then centrifuged at 5000 rpm for 5 min. Serum samples were removed and stored at −20°C.
Four weeks after final vaccination, mice were challenged intranasally with 5 × 104 plaque forming units (PFU) of A/California/07/2009 (H1N1) or 1 × 107 PFU of A/Swine/North Carolina/152702/2015 (H1N2) in a volume of 50 μl. Challenge PFUs were calculated to be ten times the LD50 for each challenge virus. For fourteen days after challenge, mice were monitored, at minimum, daily for weight loss, disease signs, and death for 14 days after infection. Three days post-infection, lungs (n = 3) were harvested, transferred to dry ice, and stored at −80°C until enumeration of viral lung titers. Individual body weights were recorded daily post-infection for each group. Any animal exceeding 20% weight loss was humanely euthanized. Surviving mice were confirmed for successful infection indicative by seroconversion. All procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals, Animal Welfare Act, and Biosafety in Microbiological and Biomedical Laboratories.
Hemagglutination inhibition (HAI) assay
The hemagglutination inhibition (HAI) assay assessed functional antibodies to the HA able to inhibit agglutination of turkey erythrocytes. The protocols were adapted from the WHO laboratory influenza surveillance manual.Citation19 To inactivate nonspecific inhibitors, sera were treated with receptor-destroying enzyme (RDE) (Denka Seiken, Co., Japan) prior to being tested. Briefly, three parts RDE was added to one part sera and incubated overnight at 37°C. RDE was inactivated by incubation at 56°C for 30 min. After heat treatment, six parts PBS were added to the RDE-treated sera. RDE-treated sera were two-fold serially diluted in V-bottom microtiter plates. An equal volume of each virus (or VLP where applicable), adjusted to approximately 8 hemagglutination units (HAU)/50 μl, was added to each well. The plates were covered and incubated at RT for 20 min, and then 50 μl RBCs were added to each well. The plates were mixed by agitation and covered, and the RBCs were allowed to settle for 30 min at RT. The HAI titer was determined by the reciprocal dilution of the last well that contained non-agglutinated RBCs. Positive and negative serum controls were included for each plate. All mice were negative (HAI < 1:10) for preexisting antibodies to currently circulating human and swine influenza viruses prior to vaccination and seroprotection was defined as HAI titer ≥1:40 and seroconversion as a 4-fold increase in titer compared to baseline, as per the WHO and European Committee for Medicinal Products to evaluate influenza vaccines;Citation40 however, we often examined a more stringent threshold of ≥1:80. Mice were naïve and seronegative at the time of vaccination, thus seroconversion and seroprotection proportions were interchangeable in this study.
Determination of viral lung titers
Whole lung samples were thawed on ice. CA/09 lung samples were weighed and per 0.1 g a volume of 1 ml of Dulbecco modified Eagle medium (DMEM) supplemented with penicillin-streptomycin (P/S) was added. For SW/NC/152702/15 lung samples, whole lungs were suspended in 1 ml of DMEM + P/S. The tissue was macerated through a 0.70 μm nylon filter (Corning Cell Strainer, Sigma Aldrich, St. Louis, MO, USA) until thoroughly homogenized. Lung filtrate was then enumerated for viral lung titers. Ten-fold serial dilutions of lung homogenate were overlaid onto MDCK cells seeded at 5 × 105 cells per well of a six-well plate. Samples were incubated for 1 h at RT with intermittent shaking every 15 min. Medium was removed, and the cells were washed twice with DMEM + P/S. Wash medium was replaced with 2 ml of L15 medium with 2 µg/ml TPCK-trypsin plus 0.8% agarose (Cambrex, East Rutherford, NJ, USA) and incubated at 37°C with 5% CO2 for 72 h and 48 h for CA/09 and SW/NC/152702/15, respectively. Agarose was removed and discarded. MDCK cells were fixed with 10% buffered formalin for 10 min and stained with 1% crystal violet for 15 min. The plates were thoroughly washed in distilled water to remove excess crystal violet, and the plaques were counted and recorded to determine the PFU per ml lung homogenate.
ELISA for elicited antibody binding
A high-affinity, 96-well flat-bottom enzyme-linked immunosorbent assay (ELISA) plate was coated with 50 ul of 2 ug/ml of recombinant hemagglutinin of either CA/09 or SW/NC/152702/15 in ELISA carbonate buffer (50mM carbonate buffer [pH 9.5] with 5 ug/ml BS), and the plate was incubated overnight at 4°C. The next morning, the plates were washed in PBS, and nonspecific epitopes were blocked with 1% bovine serum albumin (BSA) in PBS with 0.05% Tween 20 (PBST+BSA) solution for 1 h at RT or overnight at 4°C. Buffer was removed and three-fold serial dilutions of RDE treated sera were added to the plate with a highest initial dilution of 1:50. The initial dilution of SW/NC/02744/2009 was 1:110 due to sera limitations. Plates were incubated at 37°C for 90 min. The plates were washed in PBS, and goat anti-mouse IgG-HRP was added 1:4000 in PBST+BSA. Plates were incubated at 37°C for 1 hr. After washing, 2,2ʹ-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) substrate in McIlvain’s Buffer (pH 5) was added to each well, and incubated at 37°C for 15 min. The colorimetric reaction was stopped with the addition of 1% SDS in ddH2O, and the absorbance was measured at 414 nm.
Statistical analysis
Statistical significance was defined as a p-value less than 0.05. The mean of the viral lung titers and day 6 weights were analyzed by an ordinary one-way ANOVA followed by Tukey’s multiple comparisons test, with a single pooled variance. The viral lung titers were transformed by log10 and the mean calculated. The lowest limit of detection for viral lung titers was 10 PFU/ml lung homogenate, and was used for the statistical analysis of samples below that. The day 6 weights were determined by dividing the measured weight on day 6 by the pre-challenge weight on day 0, multiplied by 100. The standard deviations for viral lung titers and day 6 weights were determined. If an animal was sacrificed before day 6 due to a greater than 20% drop in original weight, a percent weight of 80 was used as the limit of detection for the statistical analysis. The HAI titers were transformed by log2 and the mean calculated. The ELISAs were performed in quadruplicate. After subtraction of the background absorbance replicates were averaged and the standard deviations plotted. Analyses were done using GraphPad Prism software.
Nucleotide sequences
The nucleotide sequences for all three human and four swine COBRA HA sequences have been reported in U.S. patent filings 14332088_1.
