ABSTRACT
Global efforts on the replacement of the in vivo rabies vaccine potency test (NIH method) with in vitro methods for quantification of immunodominant glycoprotein (GP) in rabies vaccine have made significant progress. We report here, a sandwich ELISA method based on the use of a neutralizing rabies GP site III directed human monoclonal antibody (RAB-1) and a polyclonal GP specific antibody recognizing the intact form of viral GP. The method was shown to be robust, specific, linear, precise and accurate in the quantification of intact GP in vaccine samples. The assay was able to differentiate between potent and sub-potent vaccine samples. The assay was shown to be linear over the range of 0.07–2.25 IU/mL with LOD and LLOQ values of 0.035 and 0.070 IU/mL, respectively. The assay was able to quantify the GP content of rabies vaccines derived from rabies vaccine strains, e.g., Pittman-Moore, Pasteur and Flury LEP with acceptable precision (CV < 20%) and also showed good agreement with NIH potency estimates. The binding kinetics of RAB-1 with intact and modified vaccine samples were also characterized using biolayer interferometry (BLI). The developed method may be used as an alternative to the NIH method in quality control testing of human rabies vaccines.
1. Introduction
Rabies is a zoonotic viral disease that results in acute encephalitis in both humans and animals. The causative agent is a rabies virus (RABV) belonging to the Rhabdoviridae family. Rabies is an almost completely vaccine-preventable disease using a combination of active and passive immunization strategies. As per WHO estimates, annually, more than 29 million people receive a post-bite vaccination. This is estimated to prevent hundreds of thousands of rabies deaths annually.
Potent and safer human rabies vaccines hold the key to prevention and post-exposure prophylaxis.Citation1 Presently, potency testing of inactivated human rabies vaccine is performed using vaccination-intra-cerebral challenge assay, developed by the National Institutes of Health (NIH) more than 50 years ago.Citation2,Citation3 The NIH method has limitations, including the use of animals, lethal endpoint, handling of live virus and higher variability (requires large numbers of animals for reliable estimates).Citation4 Several international agencies such as European Pharmacopoeia, Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM), and European Partnerships for Alternative Approaches to Animal Testing (EPAA) are working for the development of suitable in-vitro methods as a replacement to NIH method. Such approaches for replacement with in-vitro potency methods have been successful with other viral vaccines such as Hepatitis B and Human Papilloma Virus vaccines.Citation5,Citation6 Based on the fact that among all the proteins of the rabies virus, glycoprotein (GP) is the major antigen for inducing viral neutralizing antibodies, its intact amount in the vaccines will be critical to the antigenicity and potency of vaccines.Citation7,Citation8 In an EPAA workshop held in 2012, a scientific consensus was achieved that a standardized sandwich ELISA targeted against rabies GP would be an ideal approach for potency testing of rabies vaccine.Citation9,Citation10 Several ELISA methods based on the use of monoclonal antibodies directed against GP have been studied and reported.Citation11–Citation15 These studies also highlighted the need for rationalization in the selection of antibodies for ELISA, as they should be neutralizing in function with the ability to identify the intact or immunogenic form of GP in vaccines. The structure of rabies GP is well studied and known to harbor five antigenic sites and conformational epitopes of antigenic site II (denoted by amino acid residues 34–42 and 198–200) and antigenic site III (denoted by residues at 330–338) which are the major targets of virus-neutralizing antibodies. Thus, the use of site II and/or site III directed antibodies in ELISA will be important to capture the intact form of GP in the vaccines. In the EPAA workshop held in the year 2015, one of these sandwich ELISA methods involving the use of site II and III monoclonal antibodies D1-125 and WI 1112 mAbs was selected for international collaborative study. The workshop also invited other potential ELISA methods from different vaccine manufacturers including the assay developed at Serum Institute of India using clinically proven site III human monoclonal antibody (RMab/RAB-1).Citation16
Rabies monoclonal antibody (RMab/RAB-1) is the world’s first human therapeutic antibody clinically shown to possess all the features needed for replacing Human Rabies Immunoglobulin (HRIG) as a single antibody. RAB1 reportedly has the singular ability to neutralize viruses representing isolates from numerous species and geographical locations.Citation17 The antibody (RAB-1) interacts with an epitope which includes amino acids 336–342 of the GP (antigenic site III). Preclinical studies have shown the antibody to be protective in a well-established in-vivo model (hamsters) of post-exposure prophylaxis in the presence and absence of the vaccine.Citation18 RAB-1 has also been proven effective in clinical trials and is currently licensed in India for human use.Citation19,Citation20
We describe here the development and validation of a novel sandwich ELISA for quantification of rabies GP content using RAB-1 as a capture antibody to monitor the quality of the vaccine batches. The assay shows acceptable linearity, precision, and accuracy in the range of 0.07–2.25 IU/ml and can differentiate between potent and sub-potent vaccine samples. The assay can quantify the GP content of different vaccines derived from the Pitman-Moore (PM), Pasteur, and FluryLEP strains. The assay also shows a satisfactory agreement with NIH and single radial immunodiffusion (SRID) potency test for the detection of the intact and degraded form of GP in vaccine samples.
