ABSTRACT
Respiratory Syncytial virus (RSV) is a major threat to many vulnerable populations. There are currently no approved vaccines, and RSV remains a high unmet global medical need. Here we describe the employment of a novel synthetic DNA-encoded antibody technology platform to develop and deliver an engineered human DNA-encoded monoclonal antibody (dMAbTM) targeting the fusion protein (F) of RSV as a new approach to prevention or therapy of at risk populations. In in vivo models, a single administration of synthetic DNA-encoding the single-chain fragment variable-constant fragment (scFv-Fc) RSV-F dMAb resulted in robust and durable circulating levels of a functional antibody systemically and in mucosal tissue. In cotton rats, which are the gold-standard animals to model RSV infection, we observed sustained scFv-Fc RSV-F dMAb in the sera and lung-lavage samples, demonstrating the potential for both long-lasting immunity to RSV and effective biodistribution. The scFv-Fc RSV-F dMAb harbored in the sera exhibited RSV antigen-specific binding and potent viral neutralizing activity. Importantly, in vivo delivery of synthetic DNA-encoding, the scFv-Fc RSV-F dMAb protected animals against viral challenge. Our findings support the significance of dMAbs as a potential platform technology for durable protection against RSV disease.
Introduction
RSV is a major global health burden. RSV complications associated with lower respiratory infections are the major cause of hospitalization for infants in the US resulting in between 58 and 132 K hospitalizations per year, 1.5 M-1.7 M office visits, and 400–500 K emergency room visits in children under 5 years of ageCitation1. Globally, for the year 2015, 59,600 children 5 years or younger were estimated to have succumbed to RSV-related acute lower respiratory infections. There are close to 180 K hospitalizations in the elderly population per year in the US.Citation2
Conventionally, vaccination has been the approach to provide sustained immunity to an infectious agent. However, a RSV vaccine remains an unmet need, and there has been a long track record of vaccine candidates failing to elicit consistent immune responses which translate to a sufficient levels of protection across the population.Citation3 Furthermore, serious vaccine enhanced disease (VED) was observed in some infants immunized with the formalin-inactivated RSV vaccine.Citation4 Thus, a strategy which bypasses the requirement to engage the host immune system would be very attractive. Passive immunizations with immunoprophylactic anti-RSV-F antibodies have proved successful in reducing disease in vulnerable pediatric patients.Citation5-Citation7 In the US, the FDA-approved recombinant monoclonal antibody, Palivizumab (Synagis) is a standard-of-care prophylaxis for lower respiratory disease (LRD) in high-risk infants, reducing hospitalizations of infected infants by 50%. However, the cost burden associated with Palivizumab restricts its use to limited populations even in high-income countries. Furthermore, the short biological half-life of the recombinant mAb means multiple rounds of dosing are required to maintain effective circulating concentrations of Palivizumab throughout the RSV-season.Citation8 These burdens and the complexities involved in development, manufacture, cold chain distribution, and often lack of suitable dosing methods which can be employed in the field, have hindered patient access not just to RSV mAbs, but too many other effective mAb-based prophylactics and therapeutics targeting a wide spectrum of diseases.Citation9
With the goal of permitting unrestricted global access to mAbs, new paradigms are being developed. Some of these leverage the recent developments in the in vivo delivery of genes encoding for antibodies. One highly promising technology is the in vivo delivery of a non-viral, synthetic DNA-encoding monoclonal antibody (dMAb). The synthetic DNA is delivered directly to the tissue of choice, and the encoded antibody is expressed and secreted into circulation. An electroporation (EP)-based delivery by CELLECTRA® using an optimized protocol ensures robust local tissue expression and high systemic levels of the mAb.Citation10,Citation11 Employment of this platform to endow a state of immunity against invading pathogens is termed active immunoprophylaxis. In proof-of-concept studies dMAbs have provided protection against various viral and bacterial infectious diseases, including influenza virus, pseudomonas, Lyme disease, Chikungunya virus and Ebola, Zika, and HIV, as well as efficacy in the treatment of malignancies in preclinical small and large animal models.Citation12 Citation13 Citation14 Citation15 Citation16 Citation17 Citation18 Citation19 Citation20 Citation21-Citation22 The following characteristics highlight the strength of the dMAb technology: the drug product is a non-viral, plasmid DNA (pDNA); lack of anti-vector immunity post-administration allows for continued and repeated use of dMAbs for a multitude targets; pDNA formulations are highly thermostable, allowing cold chain distribution to be bypassed; and synthetic DNA can be rapidly engineered in silico and manufactured at a cost of goods compatible with unrestricted global usage.Citation23 Importantly, with the employment of adaptive in vivo EP systems, pDNA can be efficiently delivered into highly accessible tissues sites (skeletal muscle or skin), permitting in field administration protocols lasting seconds rather than through lengthy IV administrations which is often required for recombinant mAbs. Furthermore, similar to half-life extension technologies, dMAb can be expressed in vivo at consistent levels for months, limiting the requirement for repeat dosing for seasonal coverage against pathogens such as RSV and Influenza.Citation24
In this study, we delineate the development of a human dMAb targeting the same RSV-Fusion antigen as Palivizumab. We designed a novel synthetic DNA sequence, pGX9369, encoding for RSV-F dMAb, scFv-Fc against RSV-F antigen. The scFv-Fc format has previously been shown to benefit half-life, effector-function, and biodistribution.Citation25,Citation26 Intramuscular (IM) delivery of pGX9369 led to robust in vivo levels of the functional scFv-Fc RSV-F mAb in the serum and lungs of experimental animals. Importantly, this treatment provided protection from LRD in a live virus challenge study in the cotton rat, the gold standard animal surrogate for evaluating potential RSV VED, as well as human RSV infection now that chimpanzees can no longer be used.Citation27 Our findings highlight the potential of dMAb technology as a prophylactic platform to fight infectious disease.
Materials and methods
SDS-PAGE and western blot
Lipofectamine 3000 transfection kit (ThermoFisher, Waltham, MA) was used to transfect adherent HEK 293 T cells (ATCC® CRL11268™) with plasmids encoding for human RSV IgG and scFv-Fc RSV dMAb. Media was harvested 72-h post-transfection and filtered using 0.22 µm stericup-GP vacuum filtration system (Millipore, Burlington, MA). The supernatant from scFv-Fc RSV dMAb transfected cells was purified using Protein A spin columns (ThermoFisher) by following the manufacturer’s instructions. The eluted protein was concentrated by Amicon Ultra-15 Centrifugal Filter unit (30 kDa), and quantified by Nanodrop. The supernatant from human RSV IgG transfected cells was purified using Protein G GraviTrap (GE Healthcare, Chicago, IL) using manufacturer’s instructions. The eluted protein was concentrated by Amicon Ultra-15 Centrifugal Filter unit (30 kDa). For each sample, 10 µg were loaded on a NuPAGE™ 4–12% Bis-Tris gel (ThermoFisher). Precision Plus Protein Kaleidoscope Prestained Protein Standard (Bio-Rad, Hercules, CA) was used as the standard marker. After electrophoresis, the proteins were transferred to PVDF membrane using iBlot™ 2 Transfer device (Invitrogen, Waltham, MA). The membrane was blocked with goat histology buffer (1% (w/v) BSA (Sigma, St. Louis, MO), 2% (v/v) goat serum (Sigma), 0.3% (v/v) Triton-X (Sigma) and 0.025% (v/v) 1 g/ml Sodium Azide (Sigma) in PBS) for 30 minutes at room temperature (RT). Goat anti-Human IgG-heavy and light chain monkey-adsorbed Antibody HRP Conjugated (A80-319P, Bethyl, Montgomery, TX) diluted1:5000 in goat histology buffer was added and incubated for 1 h at RT. After washing the blot three times for 5 minutes in 1X DPBS (HyClone, Logan, UT) goat anti-Human IgG-Fc Fragment Antibody HRP (A80-104P, Bethyl) diluted 1:5000 in goat histology buffer was added and incubated for 1 h at RT. After washing the blot three times for 5 minutes in 1X DPBS, the membrane was developed using ECL™ Prime Western Blotting system (GE Healthcare) and imaged using Protein Simple FluorChem System (Proteinsimple, CA).
