ABSTRACT
Hepatitis B virus (HBV) causes a potentially life-threatening liver infection that frequently results in life-long chronic infection. HBV is responsible for 887,000 deaths each year, most resulting from chronic liver diseases and hepatocellular carcinoma. Presently, there are 250 million chronic HBV carriers worldwide who are at a high risk for developing cirrhosis and hepatocellular carcinoma (HCC). HCC is the most common type of liver cancer with a strong association with HBV infection. HBV transmission through blood transfusions and perinatal transfer from infected mother to child have been common routes of infection. In the present study, we describe the development of a synthetic DNA plasmid encoding an anti-HBV human monoclonal antibody specific for the common “a determinant region” of HBsAg of hepatitis B virus and demonstrate the ability of this platform at directing in vivo antibody expression. In vivo delivery of this DNA encoded monoclonal antibody (DMAb) plasmid in mice resulted in expression of human IgG over a period of one month following a single injection. Serum antibody was found to recognize the relevant conformational epitope from plasma purified native HBsAg as well as bound HBV in HepG2.2.15 cells. The serum DMAb efficiently neutralized HBV and prevented infection of HepaRG cells in vitro. Additional study of these HBV-DMAb as a possible therapy or immunoprophylaxis for HBV infection is warranted.
Introduction
Chronic hepatitis caused by hepatitis B virus (HBV) infection represents a major health burden globally.1,Citation2 More than 250 million people are chronically infected with HBV, out of which 1 million people die each year due to life-threatening complications of this infection including liver cirrhosis and hepatocellular carcinoma.Citation3 Current therapies of chronic hepatitis include interferon-α (IFN-α) and nucleos(t)ide analogue inhibitors against viral reverse transcriptase.Citation4,Citation5 These antiviral therapies have been demonstrated to control HBV replication, but fail to eliminate infection because of the tendency of HBV to integrate into the host genome or remain latent episomally as covalently closed circular DNA (cccDNA). Moreover, these approaches are associated with the development of acquired drug resistance, resulting in low response rates, and risk of numerous side-effects.
The current commercially available vaccine for hepatitis B contains yeast-derived recombinant major surface protein HBsAg of HBV.Citation3,Citation6,Citation7 Despite its proven immunogenicity and high efficacy overall, the vaccine is unable to induce adequate immune response in approximately 5–15% of healthy adults. Such individuals remain susceptible to HBV infection.Citation2,Citation8 In this regard, hepatitis B Immune Globulin (HBIG) can provide rapid passive protection against infection after acute exposure to infection.Citation9,Citation10 HBIG is a polyclonal antibody against HBsAg derived from pooled plasma of individuals having high anti-HBs titer.Citation11,Citation12 Pooled plasma can vary in the titer of anti-HBs which is a concern for the consistency. Apart from variability in anti-HBs titer among patients, the neutralization efficiency of HBIG is largely based on protective anti-HB serum titer. In addition, the polyclonal immunoglobulin contains antibodies with different specificities which may select for HBV mutants resistant to currently used antiviral drugs and provide variability for patient management.
In the present study, we describe the design and development of a synthetic DNA plasmid-encoding human anti-HBV monoclonal antibody sequences as a novel approach to immunotherapy of HBV infection. We demonstrate that a single inoculation with this anti-HBV-DMAb generates functional anti-HBV activity for several weeks in the serum of inoculated animals. Immunoglobulins generated in vivo were properly folded and were able to neutralize HBV and prevent HBV infection of HepaRG cells. Additional study of anti-HBV DMAbs may have value as an immunoprophylaxis strategy for HBV infection.