2. Materials and methods
2.1. Vaccine samples
Vaccine samples representing different strains of the rabies virus, including PM, Pasteur, and Flurry LEP were procured from the market and used in the study. Vaccine samples representing lyophilized rabies vaccines derived from different strains, sub-potent and/or potent batches were used in the study. The details of the vaccine samples used in the study are provided in .
Table 1. Details of reference standard and vaccine samples used in the study.
2.2. Reference vaccine
The lyophilized rabies vaccine (in-house, Pitman-Moore Strain; PM) manufactured at the Serum Institute of India Pvt Ltd was used as a reference standard (RS) in the study. RS is calibrated against the 6th international NIBSC reference standard, national reference standard (NRS/2006) using validated single radial immunodiffusion (SRID) assay. A mean unitage of 5.78 IU/ml was assigned to the RS based on 10 SRID independent assays. The potency of RS was also studied using in-vivo NIH method, and a potency of 5 IU/ml was assigned to it ()
2.3. Virus-neutralizing activity of antibodies used in the method
The method uses a specific combination of virus-neutralizing human monoclonal (RAB-1) and polyclonal antibodies against the rabies viral GP (AGS). RAB-1 is commercially available and marketed by the Serum Institute of India under the trade name of RABISHIELD. AGS was developed in-house. Both these antibodies were characterized for their virus-neutralizing activities using the Rapid Fluorescent Focus Inhibition Test (RFFIT) and mouse neutralization tests.
2.3.1. Development of polyclonal antibody against rabies GP (AGS)
Anti-rabies virus GP polyclonal antibody was raised in sheep using purified GP of PM strain (supplied by Serum Institute). Briefly, GP was extracted and purified using sucrose density gradient centrifugation and detergent treatment (mulgofen, 2%). The purified GP is checked for its purity using 8–12% SDS gel stained with Coomassie blue and bands are verified for their molecular weights (40–60 kDa). GP content is determined using the SRID assay (29 IU/ml). The purified GP is stored at −70 degrees. The GP: adjuvant mixture (complete Freund’s adjuvant, Sigma-Aldrich, USA) was injected subcutaneously on day 0. Each sheep is reimmunized at 2-week intervals with GP and incomplete Freund’s adjuvant (Sigma Aldrich, USA). A total of two sheeps were used for anti-sera production. Blood samples were collected on day 56 and pooled serum is checked for its titer. Sera were aliquoted and stored in lyophilized form.
2.3.2. RFFIT protocol
Rapid Focus Fluorescent Inhibition Test (RFFIT) was used for the determination of RAB-1 and Anti-rabies virus GP polyclonal antibody neutralizing titers against the challenge virus strain CVS-11. Briefly, serial fold dilutions of RAB-1 or polyclonal antibody were incubated with 50 FFD50 (50% fluorescent focus forming doses) of the challenge rabies virus in tissue culture slides for 90 min at 37°C. Two hundred microliters of MNA cells (murine neuroblastoma) cell line sourced from Diagnostics Hybrids, USA; maintained in MEM media and 10% FBS adjusted at concentration of 5 × 105 cells/ml were then added to virus/mAB mixture and incubated for 24 h at 36 ± 1oC in humidified CO2 incubator (Thermo and 2–2.5% CO2). The slides were then acetone-fixed and cells infected with rabies virus were detected using FITC conjugated rabies monoclonal antibody (Millipore, USA). Twenty distinct microscopic fields are counted using a fluorescence microscope to score the virus-infected cells (indicated by bright green intracellular spots in the cell). The neutralization endpoint titer was defined as the highest dilution of antibody at which 50% of observed microscopic fields contained one or more infected cells. Titers were mathematically interpolated using the Reed and Muench method. Endpoint titer is then transformed into international units by calibrating against international reference serum (WHO reference standard for rabies immunoglobulin).