Results
Design of H1N1 COBRA HA genes
Previous studies from our laboratory used the COBRA methodology to design H1N1 HA sequences to account for the evolution of H1N1 influenza viruses isolated in humans over the past 100 years.Citation17 The COBRA design utilized these chronologically different eras of primarily human H1 HA sequences to account for the unique antigenic types of HA domains. Three lead candidates (P1, X3, and X6) elicited broadly protected antibody responses in immunologically naïve mice and ferrets,Citation17 as well as ferrets with pre-existing antibodies to H1N1 influenza viruses.Citation41 In this study, new H1 COBRA HA sequences were designed based exclusively on swine isolates. Examples of the COBRA sequence clustering are shown in . Primary consensus HA sequences were aligned, and the most common amino acid was chosen resulting in secondary consensus sequences. The secondary and tertiary consensus sequences were aligned sequentially, and the most common amino acid at each position was selected ultimately resulting in the final consensus sequence referred to by one of four swine H1 COBRA HA designations (SW1, SW2, SW3, SW4) (). Three of the four swine COBRA HA vaccines utilize HA amino acid sequences from H1N1 and H1N2 isolates in different configurations with the same representative years (). However, SW4 was designed using swine HA sequences from isolates collected between 1930 and 2013 ().
An unrooted phylogenetic analysis of the aligned wild-type human, swine, and COBRA HA1 amino acid sequences determined that the COBRA sequences clustered into different lineages (). One family has similarity to human seasonal influenza strains, and the other cluster is similar to the 2009 pandemic H1N1 swine strains. The alpha, beta, and gamma swine clades of the classical lineage cluster with the CA/09 pandemic-like virus, and the human seasonal viruses cluster with the delta lineage swine viruses. X3 and X6 both clustered with the human isolates most closely related to the human seasonal swine HA. P1 COBRA was most closely related to the human isolates from the 1930–1950’s which were unrelated to swine isolate HA proteins. SW-2 and SW-4 COBRAs were most closely related to each other, and then to viruses in the classical swine lineage (). Therefore, they were similar to the CA/09 HA sequence. Using p-epitope predictions, P1, SW2, and SW4 should all elicit antibodies that neutralize CA/09 infection (). In contrast, the SW1 and SW3 HA sequences, as well as X3 and X6 HA sequences, aligned closer to the human isolates and human seasonal-like swine lineage viruses (). The p-epitope values indicate that these four HA antigens are similar to each other and human seasonal H1 viruses, such as A/Brisbane/59/2007 (Bris/07) or A/Swine/North Carolina/152702/2015 (SW/NC/15) (). A BLAST search using each of the swine COBRA HA sequences revealed that each sequence was a unique sequence that has not been isolated from the environment (data not shown). The p-epitope values of the wild-type swine H1 influenza viruses indicated that classical lineage viruses may neutralize CA/09, such as A/Swine/Wisconsin/125/1997 (SW/WI/97), and that the human seasonal-like lineage viruses may protect against SW/NC/15, such as A/Swine/North Carolina/02744/2009 (SW/NC/02744/09). There were a few wild-type strains that had p-epitope values that predicted they would not bind or neutralize either classical or human seasonal-like lineage viruses, such as A/Swine/Spain/50047/2003 (SW/Spain/03).
Table 2. The p-dominant epitope based predictions for vaccine effectiveness against the challenge strains.
Predictive structures and N-linked glycosylation of the swine COBRA HA antigens
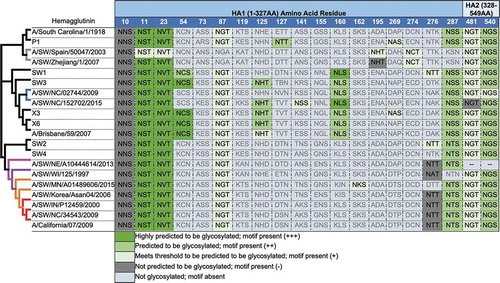
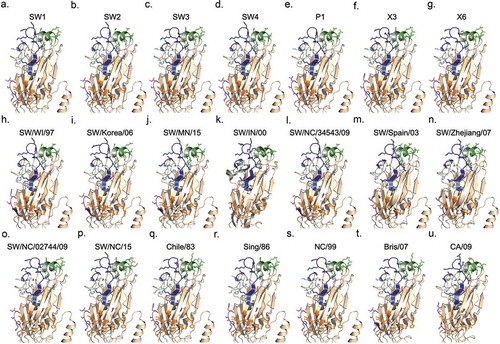
Using a predictive structural modeling of swine COBRA H1 HA sequences, three-dimensional trimerized HA proteins were designed for the COBRA HA sequences (), nine wild-type swine HA sequences (), and five wild-type human HA sequences (). Despite nearly identical predicted structures, the swine COBRA HA proteins did have subtle differences in the major antigenic antibody-binding and receptor-binding sites, more pronounced around the loop regions of the Sa and Sb sites (). For example, all the H1 HA antigens have a Lys (K) at position 154 in the Sa region. However, classical lineage H1 HA proteins, including P1, SW2, and SW4 have a Gly (G) at position 155, whereas an Asp (N) is located at position 156 (). In contrast, human or seasonal human-like swine influenza HA antigens have these amino acids transposed, with an Asp at position 155 and a Gly at 156. There are three exceptions. SW/Korea/06 HA has a glutamic acid (E) at position 155 in the Sa region (). Interestingly, the COBRA SW1 HA has the N155 and N156 in the Sa region (), and the COBRA P1 HA has G155 and G156 in the Sa site (). Both these COBRA HA antigens represents a hybrid of classical and human seasonal-like HA amino acids in the Sa antigenic region.
Figure 3. Subtle differences in the Sa and Sb regions lead to structural differences among the COBRA and wild-type predicted structures surrounding the receptor-binding site. One monomer of each predicted trimer is shown for clarity. Protein structures predicted using SWISS-MODEL. CA/09’s and SW/IN/00’s actual crystal structures were used. Antigenic sites Sa (blue), Sb (green), Ca1 (orange), Ca2 (teal), and Cb (purple) are indicated.
There was little difference in the N-linked glycosylation patterns between any of the swine or human HA antigens, except in two locations (). Human and human seasonal-like swine HA, as well as SW1, SW3, X3, and X6 COBRA HA proteins, had putative glycosylation motif (N-X-S/T, where X is any amino acid except prolineCitation42,Citation43) at amino acids 125 and 160. No classical swine or pandemic-like HA antigens had these motifs. This includes P1, SW2, and SW4 COBRA HA antigens.
