ABSTRACT
A SARS-CoV receptor-binding domain (RBD) recombinant protein was developed and manufactured under current good manufacturing practices in 2016. The protein, known as RBD219-N1 when formulated on Alhydrogel®, induced high-level neutralizing antibodies and protective immunity with minimal immunopathology in mice after a homologous virus challenge with SARS-CoV (MA15 strain). We examined published evidence in support of whether the SARS-CoV RBD219-N1 could be repurposed as a heterologous vaccine against Coronavirus Infectious Disease (COVID)-19. Our findings include evidence that convalescent serum from SARS-CoV patients can neutralize SARS-CoV-2. Additionally, a review of published studies using monoclonal antibodies (mAbs) raised against SARS-CoV RBD and that neutralizes the SARS-CoV virus in vitro finds that some of these mAbs bind to the receptor-binding motif (RBM) within the RBD, while others bind to domains outside this region within RBD. This information is relevant and supports the possibility of developing a heterologous SARS-CoV RBD vaccine against COVID-19, especially due to the finding that the overall high amino acid similarity (82%) between SARS-CoV and SARS-CoV-2 spike and RBD domains is not reflected in RBM amino acid similarity (59%). However, the high sequence similarity (94%) in the region outside of RBM offers the potential of conserved neutralizing epitopes between both viruses.
Introduction
Coronavirus disease 2019, or COVID-19Citation1, is an emerging disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Similar to SARS coronavirus (SARS-CoV), SARS-CoV-2 can cause severe respiratory illness and significant mortality among those over 60 y or with chronic conditions.Citation2 With an estimated reproductive number (R0) of 2.24–3.58,Citation3 the outbreak originated from Wuhan, China quickly spread across China and at least 25 other nations.Citation2 In addition, SARS-CoV-2 may be transmitted from infected individuals without symptoms,Citation3 which could increase the challenges for controlling the outbreak without the prospect of a vaccine.
A SARS-CoV receptor-binding domain (RBD) recombinant protein was developed and manufactured under current good manufacturing practices (cGMP) in 2016.Citation4-Citation6 The bulk drug substance has been stored frozen (−70°C to −80°C) and is under stability testing since its manufacturing, so far remaining stable. The protein known as RBD219-N1 () was expressed in yeast (Pichia pastoris X33) and purified to optimize expression yield, antigenicity, and functionality, as well as immunogenicity in mice when formulated on alum.Citation4,Citation6 Moreover, alum-adjuvanted RBD219-N1 induced protective immunity against homologous virus challenge with SARS-CoV (MA15 lethal strain), with minimal immunopathology, lessening potential safety concerns.Citation5 The high levels of protein expression in yeast, the relative ease of purification and its stability profile raise the possibility that this vaccine could be produced at a low cost for stockpiling or distribution among at-risk populations. Accordingly, we are therefore investigating whether the SARS-CoV RBD recombinant protein candidate could potentially be repurposed as a heterologous vaccine for SARS-CoV-2.
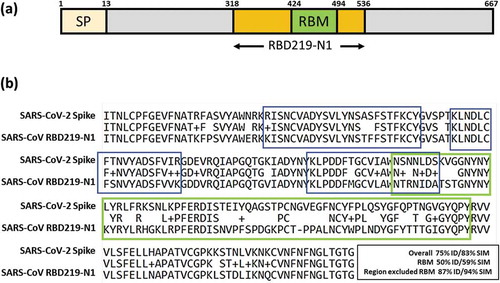
Figure 1. (a) Illustration of SARS-CoV RBD subunit S1. (b) Sequence alignment between SARS-CoV RBD219-N1 and SARS-CoV-2 spike protein. The RBM region is circled in green. An example of a neutralizing conformational epitope consisting S343–367, 373–390 and 411–428 (reported by Bian et al.) is circled in blue, indicating neutralizing epitopes do not have to be within RBM.Citation7
SARS-CoV and SARS-CoV-2
Like the SARS coronavirus, SARS-CoV-2 is closely related to bat SARS-like coronavirus.Citation8 The Receptor Binding Domain of the SARS-CoV-2 and SARS-CoV RBD219-N1 share substantial amino acid sequence similarity (75% identity, 83% similarity) () and evidence indicates that both coronaviruses use the human receptor angiotensin converting enzyme 2 (ACE2) for cell entry.Citation9,Citation10 Antibodies induced by anti-SARS vaccines can cross-neutralize bat SARS-like coronaviruses (SL-CoVs);Citation11 most importantly, serum from a convalescent SARS-CoV patient neutralized SARS-CoV-2-driven entry.Citation10 These findings suggest the possible cross-protection of using SARS-CoV RBD as the antigen against SARS-CoV-2.