2.3.2. Mouse neutralizing test
Reference and test samples (RAB-1 or AGS) were serially diluted and incubated with a fixed concentration of rabies virus (CVS-11; at a dose of 25 LD50) at 37°C for 60 min. The mixtures were inoculated in mice (NIH strain; 11–15 gms) at the dose of 30 µl/mice and were observed for 14 days for rabies specific deaths. The survivor data were transformed using the Reed and Muench method to calculate potency (IU/ml)
2.4. ELISA method
Sandwich ELISA was performed to quantify the rabies virus GP content in rabies vaccine formulations. Briefly, 96-well ELISA plates (Maxisorp; Nunc) coated with rabies monoclonal antibody (RAB-1) (100 ul/well with a target concentration of 0.5 µg per well) overnight (+4°C) using carbonate–bicarbonate buffer pH 9.6 (Sigma Aldrich, USA) were blocked with 3% bovine serum albumin (BSA; Sigma; >95% purity) at 37°C for 1 h. RS and test sample dilutions were prepared in blocking buffer and were added to washed plates and incubated for 30 min at 37°C. The GP captured by RAB-1 was detected using anti-rabies GP antibody (polyclonal antibody used at an optimized dilution of 1:4000, having an assigned potency of 964.8 IU/mL by mouse neutralization test and 749 IU/mL by RFFIT assay) after incubating at room temperature for 30 min. This is followed by incubation with anti-sheep HRP conjugated antibodies (Sigma-Aldrich, USA; 100 ul/well at optimized dilution of 1:6000) for 30 min at room temperature (18–25°C) followed by three times washing with wash buffer. The washed plates were developed using TMB (Sigma-Aldrich, USA; 100 ul/well) at room temperature for 15 min. The reaction was stopped by adding 1 M sulfuric acid (Qualigens, USA) and absorbance was measured at 450/630 nm using a plate reader (Biotek, Agilent Technologies, USA; 21 CFR compliant Gen-5 software). The GP content was calculated as IU/ml using four parametric fits.
2.5. Mouse potency assay (NIH method)
The test was performed as per standard guidance is given in WHO TRS and Indian/European Pharmacopoeia.Citation3 Mice (NIH; 11 to 15 g) were immunized with multi-dilutions of reference and test vaccine via intraperitoneal injections on days 0 and 7. On day 14, mice were challenged with an intracerebral injection of 30 µl of challenge strain (CVS11) at a dose of 25 LD50 per mouse. Mice were observed until day 28 and the number of survivors was used to estimate ED50 of vaccine using a national reference calibrated against WHO international standard (NIBSC) to obtain titer (IU/dose). All calculations are performed using CombiStats software (official computation software recommended by EP and followed by various regulatory agencies).
2.6. SRID method
Briefly, RS and test samples are treated with detergent (mulgofen) for 60 min at room temperature to release the GP. Three dilutions of test samples and RS (20 µl) were distributed (3 replicate each) in the wells punched in 1% agarose gel (Thermofisher, USA) containing immune sheep anti-GP serum raised against PM strain GP (7.5 ul/ml agarose gel). The gel is incubated for 48 h at 20–25°C and washed with phosphate-buffered saline, dried and stained with 0.05% Coomassie blue (Sigma Aldrich, USA) for detection of antigen-antibody precipitation zones. The diameters of resulting precipitating zones were plotted against concentration and analyzed using linear regression to obtain the content in IU/dose using validated Microsoft excel sheet functions.
2.7. Biolayer interferometry
Biolayer Interferometry (BLI) is a real-time and label-free technology that enables direct measurement of biomolecular interactions in solution. BLI as compared to other label-free biosensor technologies such as SPR (surface plasmon resonance) based Biacore offers simplicity in detection and sample delivery.Citation21 BLI uses a simple Dip-and-Read™ format in which biosensors are dipped into microplate wells containing purified or complex samples, which provide a highly parallel, user-friendly technique to study molecular interactions. The kinetics of binding of the RAB-1 to rabies vaccine was studied using a Forte Bio Octet instrument (Pall Life Science).Citation22 Briefly, protein A Biosensors (Pall Life Science) were loaded with RAB-1diluted in HEPES buffer (GE Healthcare Life Sciences). After capture, a 1-min wash in loading buffer was used to remove excess unbound RAB-1 to establish a new baseline signal. The sensors were dipped into wells containing rabies vaccine at different dilutions ranging from 0.612 to 10 ug/mL in loading buffer. The sensors were then dipped into wells containing loading buffer to measure the (off-rate) dissociation rate. Data analysis was performed using ForteBio Data Analysis software package 9.0.