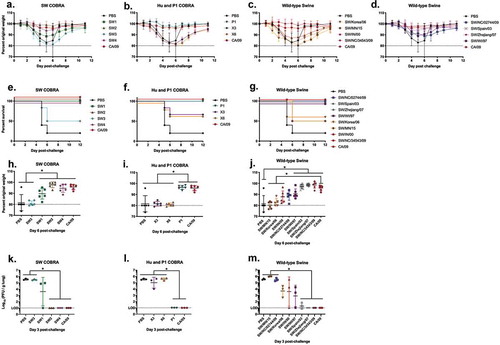
Vaccinated mice challenged with pandemic H1N1 influenza virus
BALB/c mice (n ≥ 9) were vaccinated three times at 4-week intervals via intramuscular injection with purified VLP vaccines expressing either COBRA HA, wild-type swine HA, or human HA plus an oil-in-water based adjuvant. To determine the protective efficacy of each swine COBRA HA vaccine, vaccinated mice were challenged with CA/09 influenza virus (5 x 104 PFU/50 µl) 4 weeks after the third vaccination (). Mice vaccinated with SW2, SW4, P1 or CA/09 HA VLP vaccines experienced little weight loss (,) and all survived challenge (). Mock-vaccinated animals rapidly lost weight and many mice reached experimental endpoints (80% of original body weight) between 4–6 days post-infection (dpi). Mice vaccinated with SW3, X3, or X6 had similar morbidity and mortality results as mock-vaccinated mice (). Mice vaccinated with SW1 lost on average ~10% of their original body weight by day 5 post-infection, but rapidly returned to original weight (). Some groups of mice vaccinated with wild-type swine VLP vaccines (SW/NC/34543/09, SW/WI/97) were protected against CA/09 challenge with little weight loss (). In contrast, mice vaccinated with VLPs expressing any of the other six wild-type swine HA antigens, lost between 10–15% of their body weight by day 6 post-infection with ~50% of the mice in the SW/Korea/06 and SW/MN/15 groups succumbing to infection (). The range of weight loss observed at day 6 post-infection is shown in per individual mouse for each vaccine group. Mice vaccinated with SW2, SW4, and P1 COBRA HA vaccines had little weight loss per group, which was consistent with the lack of detectable CA/09 virus in the lungs of mice collected at day 3 post-infection (). No virus was detected in mice vaccinated with SW2, SW4, P1, SW/Zhejiang/07, SW/NC/34543/09 or CA/09 VLP vaccines ().
Figure 5. A/California/07/2009 H1N1 viral challenge of vaccinated BALB/c mice. Mice vaccinated with either COBRA or wild-type HA VLP vaccines were challenged three weeks after final boost with 5 × 104 PFU of CA/09. Weight loss (a–d) and survival proportions (e-g) were monitored during the course of infection. The peak weight loss observed at six days post-infection was analyzed for statistical difference to the CA/09 group (h-j). The viral load of CA/09 was enumerated from challenged mice three days post-infection (n = 3) (k-m).
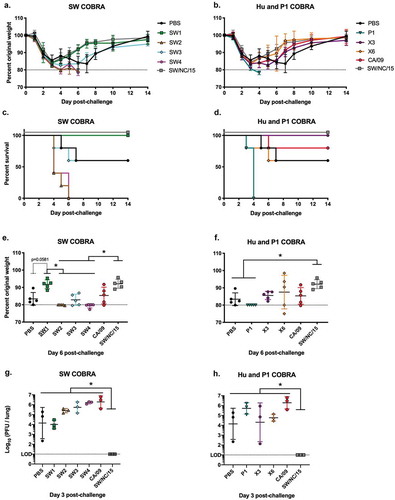
Vaccinated mice challenged with swine H1N2 influenza virus
To determine if these COBRA HA antigens would elicit protective immune responses against a human seasonal-like swine virus, vaccinated mice were challenged with SW/NC/152702/15 (H1N2) influenza virus (1 x 107 PFU/50 µl) 4 weeks after the third vaccination (). All mice vaccinated with SW1, X3, and the homologous SW/NC/15 HA VLP vaccines quickly recovered from early weight loss () and all survived challenge (). Mice vaccinated with SW3, X6 or CA/09 HA VLP vaccines had similar morbidity and mortality results as mock-vaccinated mice. By day 6 post-infection, mice vaccinated with SW1 or SW/NC/15 lost on average ~8% of their original body weight by day 6 (), but rapidly returned to their original weight. In contrast, mice vaccinated with VLPs expressing CA/09 or any of the other six COBRA HA antigens, lost between 12–20% of their body weight by day 6 post-infection (). However, only mice vaccinated with SW/NC/15 HA VLPs had no detectable virus in their lungs at day 3 post-infection (). Mice vaccinated with SW1, X3, X6, or mock had lung viral titers between 104 to 105 PFU/lung. All the other groups had lung viral titers greater than 105 PFU/lung.
Figure 6. A/Swine/North Carolina/152702/2015 H1N2 viral challenge of COBRA vaccinated BALB/c mice. Mice vaccinated with COBRA HA VLP vaccines (n = 11) were challenged three weeks after final boost with 1 × 107 PFU of A/Swine/North Carolina/152702/2015 H1N2. Survival proportions (a, b) and weight loss were monitored during the course of infection (n = 5). The peak weight loss observed at day six was analyzed for statistical difference to the SW/NC/15 group (c, d). The viral lung titer was determined from lungs (n = 3) harvested 3 days post-challenge (e, f).
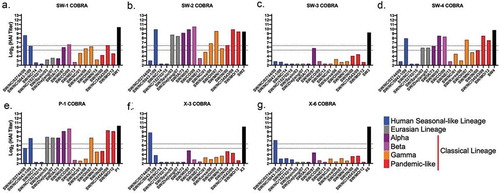
VLP vaccines with swine H1 HA antigens elicit antibodies with HAI activity against swine H1 strains, but not human H1 viruses
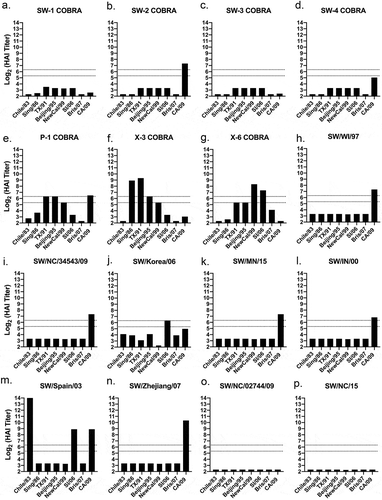
Serum was collected from vaccinated mice on day 56, following the third vaccination (). Mice vaccinated with the SW1 VLP vaccine elicited antibodies with HAI activity exceeding 1:40 against 50% (2/4) of the human seasonal-like swine H1 lineage isolated from 2009 to 2015 and HAI activity against 45% (5/11) of the classical swine H1 lineage viruses in the panel (). Mice vaccinated with SW2 and SW4 had similar patterns of HAI activity against the viruses in the panel. Mice vaccinated with SW2 had antibodies with high HAI activity against one of four human seasonal-like swine H1 viruses and recognized all the classical swine H1 viruses (). SW4 had similar responses and recognized the same human seasonal-like swine virus (SW/NC/5043–1/09). However, these induced antibodies had lower HAI activity, and all the SW4 vaccinated mice did not detect SW/Korea/06 and SW/MN/15 human classical H1 viruses to such a degree as SW2 (). The HAI activities induced by SW2 and SW4 antibodies were similar to antibodies induced by P1 COBRA HA (), which was designed using both human and swine H1 sequences. Mice vaccinated with SW3 recognized only one of seventeen swine viruses in the panel (), which was similar to HAI activities by antibodies induced by the X3 and X6 COBRA HA antigens ().