Animals
Female BALB/c mice (Jackson Laboratory, ME) between 4 and 6 weeks of age and cotton rats (Jackson Laboratory, ME) between 6 and 8 week of age were group-housed with ad libitum access to feed and water. Husbandry was provided by Acculab (San Diego) and all procedures were in compliance with the standards and protocols of the Institutional Animal Care and Use Committee (IACUC) at Acculab.
Animal treatments
To deplete T cell populations BALB/c mice received one i.p. injection of 500ug of anti-mouse CD4 (, BE0003-1, BioXCell, West Lebanon, NH) and 500 µg of anti-mouse CD8α (BioXCell, BE0117) in 300 µl of 1X PBS. DMAb pDNA was formulated with 117.8–128.5 U/ml of human recombinant hyaluronidase (Hylenex®, Halozyme, San Diego, CA). Mice received 30 µl and cotton rats 200 µl IM injections of DNA formulation into tibialis anterior (TA) and quadriceps muscles. Mice received a pDNA formulation of 1.6 mg/ml and cotton rats received 2 mg/ml. The optimal concentration was determined in previous dosing studies (data not shown). EP was delivered at the injection site with the CELLECTRA-3P® adaptive in vivo EP system (Inovio Pharmaceuticals, PA). An array of three needle electrodes with 3 mm insertion depth was used for mice and 6 mm depth for cotton rats. The CELLECTRA® EP delivery consists of two sets of pulses with 0.1 Amp constant current, each composed of two 52 ms pulses, a 198 ms delay between pulses within a single set and 3 sec between the two sets. 50 µl Synagis (MedImmune, Gaithersburg, MD) was injected IM at a dose of 15 mg/kg.
Immunofluorescence staining
In vivo expressed dMAb: 7 days after IM delivery of 50 µg pGX9369 into TA muscle of mice and 400 µg of pGX9369 into cotton rat TA, muscle tissues were harvested, fixed in 10% Neutral-buffered Formalin (BBC Biochemical, Stanford, MA) and immersed in 30% (w/v) sucrose (Sigma) in D.I. water for in vivo staining of dMAb expression. Tissues were then embedded into O.C.T. compound (Sakura Finetek, Torrance, CA) and snap-frozen. Frozen tissue blocks were sectioned to a thickness of 18 µm. In vitro expressed dMAb: For staining of dMAb expression in transfected 293 T cells were cultured on chamber slides and fixed 3 days after transfection with 10% Neutral-buffered Formalin (BBC Biochemical) for 10 min at RT and then washed with PBS.
Slides were incubated with Blocking-Buffer (0.3% (v/v) Triton-X (Sigma), 2% (v/v) donkey serum in PBS) covered with Parafilm, and incubated at RT for 30 min. Goat anti-human IgG-Fc antibody (Bethyl) was diluted 1:100 in incubation buffer (1% (w/v) BSA (Sigma), 2% (v/v) donkey serum, 0.3% (v/v) Triton-X (Sigma) and 0.025% (v/v) 1 g/ml Sodium Azide (Sigma) in PBS). Staining solution at 150 µl was added to each slide and incubated for 2 h. Slides were washed for 5 min each with 1xPBS three times. Donkey anti-goat IgG AF488 (Abcam, Cambridge, UK) was diluted 1:200 in incubation buffer and 50 µl was added to each section. Slides were washed after 1 hr incubation and mounted with DAPI-Fluoromount (SouthernBiotech, Birmingham, AL) and covered.
In vivo and in vitro dMAb constructs production were imaged with a BX51 Fluorescent microscope (Olympus, Center Valley, PA) equipped with a Retiga3000 monochromatic camera (QImaging, Surrey, Canada).
Quantification of human RSV-F dMAb in animal serum and bronchoalveolar lavage (BAL) fluid
96-well assay plates (Thermo Scientific) were coated with 1ug/well goat anti-huIgG Fc fragment antibody (Bethyl) in 1x DPBS (ThermoFisher) overnight at 4°C. Next day plates were washed with 0.2% (v/v) TWEEN-20 in 1xPBS wash buffer and blocked with 10%(v/v)FBS in 1xDPBS for 1 hr at RT. The serum samples were diluted in 1% (v/v) FBS in 0.2% (v/v) TWEEN-1xPBS and 100 µl of this mix was added to the washed assay plate. Additionally, standard dilutions of purified human single-chain antibody was prepared as 1:2 serial dilutions starting at 500 ng/ml in dilution buffer and added in duplicates to each assay plate. Samples and standard were incubated for 1 hr at RT. For quantification of Palivizumab standard dilution of purified human kappa light chain (Bethyl) were used. After washing, the plates were incubated with a 1:10,000 dilution goat anti-human IgG-Fc antibody HRP (Bethyl, A80-104P) for 1 hr at RT. For detection SureBlue Substrate solution (Seracare, Milford, MA) was added to the washed plates. The reaction was stopped after 6 min by adding TMB Stop Solution (Seracare, Milford, MA) to the assay plates. The O.D. were read at 450 nm. The serum-level expression was interpolated from the standard curve using a sigmoidal four parameter logistic curve fit for a log of the concentration using GraphPad Prism8 (GraphPad Software, Inc; CA).
Antigen binding ELISA
Assay plates were coated with 1 µg/well of human RSV F glycoprotein (Sino Biological, Wayne, PA) diluted in 1x DPBS (ThermoFisher) overnight at 4°C. Plates were washed with 1xPBS buffer with 0.05% TWEEN. Two hundred and fifty µl/well of 3% (w/v) BSA in 1x PBS with 0.05% TWEEN were added and incubated for 1 hr at RT. Serum samples were diluted in 1% (w/v) BSA in 1x PBS with 0.05% TWEEN. After washing the assay plates serum samples were added in threefold serial dilutions. Plates were incubated 1 hr at RT. After washing, 100 µl of 1:10000 diluted goat anti-human IgG-Fc fragment antibody HRP (Bethyl, A80-104P) was added and incubated for 1 hr at RT. For development, the SureBlue/TMB Stop Solution (Seracare) was used and O.D. was recorded at 450 nm.