2.8. Generation of sub-potent vaccine samples
The following treatments were used to generate sub-potent vaccine batches. The conditions were selected based on previous studies and known to induce significant degradation in the structure of rabies GP.Citation11 These samples were used for (a) comparability studies and (b) qualification of monoclonal antibody used in the assay. Details are presented in .
2.8.1. Heat treatment of vaccine batches
Vaccine lots (lyophilized) were reconstituted in diluents and were exposed to 60°C for 8 h.
2.8.2. Inactivation using dithiothreitol (DTT)
Reconstituted lyophilized vaccine batches were treated with DTT (100 mM; Sigma-Aldrich, USA). The resulting solutions were incubated at room temperature for 1 h. The mixture was filtered using 10 kDa cut off sample cartridges and stored at 4°C until further analysis.
2.8.3. Inactivation by SDS (sodium dodecyl sulfate)
Vaccine lots (lyophilized) were reconstituted in diluents and were exposed to 10% SDS for 1 h at room temperature. The mixture was filtered using 10 kDa cut off sample cartridges and stored at 4°C until further analysis.
2.8.4. Inactivation by sodium dodecyl sulfate (SDS) + heat
Vaccine batches (lyophilized) were reconstituted in diluents and were exposed to 10% SDS for 1 h at room temperature and followed by incubation at 90°C for 5 min.
2.8.5. Inactivation by DTT + heat
Reconstituted lyophilized vaccine batches were treated with dithiothreitol (100 mM) and followed by incubation at 90°C for 5 min.
2.9. Method validation
The method was validated according to the ICH and EMEA guidance.Citation23,Citation24 Specificity, linearity, accuracy, and precision of the method were assessed. The specificity of the assay was demonstrated using the spike recovery method using reference antigen in the vaccine formulations. Linearity is demonstrated using the calibration curve, wherein concentration and response relationship is fitted using the 4-PL curve fit. Intraday and Interday precision were determined by performing the assays on the same and different days, respectively. Accuracy was demonstrated by the method of spike recovery in the sample and degraded sample matrices.
2.10. Method applicability
The method applicability was demonstrated by monitoring the production of GP content of 24 batches of PM-derived human-inactivated rabies vaccines were determined using SRID and ELISA method. The potency of these batches was also analyzed using the NIH potency method. All the batches were evaluated against NIH potency specifications of ≥2.5 IU per single human dose.
3. Results
We report here the development and validation of sandwich ELISA for the estimation of the rabies virus GP using a specific combination of virus-neutralizing monoclonal and polyclonal antibodies. provides the overall strategy followed for the qualification and validation of the ELISA method.
3.1. Virus-neutralizing activities of antibodies
RAB-1 recognizes the antigenic site III of GP. AGS is a polyclonal antibody and is predicted to recognize both sites II and III of GP. The virus-neutralizing activities of RAB-1 and AGS were demonstrated using RFFIT and mouse neutralization potency methods using a standard challenge strain CVS11 (). AGS demonstrated higher neutralizing activity, which was expected as it is a high titer serum developed by repeated immunizations with purified GP.
Table 2. Characterization data for antibodies used in the assay.
3.2. Potential of RAB-1 to capture the intact form of viral GP – biolayer interferometry analysis
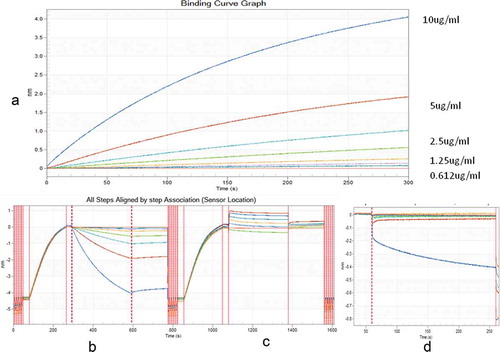
BLI is a widely used method for studying antibodies affinity and their association with the antigen in the subnanomolar range in the solution phase. (– shows the apparent binding affinities of RAB-1 to different concentrations of intact and degraded rabies antigens. RAB-1 showed a dose-dependent association with intact rabies antigen (). However, no dose-dependent response with degraded antigens was detected, indicative of RAB-1 potential to differentiate between an intact and degraded form of rabies GP in vaccine samples ().