Anti-SARS-CoV RBD neutralizing monoclonal antibodies
It is known that the blockage of the receptor-binding motif (RBM) within the RBD and the ACE2 association site, is a major mechanism of SARS-CoV neutralization. However, despite the overall high level of homology between the two SARS-CoV RBDs, it has been noted that the similarity of 70 amino acids in the RBM (S424−494) between SARS-CoV and SARS-CoV-2 is only 59%, and thus, the neutralizing antibodies (mAbs) raised from the RBM of SARS-CoV may have limited cross-reactivity to SARS-CoV-2. Nevertheless, several groups have shown (typically via a neutralization assay in Vero E6 cells) that neutralizing antibodies recognize epitopes other than in the RBM region. Using RBD proteins expressed in mammalian cells (293 T cells), He et al. reported on 27 anti-SARS-CoV RBD mAbs with 23 of them showing neutralizing activity (), and of these, some interfered with virus binding to the ACE2 receptor by affecting the binding of the RBM, but many achieved virus neutralization by recognizing epitopes outside of the RBM.Citation12 In some cases, it is likely that these non-RBM directed mAbs caused conformational changes that indirectly affected RBM binding or through mechanisms as yet undetermined.Citation13 Among these 23 neutralizing mAbs, 5 mAbs, including 24H8, 19B2, 35B5, 33G4 and 31H12, were used for a binding study on RBD219-N1, with all 5 neutralizing mAbs against both the RBM and non-RBM regions also recognized the RBD219-N1 recombinant protein.Citation6 He et al. also found neutralizing mAbs against non-RBM domains using a baculovirus expressed RBD,Citation12 while Bian et al. further identified one conformational neutralizing epitope consisting S343–367, 373–390 and 411–428,Citation7 which was on the RBD but outside of the RBM. Additionally, CR3022, a potent human neutralizing mAb, which was derived from a single-chain variable antibody fragment (scFv) phage display library was shown to bind to the RBD domain, but outside the RBM region.Citation14
Table 1. Neutralizing monoclonal antibodies reported in He et al, 2005Citation11 were categorized based on its ability to inhibit RBM binding to the ACE2.
An important conclusion of these published studies was that virus neutralization does not depend on interference with the RBM. While the mechanism of action of these mAbs requires additional studies, some appear to bind to sites outside the RBM possibly by causing conformational changes to the RBD, while others still neutralize without directly inhibiting ACE2 binding in vitro.
In more recent studies, Tian et al.Citation15 and Wrapp et al.Citation16 have used five anti-SARS-CoV RBD neutralizing mAbs to evaluate their cross-reactivity to SARS-CoV-2 RBD (). Among these five neutralizing mAbs, the four mAbs that bound the epitopes in or close to RBM expectedly only had weak or no binding, while CR3022, which recognized the epitope outside of RBM, showed potent binding. Considering the highly conserved – a similarity of 94% – amino acid sequence of RBD region after excluding the RBM (), the possibility remains that antibodies raised from the epitopes outside of the RBM region may both show cross-reactivity and induce neutralizing antibodies.
Table 2. Binding study of anti-SARS-CoV RBD neutralizing mAb against the SARS-CoV-2 RBD
Concluding comments
The yeast-expressed SARS-CoV RBD219-N1 recombinant protein has been manufactured under cGMP and could soon enter clinical testing. Even though blockage of the RBM is a major mechanism of SARS-CoV neutralization, it was proven that neutralizing antibodies can also be raised from epitopes outside of RBM. Indeed, at least one neutralizing mAb that recognizes both SARS-CoV and SARS-CoV-2 binds to a domain outside the RBM. Despite the low amino acid similarity of RBM region between SARS-CoV and SARS-CoV-2, its high amino acid similarity with the homologous RBD from SARS-CoV-2, and its potential for raising neutralizing antibodies (when formulated on alum), especially against epitopes outside the RBM, offers the possibility that it might partially protect against COVID-19. Therefore, a logical and crucial next step would be that, in parallel to efforts to initiate the development of a homologous SARS-CoV-2 recombinant RBD protein vaccine, we accelerate the evaluation of a heterologous vaccine into safety clinical testing. Clinical data obtained from these studies would greatly assist in the development of a safe, immunogenic and effective vaccine against Coronaviruses.
Disclosure of potential conflicts of interest
The authors have developed subunit vaccines against SARS and MERS coronavirus infections. They are involved in the process of developing a vaccine against SARS-CoV-2.