RSV neutralizing antibody assay
Heat-treated sera samples were diluted 1:10 with EMEM and serially diluted further 1:4. Diluted sera samples were incubated with RSV (25–50 PFU) for 1 h at RT and inoculated in duplicates onto confluent HEp-2 monolayers in 24-well plates. After one-hour incubation at 37°C in a 5% CO2 incubator, the wells were overlaid with 0.75% methylcellulose medium. After 4 days of incubation the overlay was removed, and the cells were fixed with 0.1% crystal violet stain for 1 h and then rinsed and air-dried. The corresponding reciprocal neutralizing antibody titers were determined at the 60% reduction end-point of the virus control using the statistics program “plqrd.manual.entry” (Sigmovir Biosystems, Inc; MD). The geometric means ± standard error for all animals in a group were calculated as described previously.Citation28,Citation29
Cotton rat viral challenge study
Animals
Fifteen (15) inbred female Sigmodon hispidus cotton rats between 6 and 8 weeks of age (Source: Sigmovir Biosystems, Inc., Rockville MD) were maintained and handled under veterinary supervision in accordance with National Institutes of Health guidelines and Sigmovir Institutional Animal Care and Use Committee’s approved animal study protocol. The cotton rats were housed in clear polycarbonate cages individually and provided with standard rodent chow (Harlan #7004) and tap water ad lib.
RSV-viral challenge stock
The prototype Long strain of RSV/A (ATCC, Manassas, VA) was propagated in HEp-2 cells after serial plaque-purification to reduce defective-interfering particles. A pool of virus designated as hRSV/A/Long Lot# 021413, prepared in sucrose stabilizing media, and containing approximately 2 × 107 pfu/ml was used for this in vivo experiment. This stock of virus is stored under −80°C condition and has been characterized in vivo using the cotton rat model for upper and lower respiratory tract replication.
Procedure
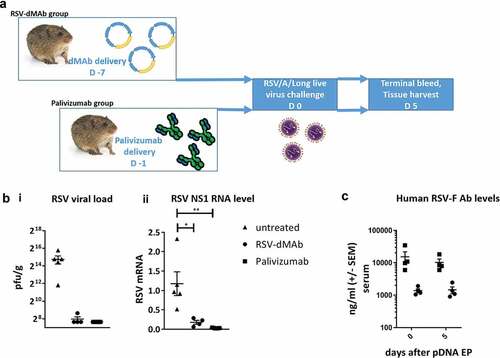
The adult female cotton rats (6–8 weeks of age) were divided into three groups of five animals each. Animals in the RSV-F dMAb group were treated with 2.4 mg pDNA on day −7 of the experiment as described in the animal treatment section. Animals in the Palivizumab (control) group were injected IM on day −1 of the experiment with 0.1 ml Palivizumab for a final concentration of 15 mg/kg. The animals in the untreated group received no injections. On day 0, all of the animals were intranasally inoculated with 105 plaque-forming units (PFU) of RSV/A/Long in a volume of 0.1 ml. All the animals were sacrificed at day 5. The lungs were harvested en bloc and trisected. The left section was used for virus titration. The lingular lobe was used for RealTime PCR.
Lung viral titration
Lung homogenates were clarified by centrifugation and diluted in EMEM. Confluent HEp-2 monolayers were infected in duplicates with diluted homogenates in 24-well plates. After one-hour incubation at 37°C in a 5% CO2 incubator, the wells were overlaid with 0.75% methylcellulose medium. After 4 days of incubation, the overlay was removed, and the cells were fixed with 0.1% crystal violet stain for 1 h and then rinsed and air-dried. Plaques were counted and viral titers were expressed as PFU per gram of tissue. Viral titers were calculated as geometric mean + standard error for all animals in a group at a given time.
Real-time PCR of lung tissue
Total RNA was extracted from homogenized lung tissue using the RNeasy purification kit (QIAGEN, Valencia, CA). One µg of total RNA was used to prepare cDNA using Super Script II RT (Invitrogen) and oligo dT primer (1 µl, Invitrogen). For the real-time PCR reactions, the Bio-Rad iQTM SYBR Green Supermix was used in a final volume of 25 µl, with final primer concentrations of 0.5 µM. Reactions were set up in duplicates in 96-well trays. Amplifications were performed on a Bio-Rad iCycler for 1 cycle of 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 15 s. The baseline cycles and cycle threshold (Ct) were calculated by the iQ5 software in the PCR Base Line Subtracted Curve Fit mode. Relative quantitation of DNA was applied to all samples. The standard curves were developed using serially diluted cDNA sample most enriched in the transcript of interest (e.g., lungs from day 4 post-primary RSV infection). The Ct values were plotted against log10 cDNA dilution factor. These curves were used to convert the Ct values obtained for different samples to relative expression units. These relative expression units were then normalized to the level of beta-actin mRNA (“housekeeping gene”) expressed in the corresponding sample. For animal studies, mRNA levels were expressed as the geometric mean ± SEM for all animals in a group at a given time.
Statistics
Statistical analysis was performed with GraphPad Prism v.7 using nonparametric two-tailed Mann–Whitney t-test.
Results
RSV-F dMAb construct design
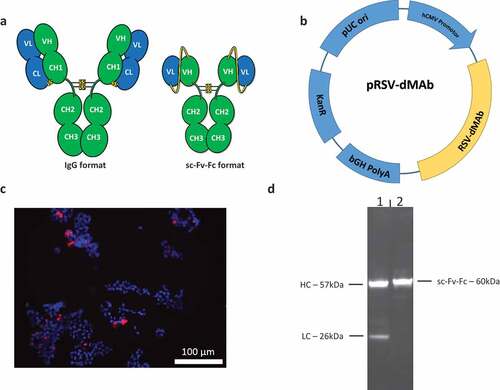
We generated a synthetic dMAb targeting RSV-fusion antigen by applying principles of antibody bioengineering. The human anti-RSV-F dMAb antigen binding fab region was designed to mimic that of the FDA-approved anti-RSV recombinant mAb Palivizumab. To assist in molecule design, models of the dMAb in both variable fragment and single-chain variable fragment (scFv) formats were generated using Discovery Studio 4.5 (Biovia, San Diego, CA). The scFv was modeled in both VH-VL and VL-VH orientations using a linker consisting of three repeats of four glycine and one serine residues ((G4 S) 3 linker). The linker as designed is the most commonly used one for scFvs and is not disruptive to Fv structure.Citation30 Based on the results of the model, the VL-VH orientation was chosen as it was predicted to cause the least perturbation to the scFv variable domains (). The synthetic pDNA molecule, pGX9369 was generated by cloning the gene for the sc-Fv-Fc RSV-F dMAb (RSV-F dMAb) into a pVAX1-based backbone, under control of the human CMV promoter (). RSV-F dMAb expression was confirmed by immunofluorescence staining of in vitro pGX9369-transfected 293 T cells with (). The staining demonstrates cytosolic expression of the RSV-F dMAb. Secretion of the RSV-F dMAb by 293 T cells was confirmed by Western-Blot analysis of cell-culture supernatant (). As expected, two bands were observed for the Palivizumab-IgG control representing heavy (57kDa) and light chain (26kDa), and a single band (60kDa) was detected for the scFv-Fc RSV-F dMAb.