Figure 1. Bioinferometric analysis (BLI) of binding kinetics of RAB-1 with the intact and degraded vaccine. RAB-1 was immobilized on protein A sensor and different concentrations of intact and degraded rabies vaccine were studied for binding kinetics. (a) depicts a binding curve of intact rabies vaccine with RAB-1. (b, c) depict a comparative overview of binding curves of intact and degraded rabies antigen, respectively. (d) depicts nonspecific binding (NSB), blank Pro A biosensor dipped in highest concentration rabies antigen.
3.3. Optimization of the ELISA method
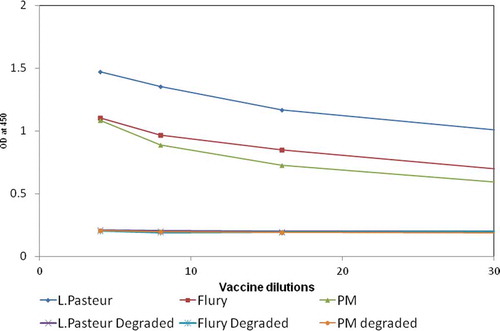
A checkerboard titration was used to determine the optimum amount of RAB-1 and AGS for the capture of GP in the assay. The highest and lowest detection limits were studied using PM strain. A coating concentration of 0.5 µg/well of RAB-1 and 1:4000 dilution of AGS resulted in the highest range with LLOQ of 0.072 IU/ml. The other steps in the method such as incubation temperature, duration of incubation, the concentration of conjugated antibody were also optimized. The temperature and duration of the incubation steps of the antigen with capture antibody and detection antibody were critical to the performance of the assay. The method conditions optimized with PM strain were extrapolated to rabies viruses of other strains. It was noted that the method did not show any bias in the capture of GP derived from Pasteur and Flury LEP strains, suggesting that optimized method conditions are robust and can be universally applied to other vaccines. depicts the dose–response curves obtained with vaccines derived from different strains and their respective sub-potent samples.
Figure 2. Depicts the potential of the method to detect concentration-dependent responses of vaccines manufactured using different strains of rabies virus such as Pasteur, Flury, and PM strain. The figure also depicts the potential of the method to detect degraded vaccine samples derived from different strains of rabies virus.
3.4. ELISA method validation
3.4.1. Calibration curve
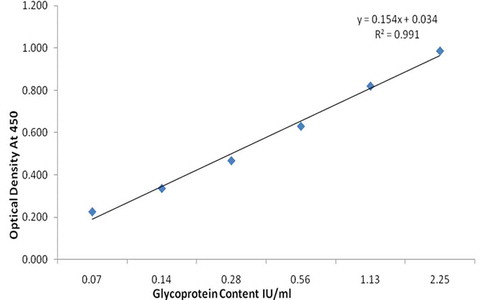
Four parametric curve back-fitted concentrations of the standard in the defined range (0.070 to 2.25 IU/ml) met the acceptance criteria of recovery within the range of 80–120% and replicate CV < 20% (). Vaccines derived from different strains were also evaluated for dilutional linearity. The back fit recovery values were observed from 97.46% to 107.55%.
3.4.2. Limits of quantification/detection
LLOQ of the assay was determined as the lowest calibration point for which the concentration can be back-calculated on the regression curve with 80–120% accuracy and a CV below 20%. The ULOQ is the upper calibration point that meets these criteria. The dynamic range thus extends from the LLOQs to ULOQs. The dynamic range of the assay was found from 0.070 to 2.25 IU/ml. Therefore, the LLOQ of the method is 0.072 IU/ml, given the sample is diluted to a minimum of 1:2 dilution before analysis. The LOD was calculated from the mean signal at background + three standard deviations. The LOD of the assay, calculated from six assays, was 0.035 IU/ml ().
Table 3. Method Validation: Summary of method linearity estimates.
3.4.3. Precision
Repeatability and intermediate precision of the method were assessed for vaccines derived from three different strains, namely LEP, PM, and Flurry. The percent CVs for intra-assay and inter-assay variations for PM-derived vaccine ranged from 2.23% to 10.17%. For Flurry LEP-derived vaccine, precision was in the range of 6.66% to 13.32%. Pasteur strain-derived vaccine showed intra-assay and inter-assay variations in the range of 3.78–10.65%. ()
Table 4. Method Validation: Summary of method precision results.