References
- World Health Organization. WHO director-general’s remarks at the media briefing on 2019-nCoV on 11 February 2020. Ed; 2020 [cited 2020 Apr 2]. Available from: https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020. doi:10.1056/NEJMoa2001316.
- Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970–71. doi:10.1056/NEJMc2001468.
- Chen WH, Chag SM, Poongavanam MV, Biter AB, Ewere EA, Rezende W, Seid CA, Hudspeth EM, Pollet J, McAtee CP, et al. Optimization of the production process and characterization of the yeast-expressed SARS-CoV Recombinant Receptor-Binding Domain (RBD219-N1), a SARS vaccine candidate. J Pharm Sci. 2017;106(8):1961–70. doi:10.1016/j.xphs.2017.04.037.
- Jiang S, Bottazzi ME, Du L, Lustigman S, Tseng CT, Curti E, Jones K, Zhan B, Hotez PJ. Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome. Expert Rev Vaccines. 2012;11(12):1405–13. doi:10.1586/erv.12.126.
- Chen WH, Du L, Chag SM, Ma C, Tricoche N, Tao X, Seid CA, Hudspeth EM, Lustigman S, Tseng CT, et al. Yeast-expressed recombinant protein of the receptor-binding domain in SARS-CoV spike protein with deglycosylated forms as a SARS vaccine candidate. Hum Vaccin Immunother. 2014;10(3):648–58. doi:10.4161/hv.27464.
- Bian C, Zhang X, Cai X, Zhang L, Chen Z, Zha Y, Xu Y, Xu K, Lu W, Yan L, et al. Conserved amino acids W423 and N424 in receptor-binding domain of SARS-CoV are potential targets for therapeutic monoclonal antibody. Virology. 2009;383(1):39–46. doi:10.1016/j.virol.2008.09.029.
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Hu Y, Song Z-G, Tao Z-W, Tian J-H, Pei -Y-Y, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269.
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273.
- Hoffmann M, Kleine-Weber H, Krüger N, Müller M, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020. Advance online publication.
- Zeng LP, Ge XY, Peng C, Tai W, Jiang S, Du L, Shi ZL. Cross-neutralization of SARS coronavirus-specific antibodies against bat SARS-like coronaviruses. Sci China Life Sci. 2017;60(12):1399–402. doi:10.1007/s11427-017-9189-3.
- He Y, Li J, Heck S, Lustigman S, Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J Virol. 2006;80(12):5757–67. doi:10.1128/JVI.00083-06.
- He Y, Lu H, Siddiqui P, Zhou Y, Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J Immunol. 2005;174(8):4908–15. doi:10.4049/jimmunol.174.8.4908.
- Ter Meulen J, van den Brink EN, Poon LL, Marissen WE, Leung CS, Cox F, Cheung CY, Bakker AQ, Bogaards JA, van Deventer E, et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3(7):e237. doi:10.1371/journal.pmed.0030237.
- Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerging Microbes Infect. 2020;9(1):382–85. doi:10.1080/22221751.2020.1729069.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;eabb2507. doi:10.1126/science.abb2507.
- Ter Meulen J, Bakker AB, van den Brink EN, Weverling GJ, Martina BE, Haagmans BL, Kuiken T, de Kruif J, Preiser W, Spaan W, et al. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363(9427):2139–41. doi:10.1016/S0140-6736(04)16506-9.
- van den Brink EN, Ter Meulen J, Cox F, Jongeneelen MA, Thijsse A, Throsby M, Marissen WE, Rood PM, Bakker AB, Gelderblom HR, et al. Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J Virol. 2005;79(3):1635–44. doi:10.1128/JVI.79.3.1635-1644.2005.
- Prabakaran P, Gan J, Feng Y, Zhu Z, Choudhry V, Xiao X, Ji X, Dimitrov DS. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J Biol Chem. 2006;281(23):15829–36. doi:10.1074/jbc.M600697200.
- Zhu Z, Chakraborti S, He Y, Roberts A, Sheahan T, Xiao X, Hensley LE, Prabakaran P, Rockx B, Sidorov IA, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A. 2007;104(29):12123–28. doi:10.1073/pnas.0701000104.
- Hwang WC, Lin Y, Santelli E, Sui J, Jaroszewski L, Stec B, Farzan M, Marasco WA, Liddington RC. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J Biol Chem. 2006;281(45):34610–16. doi:10.1074/jbc.M603275200.
- Rockx B, Corti D, Donaldson E, Sheahan T, Stadler K, Lanzavecchia A, Baric R. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J Virol. 2008;82(7):3220–35. doi:10.1128/JVI.02377-07.