Figure 1. Design and in-vitro testing of scFv-Fc RSV-F dMAb construct: (a) Illustration of human IgG1 and scFv-Fc formats. The complete hinge and Fc portions of the molecule are retained in the scFv-Fc format. Yellow line connecting VH and VL of the scFv-Fc molecule represents the (G4 S) 3 linker (b) Plasmid map of RSV-F dMAb (bGH PolyA – bovine growth hormone polyadenylation signal, KanR – kanamycin resistance gene, pUC ori – pUC origin of replication, hCMV promotor – human cytomegalovirus promotor) (c) In vitro expression of RSV-F dMAb. Immuno-fluorescence staining was performed on HEK293 T cells 3 days after in vitro transfection (DAPI – blue, RSV-F dMAb – red) (d) Western-blot analysis of cell-culture supernatant of RSV-F dMAb in vitro transfected HEK293 T cells harvested on day 3 post-transfection (lane 1: IgG-RSV dMAb, lane 2: scFv-Fc RSV-F dMAb).
dMAb in vivo expression model
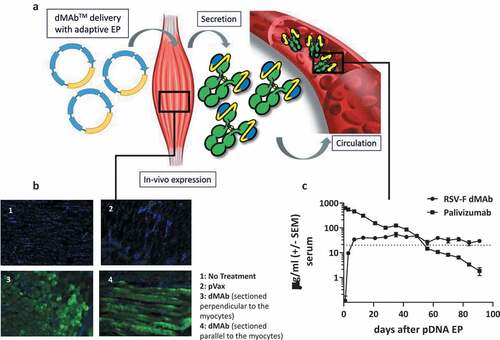
For optimal in vivo expression of dMAb we have developed an enhanced pDNA delivery protocol. The dMAb-encoding pDNA is formulated with the extra-cellular matrix disrupting enzyme hyaluronidase and administered to the muscle followed by in vivo EP. The schematic presented in outlines the pDNA delivery and in vivo expression of the dMAb. dMAb expression at the site of IM delivery of 50 µg pDNA can be visualized by staining for human IgG in myocytes 3 days post-delivery ()). The myocytes secrete the dMAb product into the circulating blood volume. Pharmacokinetic studies in animals indicate the dMAb’s expression after a single administration can be sustained about 20 µg/ml throughout the duration of a typical RSV season (). Circulating levels of the recombinant mAb, Palivizumab are not sustainable after a single bolus administration, and drop below the protective level of before the conclusion of a typical RSV season with durations reported between 6 and 23 weeks.Citation31
Figure 2. Schematic of RSV-F dMAb in-vivo delivery platform: (a) In vivo EP facilitates the delivery of the dMAb to the muscle tissue, myocytes express (b) and secret the encoded mAb (c). mAb is distributed systemically. (b) Example of expression of dMAb in myocytes 3 days after delivery of 50 µg dMAb or pVax (empty expression vector control) to mouse TA (DAPI – blue, dMAb – green). (c) Serum concentration of RSV-F dMAb and Palivizumab after delivery of 200 µg RSV-F dMAb plasmid or 15 mg/kg (clinical dose) Palivizumab measured by ELISA (± SEM, n = 6) dotted line indicates the level of protection (20 µg/ml) for Palivizumab.
In vivo expression of RSV-F dMAb in BALB/c mice
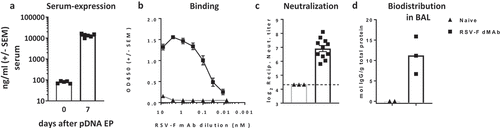
The expression and activity of RSV-F dMAb were assessed in vivo. BALB/c mice were dosed with 200 µg of pGX9369. The day 7 serum concentration of RSV-F dMAb averaged 7500 ng/ml (). As expected, there was a decline in serum concentration after day 7 which was associated with a host anti-drug antibodies response raised in WT BALB/c against the xenogeneic human antibody (data not shown). Binding of RSV-F dMAb to RSV-F antigen was confirmed by ELISA of serum samples from treated animals (). Serum from animals treated with pGX9369 displayed a serum-dilution dependent binding signal, exhibiting a ‘hook-effect’ at higher serum-concentrations. This assay-related phenomenon is the impaired ability of an antibody to bind its target effectively at high concentrations.Citation32 In vivo expressed RSV-F dMAb displayed virus-neutralization function. Serum harboring the RSV-F-dMAb reached RSV/A virus neutralizing titers of 7 log2 (). The biodistribution of the RSV-F dMAb to the site of RSV-associated pathology was confirmed by assaying lung lavage samples. Seven days after 200 µg pGX9369 administration, the level of dMAb BAL samples was 11.25 mol mAb/g total protein ±SEM 2.65 (). In summary, CELLECTRA-3P® delivery of pGX9369 resulted in the production of functional anti-RSV-F antibody, which was present in systemic circulation and at the physiologically relevant tissue site associated with viral-mediated pathology.
Figure 3. In-vivo expression of RSV-F dMAb in BALB/c mice: 200 µg RSV-F dMAb plasmid was administered IM in BALB/c mice (a-d). Serum samples were taken from treated mice before treatment (day 0) and 7 days after treatment to measure (a) Serum expression of RSV-F dMAb. scFv-Fc construct (± SEM, n = 5–8) (b) RSV-F antigen binding of dMAb. RSV-F binding signal of serum samples from treated (squares) and naïve (triangles) mice (± SEM, n = 4) (c) In vitro neutralization of RSV-A virus. Neutralization titer of serum samples from treated (squares) or naïve (triangles) mice (log2 serum dilution of 60% reduction of plaque-formation; ±SEM, n = 3–11, dotted line indicates LOD at serum-dilution of 1/20) (d) Concentration of scFv-Fc RSV-F dMAb in BAL samples 7 days after RSV-dMAb pDNA delivery in treated (squares) or naïve (triangles) mice (mol scFv-Fc dMAb per gram total protein in lavage sample, n(naïve) = 2, n(RSV-F dMAb) = 3).
In vivo expression of RSV-F dMAb in cotton rats
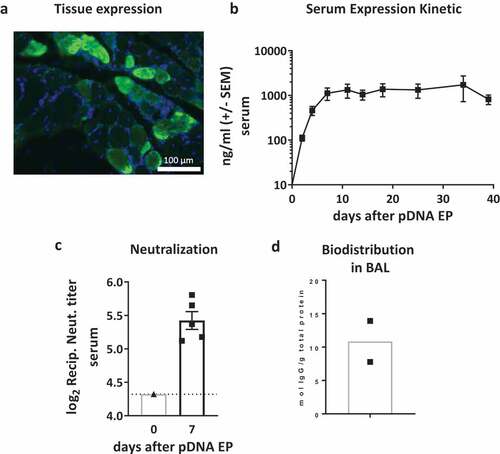
We proceeded to assess the RSV-F dMAb expression and activity in cotton rats. Cotton rats are considered the gold standard model to model RSV infection and drug interventions.Citation27 Cotton rats are susceptible to non-adapted human RSV, being 100-fold more permissive to infection than mice and display many features of the human disease pathology and display VED-associated pathology in response viral challenge after immunization with formalin-inactivated RSV.Citation33,Citation34 We initially confirmed the local expression of RSV-F dMAb in the myocytes after delivery of 400 µg pGX9369 to the TA leg muscle (). We proceed to test whether local expression led to the secretion of the RSV-F dMAb into the circulation. We measured for longer than 5 weeks greater than 1,000 ng/ml of the human RSV-F dMAb in the cotton rat sera after delivery of 800 µg pDNA (). We assessed the functionality of the expressed dMAb after delivery of a total of 2.4 mg pDNA. Neutralizing capability of cotton rat serum was tested in a RSV/A neutralization assay. In the plaque reduction assay, we measured an average reciprocal neutralization titer of 5.4 determined at 60% reduction end-point of the virus control (). The biodistribution of the dMAb to the site of RSV infection was assessed in BAL fluid. The mean level of RSV-F dMAb in BAL samples from these animals was 10.8 mol/g total protein (). In summary, in vivo delivery of pGX9369 to cotton rats led to the systemic production of functional antibodies which neutralized RSV virus and biodistributed to the lungs.