3.4.4. Accuracy
The accuracy of the method was demonstrated using two methods (A and B). In method A, the sample matrix was spiked with 0.072 IU/ml (low spike; LLOQ), 0.52 IU/ml and 2.25 IU/mL (high spike) of RS and accuracy were assessed at multiple dilutions of spiked samples. The accuracy of the measurement of spiked serum samples ranged from 88% to 114%, thus meeting the acceptance criteria (). These data showed that the sample matrix does not interfere with the method. In method B, degraded samples were spiked with intact antigen and studied using ELISA. The assay showed excellent recovery (84–91%) of intact antigen in a degraded sample matrix supporting the assay capability to capture antigenically active GP in the presence of degraded matrix.
Table 5. Summary of method accuracy data.
3.5. Agreement of ELISA method with SRID and NIH potency assays
A panel of samples representing sub-potent/degraded form of the PM strain-derived vaccine were analyzed using ELISA, SRID, and NIH methods to evaluate their ability to differentiate between intact and sub-potent form of the antigens. A decrease in ELISA values (below detection limits) indicative of reduced capture was observed for degraded samples indicating the ability of the method to detect changes in antigen integrity. Both SRID and NIH potency tests also demonstrated a significant difference between normal and degraded samples, suggesting an agreement with ELISA results. The degraded samples which were below the detection limit in the ELISA method were also not detected by SRID and NIH potency tests ( and ).
Table 7. Capture of normal and degraded samples in different methods.
The agreement between ELISA and NIH potency tests was also assessed on intact and degraded antigens derived from Flurry LEP and Pasteur strains. The samples found sub-potent by ELISA method did not show protection in NIH assay, indicating that ELISA can differentiate between potent and sub-potent batches ().
3.6. ELISA applicability in consistency monitoring of production batches
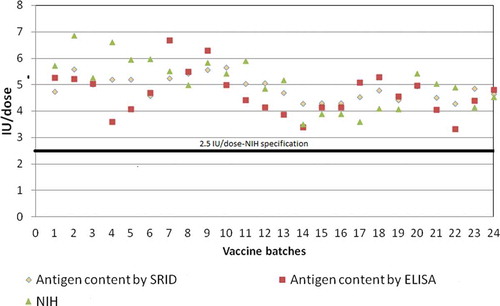
Twenty-four batches of PM derived inactivated rabies vaccine was tested for GP content using ELISA and SRID. Their potency was tested by the NIH method (). A good agreement between the three methods was observed wherein the batches showing GP content of higher than 2.5 IU/dose were found conforming to NIH potency specification of ≥2.5 IU per single human dose.
4. Discussion and conclusion
The 3Rs concept (reduction, refinement, and replacement) is currently being promoted globally in quality control testing of vaccines. Vaccine manufacturers are expected to develop and implement new methods as an alternative to in vivo animal testing. The NIH method is one such test that is being pursued under 3R global initiatives in lot release testing of human rabies vaccine. Validated in-vitro assays for quantification of rabies virus GP using virus-neutralizing antibodies have the potential to replace the NIH method. The sandwich ELISA method reported here can recognize and quantify the intact GP in vaccine samples. This capacity is demonstrated using two virus-neutralizing antibodies, RAB-1 and polyclonal anti-GP antibody in the assay. The method is novel as compared to the prior art as it uses a site III directed human monoclonal antibody with clinically proven virus-neutralizing activity. Additional data including BLI analysis were generated to support that RAB-1 is neutralizing and detects only the antigenic form of GP. Label-free technologies, such as BLI, offer means to acquire data on both binding and dissociation phases of the Ag–Ab interactions. BLI was used for measuring antigen–antibody interactions of mAbs with influenza and malaria vaccine antigens.Citation25,Citation26 This study using BLI demonstrated that the assay can detect GP even from different strains of rabies virus with high specificity, linearity, precision, and accuracy. We also attempted the evaluation of AGS as a detection antibody using BLI analysis; however, AGS, being a polyclonal antibody showed inconsistencies in coupling to BLI probes. Studies are in progress to establish BLI assay with AGS.