Figure 7. HAI titers of COBRA VLP vaccinated mice sera against swine H1 VLPs. HAI titers were determined for each group of mice (n ≥5) vaccinated at minimum twice (day 0 and 28) with wild-type HAs originating from swine or human H1 influenza viruses against a panel of seventeen swine H1 VLPs expressing their respective HA. HAIs were conducted with sera collected on day 56. Dotted lines indicate 1:40 and 1:80 HAI titer. Blue = Delta-2 human seasonal clade; Gray = Eurasian clade; Purple = alpha classical clade; Pink = beta classical clade; Orange = gamma classical clade; Red = pandemic classical clade.
Mice vaccinated with seven of the nine VLP vaccines expressing wild-type swine H1 HA antigens had antibodies with HAI activity against at least thirteen of the seventeen viruses in the panel (). In addition, the CA/09 HA VLP vaccine elicited antibodies with HAI activity against one human seasonal-like virus and all, except SW/NE/13, classical lineage viruses (). The SW/NC/15 elicited antibodies with HAI activity against itself and SW/NC/14 (). Mice vaccinated with human seasonal HA VLP vaccines elicited antibodies with HAI activity against ~73% of the viruses in the panel. Mice vaccinated with Sing/86 or Bris/07 had the same HAI pattern; detecting all viruses except two human seasonal-like viruses and two classic swine viruses (,n). None of the vaccines expressing human HA antigens detected SW/Zhejiang/07 or SW/MN/15. At least one of the four human seasonal HA VLP vaccines did not detect SW/NC/5043–1/09 (Chile/83 VLPs), SW/Spain/03, SW/Korea/06 (Sing/86, NC/99, Bris/07 VLPs), SW/OH/07 (Chile/83, NC/99 VLPs), and SW/IN/00 (NC/99 VLPs). In contrast, when swine HA antigens were used in VLP vaccination, few elicited antibodies had HAI activity against human seasonal influenza viruses isolated from 1983 to 2007 (). This same elicited antiserum from the Eurasian and classical swine lineage had HAI antibodies that recognized the CA/09 virus (). The antiserum from human seasonal-like lineage vaccinations, however, did not recognize any human virus ().
Figure 8. HAI titers of swine and human wild-type HA VLP vaccinated mice sera against swine H1 VLPs. HAI titers were determined for each group of mice (n ≥5) vaccinated at minimum 7against a panel of nineteen swine H1 VLPs expressing their respective HA. HAIs were conducted with equally pooled sera collected on day 56. Dotted lines indicate 1:40 and 1:80 HAI titer.
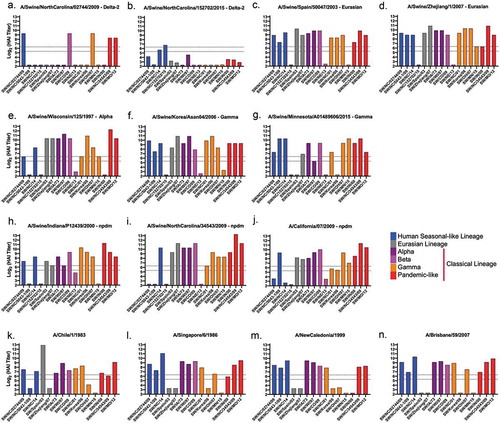
Figure 9. HAI titers of swine wild-type HA VLP vaccinated mice sera against human H1N1 viruses. HAI titers were determined for each group of mice (n ≥5) vaccinated at minimum twice (Day 0 and 28) with wild-type HAs originating from swine H1 influenza viruses against a panel of eight human H1N1 viruses. HAIs were conducted with sera collected on Day 56. Dotted lines indicate 1:40 and 1:80 HAI titer.
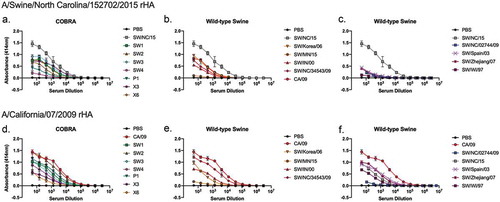
Antiserum collected from mice vaccinated with COBRA or wild-type HA vaccines had narrow HAI breadth against the panel of viruses and did not detect the human seasonal-like SW/NC/15 hemagglutinin ( and ). However, these same antibodies bound to the SW/NC/15 rHA (). Antisera elicited by SW1, SW3, X3 or X6 vaccines elicited antibodies that bound to the SW2, SW4, and P1 COBRA vaccines. The HA vaccines based upon the classical swine strains SW/Korea/06, SW/IN/00 and SW/NC/34543/09 elicited antibodies that bound to the human seasonal-like viruses (). The human seasonal-like, Eurasian, and human seasonal vaccines all had minimal antibody binding (). When vaccine antisera were tested against CA/09 rHA (), antisera collected from mice that were vaccinated with any of the COBRA HA antigens had some level of binding to the various HA proteins. The SW-2 COBRA and SW/NC/34543/09 binding curves matched the profile of antisera elicited by CA/09 HA. SW/MN/15 and SW/NC/02744/09 elicited antibodies had the lowest binding curves that correlated to the illness observed during challenge ().
Figure 10. ELISA of elicited antibodies against either SW/NC/15 or CA/09 recombinant HA protein. HA-specific IgG levels in the serum of mice vaccinated with various H1Nx VLP vaccines tested in an ELISA. (a-c) Antisera tested against rHA from SW/NC/15. (d-f) Antisera tested against rHA from CA/09. Antisera collected from mice vaccinated with (a and d) COBRA HA vaccines; (b, c, e and f) Wild-type HA vaccines.