Figure 4. Characterization of RSV-F dMAb in cotton rats: (a) Local expression of dMAb demonstrated by Immuno-fluorescence staining of RSV-F dMAb in cotton rat TA muscle tissue 7 days after delivery of 400 µg RSV-F dMAb pDNA to the tissue site (DAPI – blue; RSV-F dMAb – green; sectioned perpendicular to myocytes) (b) Levels of RSV-F dMAb (ng/ml) was measured for 39 days after IM administration of 800 µg dMAb-plasmid (±SEM, n = 5). (c) Virus neutralization function of in-vivo expressed RSV-F dMAb. Serum samples were harvested and tested 7 days after delivery of 2.4 mg scFv-Fc dMAb-pDNA: neutralizing titer (±SEM, dotted line indicates LOD at serum-dilution of 1/20). (d) Concentration of RSV-F dMAb in BAL samples from treated cotton rats.
RSV-F dMAb confers protection from LRD in the cotton rat RSV/A challenge model
The presence of circulating functional RSV-F dMAb in the cotton rat led us to test the pGX9369 treatment in an active immune-prophylaxis study. Groups of cotton rats were either dosed with 2.4 mg pGX9369 (group 1) on day −7 or 15 mg/kg Palivizumab (group 2) on day −1 or remained untreated (control). All groups were challenged intranasally with 105 pfu of RSV/A/Long on day 0. Five days after challenge lung tissue was harvested and viral loads measured (). Both group 1 and 2 animals were protected from LRD associated with RSV infection. Five days after the intranasal challenge, the lungs of pGX9369-treated contained mean viral load of 250 pfu/g (SEM±50) and the lungs of Palivizumab-treated cotton rats contained ≤ 200 pfu/g (SEM = 0). In comparison lungs of control cotton rats contained high viral load of 27,520 pfu/g (SEM = 8,408.9) (). Significantly reduced viral genome copy number in the lower respiratory tissue was measured with real-time PCR using primer targeting RSV Nonstructural Protein 1. Mean RSV-mRNA levels in pGX9369 (mean RSV mRNA = 0.181, SEM = 0.050) and Palivizumab- (mean RSV mRNA = 0.032, SEM = 0.004) treated cotton rats were reduced in comparison to untreated animals (mean RSV mRNA = 1.175, SEM = 0.306) (). Of note, pGX9369-treated cotton rats were protected from LRD, even though human RSV-F dMAb serum levels were approximately 10-fold lower than those measured for Palivizumab (). In summary, the data show successful active immune-prophylaxis using an anti-RSV-F dMAb to protect against viral challenge in the cotton rat, a gold standard RSV disease model.
Figure 5. RSV-F dMAb confers protection against LRD after RSV/A challenge of cotton rats: (a) Schematic of cotton rat challenge study: Animals were treated with 2.4 mg RSV-dMAb 7 days before challenge or with an IM injection of 15 mg/kg Palivizumab 1 day before challenge (b) Viral load of cotton rat lung tissue (pfu/g) harvested 5 days after intra-nasal live virus challenge with RSV/A/long (± SEM n = 4–5) (i). RSV Nonstructural protein-1 (NS-1) mRNA levels (log2 and normalized to beta-Actin) of cotton rat lung tissue 5 days after intra-nasal live virus challenge with RSV/A/long (± SEM, n = 4–5, Mann–Whitney non-parametric t-test: p (untreated vs RSV-dMAb) = 0.0159; p (untreated vs Palivizumab) = 0.0079) (ii). (c) Serum levels of Palivizumab and scFv-Fc RSV-F dMAb in cotton rats at the day of challenge (day 0) and 5 days after challenge (± SEM, n = 4–5).
Discussion
There is a pressing need for the development of novel platform technologies which can be employed to increase patient access to prophylactic and therapeutic monoclonal antibodies, especially those targeting infectious diseases in low and middle-income countries. The work presented here highlights a viral-vector independent antibody engineered synthetic DNA technology platform, with this potential. We apply this technology to the RSV target using a synthetic antibody DNA sequence based on the FDA-approved recombinant mAb, Palivizumab. Our findings demonstrate the successful in vivo expression of the dMAb in multiple preclinical models, and the ability to confer host protection against virus challenge (). Data presented here and in other recent reports highlight the characteristics and therapeutic potency of the dMAb platform as a cost-effective and quickly deployable prophylactic option for a myriad of infectious diseases.Citation12-Citation15,Citation24
The success of dMAb technology is dependent on the effective delivery of the antibody of interest to the host. The in vivo pDNA delivery described here targets the long-lived host myocytes in skeletal muscle, which is considered an endocrine organ that is very adept at synthesis and secretion of multiple factors, and represents an ideal biological factory for high-level transgene expression which can persist for some time after delivery.Citation35 Citation36-Citation37 However, to achieve robust local tissue transgene expression a highly efficient method of in vivo pDNA delivery is required. To optimize pDNA delivery to the muscle tissue we employ EP and hyaluronidase formulation. This enzyme modifies the extra-cellular matrix and used “on label” for IM injection in the clinic.Citation38 IM injection of pDNA formulated with human recombinant hyaluronidase (Hylenex) is used in combination with EP in the clinical trial of a Zika dMAb (NCT03831503). Together this delivery protocol leads local tissue expression 1,000 fold higher than traditional needle-based injection methods. In the absence of an adaptive host immune response against the xenogeneic dMAb we have demonstrated 13-week long-expression in BALB/c mice (). In the same experiment, we observed the steady decline of the recombinant Palivizumab after administration of the clinical dose. Protective levels could only be sustained by re-dosing the animals with the Palivizumab (data not shown) which is reflected in the clinical protocol for the drug. Administration of Palivizumab is ideally initiated prior to the RSV-season onset, which typically lasts from November through April with deviation by regions.Citation39 It is dosed at 15 mg/kg and must be administered monthly by IM injection throughout the RSV season. The serum-half life has been reported as 20 days.Citation6,Citation40 On the other hand, of Palivizumab-based scFv-Fc dMAb is expressed and secreted, and an efficacious circulating level of the antibody can be maintained as shown in . Circulating RSV-dMAb levels are well maintained above the level required for protection after one administration, indicating that there is not a need for re-delivery. Similar in essence to half-life extension technology, the dMAb approach can allow for the longer biological presence of the delivered antibody-like molecule.Citation41 Citation42-Citation43 In the presence of an intact immune system in wild type cotton rats we show continued expression of the human dMAb construct. This result was contrary to earlier studies with the construct in mice with intact T cell compartments, in which strong ADAs were observed. In the cotton rat experiment, we measured RSV-dMAb expression for a time period of 39 days following administration (). However, as this antibody is xenogeneic to the cotton rat, we would expect to observe similar or even more durable expression using a species-matched antibody. Sustained expression is an extremely important component for an immune-prophylactic modality, and a characteristic which is missing with conventional recombinant mAbs. A single administration of dMAb-encoding plasmid could provide protective levels of mAbs throughout a RSV-season. For example, children are at the highest risk of morbidity when infection occurs during the first 6 months of life. A single injection scFv-Fc RSV-F dMAb could protect children through this most critical time. In addition to RSV-F dMAb, we have observed extended serum-concentrations for anti-EBOV, influenza, and PD-1 dMAbs compared their respective recombinant mAbs in a mouse model.Citation24