Few studies have reported variable correlations of ELISA methods with NIH; however, there is a consensus that such correlations will be challenging owing to inherent variabilities of NIH method.Citation11,Citation24 Therefore, in this study agreement with NIH method was studied using two approaches. In the first approach, a panel of samples derived from different strains and exposed to different stress conditions were evaluated by ELISA and NIH method. In the second approach, agreement of NIH, SRID and ELISA methods was studied using 24 different batches of PM strain derived inactivated vaccine. All the batches that were found complying with NIH (≥2.5 IU per single human dose) also gave confirming results with the ELISA method. Both the methods showed agreement in discriminating between potent and degraded samples. This could be attributed to the use of RAB-1 as the capture antibody as the majority of the neutralizing antibodies are directed against the G-protein domain III during the infection or virus challenge in the NIH method.Citation27
The study characterized the activity and role of RAB-1 in terms of capture and neutralizing activities. The limitations of the study include that it does not address the possible role of AGS in the performance of the method, especially in terms of its activity concerning antigenic regions of GP. Competitive inhibition studies using different monoclonal antibodies directed against sites II and III are in progress to characterize the role of AGS in the method. Additionally, the study used different stress methods to completely degrade the rabies human vaccine, which was very well detected by the assay. It will be worthwhile to evaluate the performance of the assay with partially degraded antigens vis a vis NIH method results in further support of the assay.
In conclusion, the method described here provides a simple and efficient tool for quantification of GP in vaccine preparations. The method uses a novel combination of neutralizing antibodies, which enables reliable detection of the natively folded Rabies virus GP antigen in vaccine preparations. With an LLOQ of 0.072 IU/ml, the method can detect even slight changes in the antigen content, which will be useful for tracking the stability of vaccine batches and routine monitoring of vaccine batches for consistency. Overall the data suggest that the method meets all the requirements to be used as a possible alternative to NIH test in lot release testing of human rabies vaccines.
Disclosure of potential conflicts of interest
The authors declare no conflict of interest. The opinions expressed in this article are of authors and do not necessarily express opinions of institute/company.
Acknowledgments
The authors are grateful to Serum Institute of India Private Limited for providing all the facilities to carry out this study.
References
- WHO Fact Sheet on Rabies. 2019 Sept 27 [Assessed 2019 Nov 3]. https://www.who.int/news-room/fact-sheets/detail/rabies.
- Recommendations for an inactivated rabies vaccine for human use produced in cell substrates and embryonated eggs. WHO TRS941 (Annex II); 2007.
- Rabies Vaccine Monograph for Human use, Ed. Ph Eur. Monograph 2018. Strasbourg (France): Council of Europe; 2015.
- Barth R, Diderrich G, Weinmann E. NIH test, a problematic method for testing potency of inactivated rabies vaccine. Vaccine. 1988 Aug;6(4):369–77. doi:10.1016/0264-410X(88)90185-5.
- Shank-Retzlaff M, Wang F, Morley T, Anderson C, Hamm M, Brown M, Rowland K, Pancari G, Zorman J, Lowe R, et al. Correlation between mouse potency and in vitro relative potency for human Papillomavirus Type 16 virus-like particles and gardasil (R) vaccine samples. Hum Vaccin. 2005;1:191–97. doi:10.4161/hv.1.5.2126.
- Giffroy D, Mazy C, Duchêne M. Validation of a new ELISA method for in vitro potency assay of hepatitis B-containing vaccines. Pharmeuropa Bio. 2006;2006:7–14.
- Perrin P, Thibodeau L, Sureau P. Rabies immunome (subunit vaccine) structure and immunogenicity. Pre- and post-exposure protection studies. Vaccine. 1985;3:325–32.
- Celis E, Ou D, Dietzschold B, Koprowski H. Recognition of rabies and rabies-related viruses by T cells derived from human vaccine recipients. J Virol. 1988;62:3128–34.
- Stokes W, McFarland R, Kulpa-Eddy J, Gatewood D, Levis R, Halder M, Pulle G, Kojima H, Casey W, Gaydamaka A, et al. Report on the international workshop on alternative methods for human and veterinary rabies vaccine testing: state of the science and planning the way forward. Biologicals. 2012;40(5):369–81. doi:10.1016/j.biologicals.2012.07.005.
- Isbrucker R, Levis R, Casey W, McFarland R, Schmitt M, Arciniega J. Alternative methods and strategies to reduce, refine, and replace animal use for human vaccine post-licensing safety testing: state of the science and future directions. Proc Vaccinol. 2011;5:47e59. antibodies. Microbiol Immunol 1998;42:187e93. doi:10.1016/j.provac.2011.10.004.
- Luo TR, Minamoto N, Hishida M, Yamamoto K, Fujise T, Hiraga S, Ito N, Sugiyama M, Kinjo T. Antigenic and functional analyses of the GP of rabies virus using monoclonal antibodies. Microbiol Immunol. 1998;42:187e93.
- Wiktor TJ, Gyorgy E, Schlumberger D, Sokol F, Koprowski H. Antigenic properties of rabies virus components. J Immunol. 1973;110:269e76.