Discussion
In the past 70 years, pig farming practices have changed from simple small-scale herds to immense herds in large, co-operate settings. These large herds are maintained by a constant introduction of young swine, leading to a constant supply of pathogen-susceptible animals and changes in farming practices.Citation44,Citation45 Cross-species transmission and reassortment of avian and human influenza viruses in pigs has been documented.Citation4,Citation46-Citation48 These reassortment events result in emergence of genetically and antigenically diverse influenza viruses within the pig population with pandemic potential.Citation21,Citation49-Citation51 In 2009, a pandemic-like virus of swine origin, originated in Mexico and quickly spread to the United States and ultimately leading to a worldwide pandemic.Citation52 With the diversity of circulating swine-like H1 viruses, there is an urgent unmet need to develop universal pandemic vaccine strategies to control spread of swine influenza viruses in pig populations, as well as, to control the potential spread of these viruses to humans.
Despite the economic importance of swine influenza viruses, immune mechanisms of protection induced by natural infection or vaccination have been understudied in pigs compared to small animal models or humans. Neutralizing antibodies with HAI activity correlate with improved protection in humans and suggests that similar antibodies could protect against swine influenza viruses in pigs. However, immune mechanisms, such as cell-mediated immunity through direct killing, antibody dependent cellular cytotoxicity (ADCC), or induction of mucosal IgA may also contribute protection against swine influenza viruses in pigs. The administration of influenza vaccines that provide more “universal” protection against these viruses in both pigs and humans is needed.
The COBRA method uses multiple rounds of consensus building based upon not only the phylogenetic sequence of each isolate, but also the outbreak and specific time that each isolate was collected, thereby eliminating the bias in the number of sequences uploaded to online databases. Previously, our group has demonstrated the effectiveness of the COBRA HA antigens for H5N1,Citation14,Citation15 H3N2,Citation53-Citation55 and H1N1 against human seasonal and pandemic influenza viruses.Citation17 These H1N1 COBRA HA proteins (X3, X6, and P1) elicited antibodies with HAI activity against a range of human H1N1 viruses spanning the last 100 years. However, when antibodies elicited by these same three COBRA HA vaccines were tested against swine H1 influenza viruses, the effectiveness of these vaccine antigens decreased. The X3 and X6 H1N1 HA COBRA proteins were designed from predominantly human seasonal H1 influenza HA sequences.Citation17 The antibodies, while effective against human seasonal H1N1 influenza viruses, had little or no HAI activity against either classical or human seasonal-like swine influenza viruses (). And the P1 COBRA HA antigen, a hybrid derivative HA designed using both human and swine influenza HA input sequences,Citation17 while more effective at eliciting antibodies with HAI activity classical swine influenza viruses, was less effective against human seasonal-like swine influenza viruses.
In this study, we used the COBRA approach to design and develop HA vaccine candidates solely based upon swine H1 HA input sequences. Four new COBRA HA antigens (SW1-4) were developed following multiple rounds of sequence alignment to generate a single unique swine H1 HA sequence only. The swine specific COBRA HA antigens, SW1 and SW3, elicited antibodies with HAI activity, but did not protect against classical swine influenza virus infections, which was similar to the previously published X3 and X6 COBRA HA antigens.Citation17 However, SW1 was as effective at protecting mice against the H1N2 human seasonal-like swine influenza virus, SW/NC/15 (). Furthermore, SW2 and SW4 COBRA HA antigens elicited antibodies with similar HAI activity as the P1 COBRA HA. In addition, all three of these antigens protected against the CA/09 pandemic-like influenza virus, but none of them elicited protective immunity against the human seasonal-like swine virus. These results indicate that, while similar, the swine COBRA HA antigens have epitopes that allow for the elicitation of antibodies with HAI activity against subsets of swine influenza viruses, but are missing epitopes that allow for elicitation of antibodies against human seasonal H1 influenza viruses. Concurrently, the human seasonal COBRA HA antigens, X3 and X6, elicit antibodies specific for human seasonal H1N1 influenza viruses only. However, since the COBRA P1 HA incorporates both swine and human specific epitopes, this HA antigen elicits a broader breadth of HAI activity against both human and swine H1 influenza viruses. However, the antibody profile indicates that P1 elicited antibodies are still missing some critical epitopes found in both swine and human H1 influenza viruses, since P1 elicited antibodies do not have HAI activity against some important H1 influenza viruses, such as A/Brisbane/59/2007.Citation17 Future studies using monoclonal antibodies will need to determine the specific epitopes on each of these COBRA antigens (alone or in combination) that elicit specific protective antibodies against the panel of swine and human viruses.
Analysis of amino acids within the antigenic sites indicated a pattern that strongly suggests an existence of shared epitopes amongst swine and human influenza viruses (). SW1 and SW3 have a more human seasonal-like set of amino acids in the antigenic sites and have similar predicted HA structures (). Both these HA vaccines sequences were generated exclusively or predominately from H1N2 swine influenza viruses, which implies that the HA sequences in many H1N2 viruses are more closely related to the HA in human seasonal influenza viruses. This is consistent with the observation of cross-reactivity of human H1N1 antisera against swine H1 viruses.Citation56 In contrast, the SW2 and SW4 HA sequences were developed using primarily HA sequences from H1N1 influenza viruses, which biases the HA to a classical or pandemic-like phenotype. Indeed, the major antigenic sites in SW2 and SW4 have many identical amino acids as the CA/09 and P1 HA sequences. Structural analysis of the antigenic sites showed similarity especially around the Sa antigenic site that could potentially determine vaccine effectiveness.
Figure 11. Comparison of antigenic site residues of CA/09 and SW/NC/152702/15. The order from left to right is based on decreasing similarity to CA/09 antigenic residues (red). For each SW/NC/15 residue (blue) a penalty was scored (−1). Completely different amino acid residues (white) scored no penalty. The narrowed antigenic site residues were used for the comparison. Sites where CA/09 and SW/NC/152702/15 were identical are not shown. The color legend for each antigenic site is associated with the matching PYMOL site.
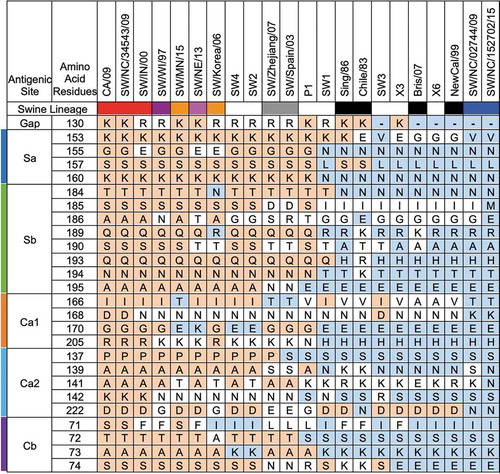
There was little difference in the N-linked glycosylation patterns between any of the swine or human HA antigens (), except in two locations at amino acids 125 and 160. No classical swine or pandemic-like HA antigens had these motifs, including P1, SW2, and SW4 COBRA HA antigens. Glycosylation in HA is important for the folding and stability of the protein,Citation57 and, in some cases, significantly affects receptor binding and cleavage of the precursor HA0 protein, influencing the virulence and antigenicity of the virus.Citation58 Our group and others have shown that glycosylation on the HA globular head domain physically shields the antigenic sites, preventing antibody recognition and leading to viral evasion from antibody-mediated neutralization.Citation59-Citation63 HA antigens acquire increased glycans as influenza viruses circulate in people,Citation64 therefore it is not surprising that human-like wild-type or COBRA H1 HA antigens have additional glycans that classical swine viruses do not. More analysis aimed at elucidating the mechanism(s) of COBRA HA vaccines is needed to generate next-generation vaccine designs. An approach that maps out the antigenic epitopes on theses vaccine candidates, as well as addressing effect of glycosylation, will provide a better understanding how these vaccines operate.