A characteristic of sc-Fv antibodies is their excellent biodistribution. Due to their small size, they can efficiently penetrate tissue. However, their effectiveness is limited by a serum half-life of just several hours. The fusion to a Fc protein increases the serum half-life of the resulting scFv-Fc molecule from hours to multiple days.Citation26 Although it does not exceed the half-life of a full-length IgG molecule, it still exhibits superior biodistribution because of its smaller size.Citation44 The increased ability of the smaller scFv-Fc molecule to distribute to the site of RSV-infection may explain the similar levels of protection from LRD () despite 10-fold lower serum-concentration () compared to Palivizumab. We successfully detected the RSV-F dMAb in the lungs of mice and cotton rats ( and ). The importance of biodistribution of mAbs in preventing LRD after RSV challenge has been previously reported. Wu et al. report lower in vivo efficacy for motavizumab, a variant of Palivizumab, despite high in-vitro neutralizing ability due to poor biodistribution.Citation45 Tiwari et al. reported results by targeting the lung-tissue directly for the expression of mRNA-encoded RSV-Ab.Citation46 Citation47
The unique mechanism of the dMAb technology platform by which the antibody molecule is constantly replenished due to secretion from expressing myocytes counteracts potential loss of serum half-life of the scFv-Fc format. To allow a more accurate comparison of the two molecules and their biodistribution, future experiments will be designed for both antibodies to be at equal concentrations in the animal serum at the time of viral challenge.
The threefold increase of pDNA dose used in the cotton rat viral challenge (compare 2.4 mg, versus 0.8 mg, ) did not result in a proportional increase of serum-level expression as we would have expected from previous studies in mouse models. This could be attributed to differences between the two animal models and the adaptions of the mouse EP-delivery procedure that were made to accommodate the larger cotton rat model.
While other antibody gene delivery platforms have been employed, antibody-encoding DNA plasmid possess multiple advantages over other nucleic acid-based monoclonal antibody technologies, including viral-based vector and mRNA-based delivery of antibodies. Firstly, the injected drug product is a pure optimized DNA plasmid, and devoid of any anti-vector immunity properties, which limits the reuse of adenovirus or recombinant adeno-associated viral-based vectors.Citation48 Secondly, the DNA does not require potentially complex chemical delivery formulations often used with some nucleic acid-based deliveries of biologics, which generally require lipid nanoparticle-encapsulation.Citation49 Citation50-Citation51 As discussed above and highlighted in the data presented in this study there is a sustained, but finite expression profile for dMAbs as the designed construct delivered is non-integrating. Thirdly, unlike many recombinant and mRNA-based antibody candidates which are delivered IV and require significant infrastructure in place to dose patients, DNA plasmids can be administered to local accessible sites rapidly in the field. Viral vectored delivery of RSV mAbs has also been described.Citation52 However, in addition to anti-vector serology concerns, such vectors are not temperature stable and complex to manufacture. We have developed plasmid formulations that are highly temperature stable without the need for lyophilization, and the product storage and distribution does not require a cold chain.Citation53
In summary, applying the dMAb platform to the target of the RSV in established disease models, we demonstrate biological significant circulating levels of the RSV antibodies in the serum and lungs of animals. Furthermore, the in vivo expressed RSV-dMAb was functionally active and provided protection from RSV-mediated respiratory disease in a viral challenge study (), even at a lower concentration than the comparator recombinant antibody standard used in this study. Our findings support the significance of the dMAb technology, and its potential to fight global mortality caused by infectious diseases such as RSV.
The current study describes the proof of concept for in vivo launched mAb constructs targeting RSV. However, other studies will be needed to fully define the potency of the RSV-dMAb compared to Palivizumab, and the scalability of this technology. We postulate above that RSV-dMAb may exhibit improved biodistribution to Palivizumab. Future studies will further address this by studying the biodistribution and level of disease protection at equivalent serum concentrations of these drugs. The current body of work describes the expression and function of RSV-dMAb in 2 small animal models. Whilst the cotton rat model is considered a gold standard model for RSV pathology and drug testing, and the preclinical studies used to develop Palivizumab were largely dependent upon this model, we plan to investigate RSV-dMAb expression and function in larger animal models, including the pig, sheep, and NHP. Importantly, these models are compatible with our clinical in vivo delivery devices and will assist in determining appropriate dosing for human testing.
References
- Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet (London, England). 2017;390:946–58. doi:10.1016/S0140-6736(17)30938-8.
- Amand C, Tong S, Kieffer A, Kyaw MH. Healthcare resource use and economic burden attributable to respiratory syncytial virus in the United States: a claims database analysis. BMC Health Serv Res. 2018;18:294–294. doi:10.1186/s12913-018-3066-1.
- Noor A, Krilov LR. Respiratory syncytial virus vaccine: where are we now and what comes next? Expert Opin Biol Ther. 2018;18:1247–56. doi:10.1080/14712598.2018.1544239.
- Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–34. doi:10.1093/oxfordjournals.aje.a120955.
- O’Brien KL, Chandran A, Weatherholtz R, Jafri HS, Griffin MP, Bellamy T, Millar EV, Jensen KM, Harris BS, Reid R, et al. Efficacy of motavizumab for the prevention of respiratory syncytial virus disease in healthy Native American infants: a phase 3 randomised double-blind placebo-controlled trial. Lancet Infect Dis. 2015;15:1398–408. doi:10.1016/s1473-3099(15)00247-9.
- Griffin MP, Khan AA, Esser MT, Jensen K, Takas T, Kankam MK, Villafana T, Dubovsky F. Safety, tolerability, and pharmacokinetics of MEDI8897, the Respiratory syncytial virus prefusion F-targeting monoclonal antibody with an extended half-life, in Healthy adults. Antimicrob Agents Chemother. 2017;61. doi:10.1128/aac.01714-16.
- Ambrose CS, Chen X, Kumar VR. A population-weighted, condition-adjusted estimate of palivizumab efficacy in preventing RSV-related hospitalizations among US high-risk children. Hum Vaccines Immunother. 2014;10:2785–88. doi:10.4161/hv.32082.