Materials and methods
Antibody plasmid construction
A synthetic DNA cassette was designed that encoded the variable heavy (VH) and light (VL) chain sequences of the anti-HBV MAb ADRI-2 F3 based on sequences from a published description.Citation11 We utilized this information and designed optimized synthetic DNA expression cassettes encoding full-length IgG (Ig) which encodes for both an anti-HBV-VH and VL. We engineered the VH chain and VL chain domain constructs to be expressed at high levels using our recently described modification/optimization strategiesCitation13−17,Citation14 which can lead to drastic increases in DMAb expression levels. The final construct is named HBV-DMAb and the control plasmid backbone is pVax1. Both were synthesized by Genscript and cloned into modified mammalian expression vectors under the control of the human cytomegalovirus immediate-early promoter.Citation13
Cell lines and reagents
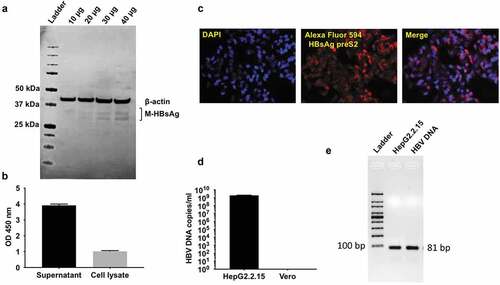
HepG2.2.15 cells (a kind gift from Dr. Charles Rice, The Rockefeller University, NY) were used as a source of HBV for infection experiments.Citation15 The cells are stably transfected with complete genome of HBV (adw2 subtype) and are able to support replication of HBV-DNA and intact virus particles. Production of HBV by HepG2.2.15 cell line was done by performing western blot of HepG2.2.15 cell lysates to determine presence of M-HBsAg (). s-HBsAg production by HepG2.2.15 cells was detected in the supernatant and cell lysates using Bio-Rad GS HBsAg ELISA kit () and immunofluorescence for detection of HBsAg preS2 antigen ().
Figure 1. HBV virus amplification & characterization from HepG2.2.15 cells.
HepG2.2.15 cells and human 293 T cells (ATCC) and were cultured using Dulbecco’s Modified Eagle Medium containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. For HBV neutralization assay, terminally differentiated No spin HepaRG cells (Lonza) were used.Citation15 The cells were plated and cultured according to supplier’s instructions. Nabi-HB (Hepatitis B Immune Globulin (HBIG), >312 IU/ml) was purchased from Biotest Pharmaceuticals Corporation, USA.
Animals and DMAb immunizations
Female, 6–8-week-old B6.Cg-Foxn1nuJ and BALB/c mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed in the animal facilities at the Wistar Institute. Mice were injected with 100 μg and 400 μg of pMV101 empty vector or HBV-DMAb plasmids were formulated in sterile water, by IM injection in the anterior tibialis (TA) muscle as previously described.Citation16,Citation17 Serum levels of DMAbs were monitored following administration. Animal experiments were approved by the Institutional Animal Care and Use Committee at The Wistar Institute.
Transfection and Western blot
One day prior to transfection, 293 T cells were plated at a density of 0.5 × 106 cells in a 6-well plate and transfected with 1 μg plasmid DNA using Gene Jammer (Agilent Technologies). Forty-eight hours post transfection, culture supernatants were collected, and cells were lysed using cell lysis buffer (Cell Signaling) containing protease inhibitor cocktail (Cell Signaling). Approximately 50 μg of culture supernatants and cell lysates were run with an Odyssey Protein Molecular weight ladder (Licor) on 4–12% pre-cast bis-tris gel (Invitrogen). The separated peptides were transferred to PVDF membrane (iblot 2, Thermo Fisher). The membrane was blocked with Odyssey blocking buffer (Licor) for 1 h at room temperature. Heavy and light chains were detected using IRDye 680RD goat anti-human IgG (H + L) (Licor). The blot was scanned using Odyssey CLx (Licor).
Enzyme-Linked Immunosorbent Assay (ELISA)
To assess in vitro binding of HBV-DMAb to HBsAg, 96 well maxisorp plate (Nunc) were coated with 1 μg/ml plasma purified HBsAg subtype ad (Fitzgerald) for overnight at 4ᵒC. Wells were blocked with PBS containing 10% FBS at room temperature for 1 hour. Serially diluted mouse sera were added to the wells. Bound antibodies were detected using 1:10000 diluted goat anti-human IgG lambda light chain HRP conjugate (Bethyl Laboratories). Plates were developed using SigmaFast OPD substrate (Sigma Aldrich) for 30 min. Absorbance was measured at 492 nm using Synergy2 plate reader (Biotek). Measurement of HBsAg secreted in the culture supernatant was done using GS HBsAg 3.0 ELISA kit (Bio-Rad) according to manufacturer’s instructions. Absorbance was measured using Synergy2 plate reader (Biotek).