The P1, SW2, and SW4 COBRA HA antigens elicited antibodies with HAI activity against Eurasian H1 isolates. In Europe, an avian H1N1 influenza virus was first detected in pigs in Belgium in 1979, referred to as ‘avian-like’ swine H1N1 lineage.Citation65 This lineage slowly established itself in the pig population eventually replacing classical swine H1N1 influenza viruses and also reassorted in pigs with human H3N2 viruses (A/Port Chalmers/1/1973-like).Citation66 Importantly, to date, there are no notable cases of Eurasian avian-like swine H1N1 influenza viruses circulating in North America. However, the potential of introduction of viruses to North American herd is plausible, therefore, a vaccine that efficiently elicits protective immune responses against all H1 influenza viruses is highly desired. The SW2 and SW4 vaccines were effective in eliciting antibodies with HAI activity against the Eurasian derived swine viruses (). However, it was surprising that wild-type HA antigens isolated from swine viruses induced antibodies with broad HAI activity. In particular, mice vaccinated with VLPs expressing wild-type Eurasian HA were the most efficient at eliciting broadly reactive antibodies against both Eurasian and North American isolates. This strongly supports the possibility of making a universal vaccine with worldwide application.
Despite the similarities between the human seasonal-like HA antigens and, for example, COBRA SW1 or SW3, these vaccines were not as effective as the SW/NC/15 HA homologous vaccine (). There is no clear explanation for why only mice vaccinated with SW/NC/15 HA VLP vaccine were protected when the antigenic sites of SW1 and SW3 HA antigens are highly similar to the SW/NC/15 HA or another virus, SW/NE/13, with similar predicted glycosylation patterns and similar predicted three-dimensional structures, but these viruses may be antigenically distinct from other contemporary swine influenza strains. Even though SW1 HA VLP vaccinated mice had detectable virus in the lung (day 3 p.i.), mice vaccinated with the SW1 vaccine all survived challenge and had the lowest viral lung titers, other than the homologous vaccine. The SW1 vaccine antisera was, however, able to bind to SW/NC/15 rHA implying that the SW1 COBRA was able to elicit antibodies against the HA, but these were not receptor site binding. Antibodies elicited by vaccination may have broader activity than just receptor binding site inhibiting, as seen by the lack of HAI titer, but the presence of antibody binding (). Future studies will need to tease out the specific epitopes on these HA antigens that elicit the most effective neutralizing antibodies.
It is estimated that more than 100,000 workers work in swine barns with live pigs, and in just Iowa alone – the leading swine-producing US state – more that 25 million hogs per year are reared (a ratio of more that 9 swine per human resident).Citation56 Therefore, it is likely that many people around the world have been exposed to multiple swine influenza virusesCitation67 and have a B cell memory pool and antibody repertoire with HAI activity against multiple swine influenza viruses, but also historical human strains that represent various antigenic types.Citation68,Citation69 Our group and others have demonstrated that ferrets pre-immune to H1N1 influenza viruses elicited antibodies with broader HAI activity against more antigenic variants following COBRA HA vaccination than using wild-type HA antigens.Citation70-Citation72 Therefore, we would expect these COBRA vaccines to be even more effective in people pre-existing memory B cells to previous swine influenza exposures. However, the vaccines used in this study have limitations since they did not elicit antibodies with HAI activity against all swine and human strains. Using a divalent or multivalent COBRA HA-based vaccine may elicit broader HAI activity against viruses in this H1 panel. There may be other factors that play a role in protection in pigs beyond HAI demonstrated in the vaccinated mouse model. Therefore, until these vaccine immunogens are tested in humans or a pig vaccination and challenge model, the overall effectiveness of these vaccines to prevent influenza virus induced illness and transmission is not known. Overall, this study demonstrated that computationally optimized HA antigens are a viable way of designing vaccines with broader application for both human and swine population, which is important given the zoonotic potential of influenza viruses. vaccines that are aimed at protecting both humans and swine population.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Supplemental Material
Download Zip (54.9 KB)Acknowledgments
The authors would like to thank Dr. Ying Huang, Z. Beau Reneer, and James D. Allen for technical assistance, and Dr. Mark Tompkins’ laboratory for graciously providing the A/Swine/North Carolina/152702/2015 virus. Furthermore, we thank both Drs. Mark Tompkins and Constantinos Kyriakis for helpful discussions. Certain H1N1 and H1N2 influenza A viruses were obtained through the Influenza Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA, USA. The authors would like to thank the University of Georgia Animal Resource staff, technicians, and veterinarians for animal care.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Choi YK, Goyal SM, Joo HS. Prevalence of swine influenza virus subtypes on swine farms in the United States. Arch Virol. 2002;147(6):1209–20. doi:10.1007/s00705-002-0788-4.
- Olsen CW. The emergence of novel swine influenza viruses in North America. Virus Res. 2002;85:199–210.
- Thacker E, Janke B. Swine influenza virus: zoonotic potential and vaccination strategies for the control of avian and swine influenzas. J Infect Dis. 2008;197(Suppl 1):S19–S24. doi:10.1086/524988.
- Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. Swine influenza viruses a North American perspective. Adv Virus Res. 2008;72:127–54.
- Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes. 2009;39(2):176–85. doi:10.1007/s11262-009-0386-6.
- Karasin AI, Landgraf J, Swenson S, Erickson G, Goyal S, Woodruff M, Scherba G, Anderson G, Olsen CW. Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J Clin Microbiol. 2002;40(3):1073–79. doi:10.1128/JCM.40.3.1073-1079.2002.
- Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res. 2004;103(1–2):67–73. doi:10.1016/j.virusres.2004.02.015.
- Ma W, Vincent AL, Lager KM, Janke BH, Henry SC, Rowland RR, Hesse RA, Richt JA. Identification and characterization of a highly virulent triple reassortant H1N1 swine influenza virus in the United States. Virus Genes. 2010;40(1):28–36. doi:10.1007/s11262-009-0413-7.
- Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, et al. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere. 2016;1(6). doi:10.1128/mSphere.00287-16.
- Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, Burke DF, Dudas G, Fonville JM, Fouchier RA, et al. The global antigenic diversity of swine influenza A viruses. Elife. 2016;5:e12217. doi:10.7554/eLife.12217.
- Gibbs AJ, Armstrong JS, Downie JC. From where did the 2009 ‘swine-origin’ influenza A virus (H1N1) emerge? Virol J. 2009;6:207. doi:10.1186/1743-422X-6-207.
- Crevar CJ, Ross TM. Elicitation of protective immune responses using a bivalent H5N1 VLP vaccine. Virol J. 2008;5:131. doi:10.1186/1743-422X-5-131.
- Giles BM, Bissel SJ, DeAlmeida DR, Wiley CA, Ross TM. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin Vaccine Immunol. 2012;19(2):128–39. doi:10.1128/CVI.05533-11.
- Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, Ross TM. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis. 2012;205(10):1562–70. doi:10.1093/infdis/jis232.
- Giles BM, Ross TM. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine. 2011;29(16):3043–54. doi:10.1016/j.vaccine.2011.01.100.
- Monto AS. Seasonal influenza and vaccination coverage. Vaccine. 2010;28(Suppl 4):D33–D44. doi:10.1016/j.vaccine.2010.08.027.
- Carter DM, Darby CA, Lefoley BC, Crevar CJ, Alefantis T, Oomen R, Anderson SF, Strugnell T, Cortés-Garcia G, Vogel TU, et al. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J Virol. 2016;90(9):4720–34. doi:10.1128/JVI.03152-15.
- Ross TM, Xu Y, Bright RA, Robinson HL. C3d enhancement of antibodies to hemagglutinin accelerates protection against influenza virus challenge. Nat Immunol. 2000;1(2):127–31. doi:10.1038/77802.
- Organization WH, Network WGIS. Manual for the laboratory diagnosis and virological surveillance of influenza. Geneva (Switzerland): World Health Organization; 2011.
- Anderson TK, Campbell BA, Nelson MI, Lewis NS, Janas-Martindale A, Killian ML, Vincent AL. Characterization of co-circulating swine influenza A viruses in North America and the identification of a novel H1 genetic clade with antigenic significance. Virus Res. 2015;201:24–31. doi:10.1016/j.virusres.2015.02.009.
- Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. Population dynamics of co-circulating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respi Viruses. 2013;7(S4):42–51. doi:10.1111/irv.2013.7.issue-s4.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–49. doi:10.1093/bioinformatics/bts199.
- Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328(5976):357–60. doi:10.1126/science.1186430.
- Xu R, Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J Virol. 2012;86(17):9221–32. doi:10.1128/JVI.00697-12.
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TA, Rempfer C, Bordoli L, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi:10.1093/nar/gky427.
- Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, Schwede T. The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res. 2017;45(D1):D313–D319. doi:10.1093/nar/gkw1132.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi:10.1186/1471-2105-10-421.
- Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9(2):173–75. doi:10.1038/nmeth.1818.
- Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-Pdbviewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–73. doi:10.1002/elps.200900140.
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdbviewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–23. doi:10.1002/elps.1150181505.
- Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27(3):343–50. doi:10.1093/bioinformatics/btq662.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi:10.1093/nar/25.17.3389.
- Gupta R, Jung E, Brunak S. Prediction of N-glycosylation sites in human proteins. 2004. http://www.cbs.dtu.dk/services/NetNGlyc/
- Deem MW, Pan KY. The epitope regions of H1-subtype influenza A, with application to vaccine efficacy. Protein Eng Des Sel. 2009;22(9):543–46. doi:10.1093/protein/gzp027.
- Winter G, Fields S, Brownlee GG. Nucleotide sequence of the haemagglutinin gene of a human influenza virus H1 subtype. Nature. 1981;292(5818):72–75. doi:10.1038/292072a0.
- Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino-acid-sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A-viruses. Virology. 1991;182(2):475–85. doi:10.1016/0042-6822(91)90588-3.
- Burke DF, Smith DJ. A recommended numbering scheme for influenza A HA subtypes. PLoS One. 2014;9(11):e112302. doi:10.1371/journal.pone.0112302.
- Pan K, Subieta KC, Deem MW. A novel sequence-based antigenic distance measure for H1N1, with application to vaccine effectiveness and the selection of vaccine strains. Protein Eng Des Sel. 2011;24(3):291–99. doi:10.1093/protein/gzq105.
- Green TD, Montefiori DC, Ross TM. Enhancement of antibodies to the human immunodeficiency virus type 1 envelope by using the molecular adjuvant C3d. J Virol. 2003;77(3):2046–55. doi:10.1128/jvi.77.3.2046-2055.2003.
- Agency EM. Guideline on influenza vaccines: non-clinical and clinical module [Draft]. In: Use CFMPFH, editor. EMA/CHMP/VWP/457259/2014. London E14 4HB (UK): European Medicines Agency; 2014.
- Carter DM, Darby CA, Johnson SK, Carlock MA, Kirchenbaum GA, Allen JD, Vogel TU, Delagrave S, DiNapoli J, Kleanthous H, et al. Elicitation of protective antibodies against a broad panel of H1N1 viruses in ferrets preimmune to historical H1N1 influenza viruses. J Virol. 2017;91(24). doi:10.1128/JVI.01283-17.
- Kim JI, Park MS. N-linked glycosylation in the hemagglutinin of influenza A viruses. Yonsei Med J. 2012;53(5):886–93. doi:10.3349/ymj.2012.53.5.886.
- Wei CJ, Boyington JC, Dai K, Houser KV, Pearce MB, Kong WP, Yang ZY, Tumpey TM, Nabel GJ. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med. 2010;2(24):24ra21. doi:10.1126/scitranslmed.3000799.
- White LA, Torremorell M, Craft ME. Influenza A virus in swine breeding herds: combination of vaccination and biosecurity practices can reduce likelihood of endemic piglet reservoir. Prev Vet Med. 2017;138:55–69. doi:10.1016/j.prevetmed.2016.12.013.
- Simon-Grife M, Martín-Valls GE, Vilar MJ, Busquets N, Mora-Salvatierra M, Bestebroer TM, Fouchier RA, Martín M, Mateu E, Casal J. Swine influenza virus infection dynamics in two pig farms; results of a longitudinal assessment. Vet Res. 2012;43:24. doi:10.1186/1297-9716-43-24.
- Nelson MI, Gramer MR, Vincent AL, Holmes EC. Global transmission of influenza viruses from humans to swine. J Gen Virol. 2012;93(Pt 10):2195–203. doi:10.1099/vir.0.044974-0.