- Subramanian KN, Weisman LE, Rhodes T, Ariagno R, Sánchez PJ, Steichen J, Givner LB, Jennings TL, Top FH, Carlin D, et al. Safety, tolerance and pharmacokinetics of a humanized monoclonal antibody to respiratory syncytial virus in premature infants and infants with bronchopulmonary dysplasia. MEDI-493 Study Group. Pediatr Infect Dis J. 1998;17:110–15. doi:10.1097/00006454-199802000-00006.
- Shepard HM, Phillips GL, Thanos CD, Feldmann M. Developments in therapy with monoclonal antibodies and related proteins. Clin Med (Lond). 2017;17:220–32. doi:10.7861/clinmedicine.17-3-220.
- Schultheis K, Smith TRF, Ramos S, Schommer N, Jian J, Yung B, Oh J, Yan J, Patel A, Elliott S. Molecular therapy. p. 269–269. (CAMBRIDGE, MA: CELL PRESS).
- McMahon JM, Signori E, Wells KE, Fazio VM, Wells DJ. Optimisation of electrotransfer of plasmid into skeletal muscle by pretreatment with hyaluronidase – increased expression with reduced muscle damage. Gene Ther. 2001;8:1264–70. doi:10.1038/sj.gt.3301522.
- Elliott STC, Kallewaard NL, Benjamin E, Wachter-Rosati L, McAuliffe JM, Patel A, Smith TRF, Schultheis K, Park DH, Flingai S, et al. DMAb inoculation of synthetic cross reactive antibodies protects against lethal influenza A and B infections. NPJ Vaccines. 2017;2:18. doi:10.1038/s41541-017-0020-x.
- Patel A, DiGiandomenico A, Keller AE, Smith TRF, Park DH, Ramos S, Schultheis K, Elliott STC, Mendoza J, Broderick KE, et al. An engineered bispecific DNA-encoded IgG antibody protects against Pseudomonas aeruginosa in a pneumonia challenge model. Nat Commun. 2017;8:637. doi:10.1038/s41467-017-00576-7.
- Wang Y, Esquivel R, Flingai S, Schiller Z A, Kern A, Agarwal S, Chu J, Patel A, Sullivan K, Wise M C, et al. Anti-OspA DNA-encoded monoclonal antibody prevents transmission of spirochetes in tick challenge providing sterilizing immunity in mice. J Infect Dis. 2018. doi:10.1093/infdis/jiy627.
- Muthumani K, Block P, Flingai S, Muruganantham N, Chaaithanya IK, Tingey C, Wise M, Reuschel EL, Chung C, Muthumani A, et al. Rapid and long-term immunity elicited by DNA-encoded antibody prophylaxis and DNA vaccination against Chikungunya virus. J Infect Dis. 2016;214:369–78. doi:10.1093/infdis/jiw111.
- Andrews CD, Luo Y, Sun M, Yu J, Goff AJ, Glass PJ, Padte NN, Huang Y, Ho DD. In vivo production of monoclonal antibodies by gene transfer via electroporation protects against lethal influenza and ebola infections. Mol Ther Methods Clin Dev. 2017;7:74–82. doi:10.1016/j.omtm.2017.09.003.
- Esquivel RN, Patel A, Kudchodkar SB, Park DH, Stettler K, Beltramello M, Allen JW, Mendoza J, Ramos S, Choi H, et al. In vivo delivery of a DNA-encoded monoclonal antibody protects non-human primates against Zika virus. Mol Ther. 2019;27:974–85. doi:10.1016/j.ymthe.2019.03.005.
- Wise MC, Xu Z, Tello-Ruiz E, Beck C, Trautz, A, Patel A, Elliott ST, Chokkalingam N, Kim S, Kerkau MG, et al. In vivo delivery of synthetic DNA-encoded antibodies induces broad HIV-1-neutralizing activity. J Clin Invest. 2019. doi:10.1172/JCI132779.
- Muthumani K, Marnin L, Kudchodkar SB, Perales-Puchalt A, Choi H, Agarwal S, Scott VL, Reuschel EL, Zaidi FI, Duperret EK, et al. Novel prostate cancer immunotherapy with a DNA-encoded anti-prostate-specific membrane antigen monoclonal antibody. Cancer Immunol Immunother. 2017;66:1577–88. doi:10.1007/s00262-017-2042-7.
- Hollevoet K, De Smidt E, Geukens N, Declerck P. Prolonged in vivo expression and anti-tumor response of DNA-based anti-HER2 antibodies. Oncotarget. 2018;9:13623–36. doi:10.18632/oncotarget.24426.
- Duperret EK, Trautz A, Stoltz R, Patel A, Wise MC, Perales-Puchalt A, Smith T, Broderick KE, Masteller E, Kim JJ, et al. Synthetic DNA-encoded monoclonal antibody delivery of anti-CTLA-4 antibodies induces tumor shrinkage in vivo. Cancer Res. 2018;78:6363–70. doi:10.1158/0008-5472.can-18-1429.
- Hollevoet K, De Vleeschauwer S, De Smidt E, Vermeire G, Geukens N, Declerck P. Bridging the clinical gap for DNA-based antibody therapy through translational studies in sheep. Hum Gene Ther. 2019;30:1431–43. doi:10.1089/hum.2019.128.
- Hollevoet K, Declerck PJ. State of play and clinical prospects of antibody gene transfer. J Transl Med. 2017;15:131–131. doi:10.1186/s12967-017-1234-4.
- Patel A, Park DH, Davis CW, Smith TRF, Leung A, Tierney K, Bryan A, Davidson E, Yu X, Racine T, et al. In vivo delivery of synthetic human DNA-encoded monoclonal antibodies protect against ebolavirus infection in a mouse model. Cell Rep. 2018;25:1982–1993 e1984. doi:10.1016/j.celrep.2018.10.062.
- Repp R, Kellner C, Muskulus A, Staudinger M, Mohseni Nodehi S, Glorius P, Akramiene D, Dechant M, Fey GH, van Berkel PHC, et al. Combined Fc-protein- and Fc-glyco-engineering of scFv-Fc fusion proteins synergistically enhances CD16a binding but does not further enhance NK-cell mediated ADCC. J Immunol Methods. 2011;373:67–78. doi:10.1016/j.jim.2011.08.003.
- Unverdorben F, Richter F, Hutt M, Seifert O, Malinge P, Fischer N, Kontermann RE. Pharmacokinetic properties of IgG and various Fc fusion proteins in mice. mAbs. 2016;8:120–28. doi:10.1080/19420862.2015.1113360.
- Boukhvalova MS, Blanco JC. The cotton rat Sigmodon hispidus model of respiratory syncytial virus infection. Curr Top Microbiol Immunol. 2013;372:347–58. doi:10.1007/978-3-642-38919-1_17.
- Smith TRF, Schultheis K, Morrow MP, Kraynyak KA, McCoy JR, Yim KC, Muthumani K, Humeau L, Weiner DB, Sardesai NY, et al. Development of an intradermal DNA vaccine delivery strategy to achieve single-dose immunity against respiratory syncytial virus. Vaccine. 2017;35:2840–47. doi:10.1016/j.vaccine.2017.04.008.