- Nagarajan T, Reddy GS, Mohana Subramanian B, Rajalakshmi S, Thiagarajan D, Tordo N, Jallet C, Srinivasan VA. A simple immuno-capture ELISA to estimate rabies viral GP antigen in vaccine manufacture. Biologicals. 2006;34:21e7.
- Gibert R, Alberti M, Poirier B, Jallet C, Tordo N, Morgeaux S. A relevant in vitro ELISA test in alternative to the in vivo NIH method for human rabies vaccine batch release. Vaccine. 2013;31:6022e9. doi:10.1016/j.vaccine.2013.10.019.
- Gamoh K, Senda M, Itoh O, Muramatsu M, Hirayama N, Koike R, Endoh YS, Minamoto N. Use of ELISA for in vitro potency test of rabies vaccines for animal use. Biologicals. 1996;24:95e101.
- Morgeaux S, Poirier B, Ragan CI, Wilkinson D, Arabin U, Guinet-Morlot F, Levis R, Meyer H, Riou P, Shaid S, et al. Replacement of in vivo human rabies vaccine potency testing by in vitro GP quantification using ELISA - Results of an international collaborative study. Vaccine. 2017 7;35(6):966–71. doi:10.1016/j.vaccine.2016.12.039.
- Sloan SE, Hanlon C, Weldon W, Niezgoda M, Blanton J, Self J, Rowley KJ, Mandell RB, Babcock GJ, Thomas WD Jr, et al. Identification and characterization of a human monoclonal antibody that potently neutralizes a broad panel of rabies virus isolates. Vaccine. 2007 12;25(15):2800–10. doi:10.1016/j.vaccine.2006.12.031.
- Wang Y, Rowley KJ, Booth BJ, Sloan SE, Ambrosino DM, Babcock GJ. G GP amino acid residues required for human monoclonal antibody RAB1 neutralization are conserved in rabies virus street isolates. Antiviral Res. 2011;91(2):187–94. doi:10.1016/j.antiviral.2011.06.002.
- Gogtay N, Thatte U, Kshirsagar N, Leav B, Moline D, Cheslock P, Kapre SV, Kulkarni PS. SII RMab author group. Safety and pharmacokinetics of a human monoclonal antibody to rabies virus: a randomized, dose-escalation phase 1 study in adults. Vaccine. 2012;30:7315–20.
- Gogtay NJ, Munshi R, Ashwath Narayana DH, Mahendra BJ, Kshirsagar V, Gunale B, Moore S, Cheslock P, Thaker S, Deshpande S, et al. Comparison of a novel human rabies monoclonal antibody to human rabies immunoglobulin for postexposure prophylaxis: a Phase 2/3, randomized, single-blind, noninferiority, controlled study. Clin Infect Dis. 2018 18;66(3):387–95. doi:10.1093/cid/cix791.
- Kamat V, Rafique A. Designing binding kinetic assay on the bio-layer interferometry (BLI) biosensor to characterize antibody-antigen interactions. Anal Biochem. 2017;536:16–31. doi:10.1016/j.ab.2017.08.002.
- Kumaraswamy S, Tobias R. Label-free kinetic analysis of an antibody-antigen interaction using biolayer interferometry. Methods Mol Biol. 2015;1278:165–82.
- Guideline on bioanalytical method validation EMEA/CHMP/EWP/192217/2009Rev. 1
- ICH M10 guideline on bioanalytical method validation.
- Carney PJ, Lipatov AS, Monto AS, Donis RO, Stevens J. Flexible label-free quantitative assay for antibodies to influenza virus hemagglutinins. Clin Vaccine Immunol. 2010;17:1407–16.
- Dennison SM, Reichartz M, Seaton KE, Dutta S, Wille-Reece U, Hill AVS, Ewer KJ, Rountree W, Sarzotti-Kelsoe M, Ozaki DA, et al. Qualified biolayer interferometry avidity measurements distinguish the heterogeneity of antibody interactions with Plasmodium falciparum circumsporozoite protein antigens. J Immunol. 2018;201(4):1315–26. doi:10.4049/jimmunol.1800323.
- Kramer RA, Marissen WE, Goudsmit J, Visser TJ, Clijsters-van der Horst M, Bakker AQ, de Jong M, Jongeneelen M, Thijsse S, Backus HH, et al. The human antibody repertoire specific for rabies virus GP as selected from immune libraries. Eur J Immunol. 2005;35:2131–45. doi:10.1002/eji.200526134.