- Lycett SJ, Baillie G, Coulter E, Bhatt S, Kellam P, McCauley JW, Wood JL, Brown IH, Pybus OG, Brown AL. Estimating reassortment rates in co-circulating Eurasian swine influenza viruses. J Gen Virol. 2012;93(Pt 11):2326–36. doi:10.1099/vir.0.044503-0.
- Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster RG. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193(1):503–06. doi:10.1006/viro.1993.1155.
- Rajao DS, Anderson TK, Kitikoon P, Stratton J, Lewis NS, Vincent AL. Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the United States. Virology. 2018;518:45–54. doi:10.1016/j.virol.2018.02.006.
- Lorusso A, Vincent AL, Harland ML, Alt D, Bayles DO, Swenson SL, Gramer MR, Russell CA, Smith DJ, Lager KM, et al. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J Gen Virol. 2010;92(4):919–30. doi:10.1099/vir.0.027557-0.
- Anderson CS, McCall PR, Stern HA, Yang H, Topham DJ. Antigenic cartography of H1N1 influenza viruses using sequence-based antigenic distance calculation. BMC Bioinf. 2018;19. doi:10.1186/s12859-018-2042-4.
- Lemey P, Suchard M, Rambaut A. Reconstructing the initial global spread of a human influenza pandemic: A Bayesian spatial-temporal model for the global spread of H1N1pdm. PLoS Curr. 2009;1:RRN1031. doi:10.1371/currents.RRN1031.
- Allen JD, Jang H, DiNapoli J, Kleanthous H, Ross TM. Elicitation of protective antibodies against 20 years of future H3N2 co-ciruculating influenza virus variants in ferrets preimmune to historical H3N2 influenza viruses. J Virol. 2018;93. doi:10.1128/JVI.00946-18.
- Allen JD, Ray S, Ross TM. Split inactivated COBRA vaccine elicits protective antibodies against H1N1 and H3N2 influenza viruses. PLoS One. 2018;13(9):e0204284. doi:10.1371/journal.pone.0204284.
- Wong TM, Allen JD, Bebin-Blackwell AG, Carter DM, Alefantis T, DiNapoli J, Kleanthous H, Ross TM. Computationally optimized broadly reactive hemagglutinin elicits hemagglutination inhibition antibodies against a panel of H3N2 influenza virus cocirculating variants. J Virol. 2017;91(24). doi:10.1128/JVI.00955-17.
- Myers KP, Olsen CW, Setterquist SF, Capuano AW, Donham KJ, Thacker EL, Merchant JA, Gray GC. Are swine workers in the United States at increased risk of infection with zoonotic influenza virus? Clin Infect Dis. 2006;42(1):14–20. doi:10.1086/498977.
- Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139(3):613–23. doi:10.1083/jcb.139.3.613.
- Zhang Y, Zhu J, Li Y, Bradley KC, Cao J, Chen H, Jin M, Zhou H. Glycosylation on hemagglutinin affects the virulence and pathogenicity of pandemic H1N1/2009 influenza A virus in mice. PLoS One. 2013;8(4):e61397. doi:10.1371/journal.pone.0061397.
- Zost SJ, Parkhouse K, Gumina ME, Kim K, Perez SD, Wilson PC, Treanor JJ, Sant AJ, Cobey S, Hensley SE. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci USA. 2017;114(47):12578–83. doi:10.1073/pnas.1712377114.
- Pentiah K, Lees WD, Moss DS, Shepherd AJ. N-linked glycans on influenza A H3N2 hemagglutinin constrain binding of host antibodies, but shielding is limited. Glycobiology. 2015;25(1):124–32. doi:10.1093/glycob/cwu097.
- Tate MD, Job E, Deng YM, Gunalan V, Maurer-Stroh S, Reading P. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses. 2014;6(3):1294–316. doi:10.3390/v6031294.
- Laursen NS, Wilson IA. Broadly neutralizing antibodies against influenza viruses. Antiviral Res. 2013;98(3):476–83. doi:10.1016/j.antiviral.2013.03.021.
- Huang Y, Owino SO, Crevar CJ, Carter DM, and Ross TM. N-linked glycans and K147 residue on hemagglutinin synergizes to elicit broadly-reactive H1N1 influenza antibodies. J Virol. 2019. in press.
- Altman MO, Angel M, Košík I, Trovão NS, Zost SJ, Gibbs JS, Casalino L, Amaro RE, Hensley SE, Nelson MI, et al. Human influenza A virus hemagglutinin glycan evolution follows a temporal pattern to a glycan limit. MBio. 2019;10(2). doi:10.1128/mBio.00669-19.
- Nelson MI, Lemey P, Tan Y, Vincent A, Lam TT, Detmer S, Viboud C, Suchard MA, Rambaut A, Holmes EC, et al. Spatial dynamics of human-origin H1 influenza A virus in North American swine. PLoS Pathog. 2011;7(6):e1002077. doi:10.1371/journal.ppat.1002077.
- Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol. 2000;74:29–46.
- Lantos PM, Hoffman K, Höhle M, Anderson B, Gray GC. Are people living near modern swine production facilities at increased risk of influenza virus infection? Clin Infect Dis. 2016;63(12):1558–63. doi:10.1093/cid/ciw646.
- Yoshida T, Mei H, Dörner T, Hiepe F, Radbruch A, Fillatreau S, Hoyer BF. Memory B and memory plasma cells. Immunol Rev. 2010;237(1):117–39. doi:10.1111/j.1600-065X.2010.00938.x.
- Paccha B, Jones RM, Gibbs S, Kane MJ, Torremorell M, Neira-Ramirez V, Rabinowitz PM. Modeling risk of occupational zoonotic influenza infection in swine workers. J Occup Environ Hyg. 2016;13(8):577–87. doi:10.1080/15459624.2016.1159688.
- Carter DM, Bloom CE, Nascimento EJ, Marques ET, Craigo JK, Cherry JL, Lipman DJ, Ross TM. Sequential seasonal H1N1 influenza virus infections protect ferrets against novel 2009 H1N1 influenza virus. J Virol. 2013;87(3):1400–10. doi:10.1128/JVI.02257-12.
- Kirchenbaum GA, Carter DM, Ross TM. Sequential infection in ferrets with antigenically distinct seasonal H1N1 influenza viruses boosts hemagglutinin stalk-specific antibodies. J Virol. 2016;90(2):1116–28. doi:10.1128/JVI.02372-15.
- Li Y, Myers JL, Bostick DL, Sullivan CB, Madara J, Linderman SL, Liu Q, Carter DM, Wrammert J, Esposito S, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med. 2013;210(8):1493–500. doi:10.1084/jem.20130212.