- Coates HV, Alling DW, Chanock RM. An antigenic analysis of respiratory syncytial virus isolates by a plaque reduction neutralization test. Am J Epidemiol. 1966;83:299–313. doi:10.1093/oxfordjournals.aje.a120586.
- Freund C, Ross A, Guth B, Pluckthun A, Holak TA. Characterization of the linker peptide of the single-chain Fv fragment of an antibody by NMR spectroscopy. FEBS Lett. 1993;320:97–100. doi:10.1016/0014-5793(93)80070-b.
- Haynes AK, Prill MM, Iwane MK, Gerber SI. Respiratory syncytial virus–United States, July 2012-June 2014. MMWR Morb Mortal Wkly Rep. 2014;63:1133–36.
- Courtenay-Luck NS, Epenetos AA, Moore R, Larche M, Pectasides D, Dhokia B, Ritter MA. Development of primary and secondary immune responses to mouse monoclonal antibodies used in the diagnosis and therapy of malignant neoplasms. Cancer Res. 1986;46:6489–93.
- Prince GA, Capiau C, Deschamps M, Fabry L, Garçon N, Gheysen D, Prieels J-P, Thiry G, Van Opstal O, Porter DD, et al. Efficacy and safety studies of a recombinant chimeric respiratory syncytial virus FG glycoprotein vaccine in cotton rats. J Virol. 2000;74:10287–92. doi:10.1128/JVI.74.22.10287-10292.2000.
- Prince GA, Jenson AB, Hemming VG, Murphy BR, Walsh EE, Horswood RL, Chanock RM. Enhancement of respiratory syncytial virus pulmonary pathology in cotton rats by prior intramuscular inoculation of formalin-inactiva ted virus. J Virol. 1986;57:721–28. doi:10.1128/JVI.57.3.721-728.1986.
- Iizuka K, Machida T, Hirafuji M. Skeletal muscle is an endocrine organ. J Pharm Sci. 2014;125:125–31. doi:10.1254/jphs.14R02CP.
- Mir LM, Bureau MF, Rangara R, Schwartz B, Scherman D. Long-term, high level in vivo gene expression after electric pulse-mediated gene transfer into skeletal muscle. Comptes rendus l’Acad Sci. 1998;321:893–99. doi:10.1016/s0764-4469(99)80003-1.
- Maruyama H, Ataka K, Gejyo F, Higuchi N, Ito Y, Hirahara H, Imazeki I, Hirata M, Ichikawa F, Neichi T, et al. Long-term production of erythropoietin after electroporation-mediated transfer of plasmid DNA into the muscles of normal and uremic rats. Gene Therapy. 2001;8:461–68. doi:10.1038/sj.gt.3301412.
- Raghavan P, Lu Y, Mirchandani M, Stecco A. Human recombinant hyaluronidase injections for upper limb muscle stiffness in individuals with cerebral injury: a case series. EBioMedicine. 2016;9:306–13. doi:10.1016/j.ebiom.2016.05.014.
- Rose EB, Wheatley A, Langley G, Gerber S, Haynes A. Respiratory syncytial virus seasonality - United States, 2014–2017. MMWR Morb Mortal Wkly Rep. 2018;67:71–76. doi:10.15585/mmwr.mm6702a4.
- Robbie GJ, Zhao L, Mondick J, Losonsky G, Roskos LK. Population pharmacokinetics of palivizumab, a humanized anti-respiratory syncytial virus monoclonal antibody, in adults and children. Antimicrob Agents Chemother. 2012;56:4927–36. doi:10.1128/aac.06446-11.
- AlQahtani AD, O’Connor D, Domling A, Goda SK. Strategies for the production of long-acting therapeutics and efficient drug delivery for cancer treatment. Biomed Pharmacother. 2019;113:108750. doi:10.1016/j.biopha.2019.108750.
- Tan H, Su W, Zhang W, Wang P, Sattler M, Zou P. Recent advances in half-life extension strategies for therapeutic peptides and proteins. Curr Pharm Des. 2018;24:4932–46. doi:10.2174/1381612825666190206105232.
- Zaman R, Islam RA, Ibnat N, Othman I, Zaini A, Lee CY, Chowdhury EH. Current strategies in extending half-lives of therapeutic proteins. J Controlled Release. 2019;301:176–89. doi:10.1016/j.jconrel.2019.02.016.
- Kontermann RE. Strategies to extend plasma half-lives of recombinant antibodies. BioDrugs. 2009;23:93–109. doi:10.2165/00063030-200923020-00003.
- Wu H, Pfarr DS, Johnson S, Brewah YA, Woods RM, Patel NK, White WI, Young JF, Kiener PA. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007;368:652–65. doi:10.1016/j.jmb.2007.02.024.
- Tiwari PM, Vanover D, Lindsay KE, Bawage SS, Kirschman JL, Bhosle S, Lifland AW, Zurla C, Santangelo PJ. Engineered mRNA-expressed antibodies prevent respiratory syncytial virus infection. Nat Commun. 2018;9:3999–3999. doi:10.1038/s41467-018-06508-3.
- Kakuk TJ, Soike K, Brideau RJ, Zaya RM, Cole SL, Zhang J-Y, Roberts ED, Wells PA, Wathen MW. A human respiratory syncytial virus (RSV) primate model of enhanced pulmonary pathology induced with a formalin-inactivated RSV vaccine but not a recombinant FG subunit vaccine. J Infect Dis. 1993;167:553–61. doi:10.1093/infdis/167.3.553.
- Shirley JL, de Jong YP, Terhorst C, Herzog RW. Immune responses to viral gene therapy vectors. Mol Ther. 2020;S1525–0016(1520)30002–30002. doi:10.1016/j.ymthe.2020.01.001.
- Pardi N, Secreto AJ, Shan X, Debonera F, Glover J, Yi Y, Muramatsu H, Ni H, Mui BL, Tam YK, et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat Commun. 2017;8:14630. doi:10.1038/ncomms14630.
- Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK, Madden TD, Hope MJ, Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Controlled Release. 2015;217:345–51. doi:10.1016/j.jconrel.2015.08.007.
- Thran M, Mukherjee J, Pönisch M, Fiedler K, Thess A, Mui BL, Hope MJ, Tam YK, Horscroft N, Heidenreich R, et al. mRNA mediates passive vaccination against infectious agents, toxins, and tumors. EMBO Mol Med. 2017;9:1434–47. doi:10.15252/emmm.201707678.
- Skaricic D, Traube C, De B, Joh J, Boyer J, Crystal RG, Worgall S. Genetic delivery of an anti-RSV antibody to protect against pulmonary infection with RSV. Virology. 2008;378:79–85. doi:10.1016/j.virol.2008.04.016.
- Tebas P, Kraynyak KA, Patel A, Maslow JN, Morrow MP, Sylvester AJ, Knoblock D, Gillespie E, Amante D, Racine T, et al. Intradermal SynCon(R) ebola GP DNA vaccine is temperature stable and safely demonstrates cellular and humoral immunogenicity advantages in healthy volunteers. The Journal of Infectious Diseases. 2019;220:400–10. doi:10.1093/infdis/jiz132.