Quantitative Enzyme-Linked Immunosorbent Assay (ELISA)
For quantification of human IgG in culture supernatant, cell lysate and mouse sera, 96 well Maxisorp plates (Nunc) were coated overnight at 4ᵒC with 10 μg/ml goat anti-human IgG-Fc fragment (Bethyl Laboratories). Plates were washed with phosphate-buffered saline containing 0.05% tween 20 (PBST) followed by blocking with PBS containing 10% FBS at room temperature for 1 h. Samples were diluted in PBST containing 1% FBS and added to the wells. Detection of bound antibodies was performed using 1:10000 diluted goat anti-human IgG lambda light chain HRP conjugate (Bethyl Laboratories). Plates were developed using Sigmafast OPD substrate (Sigma Aldrich). A standard curve was generated using purified human IgG lambda light chain (Bethyl Laboratories). Absorbance was measured at 492 nm using Synergy2 plate reader (Biotek).
Preparation of HBV stock and quantification by qPCR
HepG2.2.15 cells were cultured for production of HBV as described previously. The culture supernatants were harvested, pooled, and filtered using 0.45 μM filter to remove cell debris. The filtered supernatant was concentrated using an Amicon centrifugal filter (EMD Millipore, MWCO 100kD) and stored in aliquots at −80ᵒC. HBV DNA was extracted from 50 μl concentrated supernatant using PureLink viral DNA/RNA kit (Invitrogen) according to manufacturer’s instructions. Quantification of HBV-DNA copies was performed by qPCR using TaqMan Universal PCR master mix (Applied Biosystems) and the following primers (5ʹ-GTCCTCCAATTTGTCCTGG-3ʹ-forward primer), 5ʹ-TGAGGCAT AGCAGCAGGAT-3ʹ (reverse primer) 5ʹ-/56FAM/CT GGA TGT G/ZEN/T CTG CGG CGT TTT ATC AT/3IABkFQ/-3ʹ (probe). PCR was performed on 7900 HT Fast Real-Time PCR system (Applied Biosystems) using the following cycling conditions: 50ᵒC for 2 min, an initial denaturation at 95ᵒC for 10 min and 40 cycles at 95ᵒC for 15 s and 60ᵒC for 1 min. A standard curve composed of 101 to 106 copies of synthetic HBV DNA (ATCC VR-3232SD) was generated and HBV-DNA copies in the concentrated supernatant was determined ().Citation15,Citation18 Following qPCR, agarose gel electrophoresis was performed using the qPCR reaction system and synthetic HBV DNA (ATCC) as positive control to determine the presence of HBV specific 81 bp amplicon (). All the analyses were performed using the SDS v2.4 statistical software.
Quantification of HBV-specific mRNAs by quantitative RT-PCR
Total RNA from HBV-infected HepaRG cells was isolated using a RNeasy Mini kit (Qiagen). On-column DNase digestion of extracted RNA was performed using RNase-free DNase set (Qiagen). Quantification of extracted RNA was done using a Nanodrop 2000 spectrophotometer (Thermo Fisher). Approximately 500 ng of total RNA was reverse transcribed into cDNA using high capacity cDNA Reverse Transcription Kit (Thermo Fisher). qPCR was performed using SYBR Green PCR Master Mix (Thermo Fisher) and primers (HBV3.5 F: 5′-GAGTGTGGATTCGCACTCC-3′) and (HBV3.5 R: 5′-GAGGCGAGGGAGTTCTTCT-3′) for HBV 3.5 kB transcript, (HBVtotalF: 5′-TCACCAGCACCATGCAAC-3′) and (HBVtotalR: 5′-AAGCCACCCAAGGCACAG-3′) for total HBV-specific transcripts, (GAPDHF: CCTGCACCACCAACTGCTTA-3ʹ) and (GAPDHR: 5ʹ-AGTGATGGCATGGACTGTGGT-3ʹ) for GAPDH mRNA. PCR was performed on 7900 HT Fast Real-Time PCR system (Applied Biosystems) using the following cycling conditions: initial denaturation at 95ᵒC for 10 min and 40 cycles at 95 ᵒC for 15 s and 60ᵒC for 1 min. Levels of HBV-specific mRNAs were normalized to GAPDH and expressed relative to cells treated with Nabi-HB by the ∆∆Ct method.
Native PAGE and Western blot analysis
For native PAGE, plasma purified HBsAg (Fitzgerald) was run on pre-cast 3–12% Native PAGE Bis-Tris gel (Thermo Fisher) along with NativeMark Unstained Protein Standard (Thermo Fisher). The gel was cut into two sections. One section was stained using Biosafe Coomassie stain (Bio-Rad) to visualize the migration of HBsAg under native conditions and the remaining half was used for electroblotting of HBsAg onto PVDF membrane using an iblot 2 dry blotting system (Thermo Fisher). Following electroblotting, the membrane was blocked with 5% skimmed milk in PBS for 1 h at room temperature. The membrane was incubated with pooled mouse sera for 1 h. HBsAg was detected using 1:5000 diluted goat anti-human IgG lambda light chain HRP conjugate (Bethyl Laboratories) by chemiluminescence using LumiGLO reagent (Cell Signaling). Image was captured using ImageQuant LAS 4000 system (GE Healthcare).
Immunofluorescence assay (IFA)
To determine if DMAb binds HBV, immunofluorescence was performed as described.Citation19 HepG2.2.15 cells were grown on 4 chambered tissue culture-treated slides (BD Falcon). Cells were washed with PBS and fixed with 4% paraformaldehyde. After being permeabilized with 0.1% Triton-X 100 in PBS, nonspecific binding was blocked with 5% goat serum (Jackson Immunoresearch) at room temperature for 1 h. The cells were washed with PBS and incubated with mouse serum diluted 1:20 in 1% BSA at room temperature for 1 h. HBsAg staining was performed using Goat-anti human IgG (H + L) Alexa Fluor 594 conjugate (Thermo Fisher). Fluoroshield mounting media with DAPI (Abcam) was added to stain the nuclei of cells. Coverslips were mounted on the slides and observed under Nikon 80i Upright Microscope for fluorescence imaging. For detection of HBV production by HepG2.2.15 cells by immunofluorescence, mouse anti-hepatitis B virus preS2 antigen antibody S26 (Abcam) was used as primary antibody while goat anti-mouse IgG (H + L) Alexa Fluor 594 (Abcam) was used as secondary antibody.
Infection with HBV
Plating and culture of HepaRG cells: HepaRG cells were resuspended in Basal Medium for thawing and plating (Lonza) and viability was checked by the Trypan Blue dye exclusion. The cells were seeded in collagen-coated 24-well tissue culture plates at a density of 0.48 × 106 cells/well. Twenty-four hours post plating, the medium was changed to Basal medium for maintenance (Lonza). The cells were maintained at 37ᵒC in a 5% humified CO2 incubator.
Neutralization assay
At 96-h post plating, cells were infected with HBV at an MOI of 500 HBV-DNA copies/cell as described below. HBV inoculum was mixed either with medium alone, Nabi-HB (1.25 mg) or HBV-DMAb containing sera (10 μg) and incubated at 37ᵒC for 90 min. Following incubation, the mixture was added to HepaRG cells in the presence of 4% PEG-8000 for 24 hours at 37ᵒC. After 24 hours, the inoculum was removed, and the cells were washed thrice with fresh culture medium. Culture medium was changed every 2 days. Culture supernatant and cells were harvested 8 days post infection and viral infection were analyzed by measuring HBsAg secreted in the culture supernatant and expression of HBV-specific mRNAs in the cells by qRT-PCR.Citation18,Citation20
Statistics
Differences between the means of experimental groups were calculated using a two-tailed unpaired Student’s t-test or one-way ANOVA where two categorical variables were measured. Repeated measures were analyzed using 2-way ANOVA. Error bars represent standard error of the mean. Survival rates were compared using the log-rank test. All statistical analyses were done using Graph Pad Prism 7.0. p < .05 was considered statistically significant.
Results
HBV DMAb expresses in vitro and in vivo
Our group and others have published multiple reports regarding the immune potency of highly optimized synthetic DNA vaccines delivered by in vivo electroporation, and we have now adapted this platform to deliver monoclonal antibodies (MAb) in vivo by encoding them in DNA plasmids. We have demonstrated that this novel platform is capable of inducing sufficient MAb levels to protect mice in a number of specific infectious disease models.Citation13−17,Citation14,Citation21 Recently, we demonstrated that this approach can generate protective levels of MAb in an NHP challenge model.Citation22 We were interested in developing more enhanced levels of expression in the context of immunotherapy for HBV infection. Such an approach could provide an additional serum-free tool for protection in seronegative, at-risk persons.Citation2,Citation23,Citation24
Multiple immunotherapeutic approaches for preventing HBV infection and treatment have been evaluated over the past several decades. We focused on developing a protective antibody response against regions of the HBsAg important in immune protection from viral infection. The HBsAg contains a highly antigenic segment known as the major hydrophilic region (MHR) from amino acids 100–169.Citation25 The MHR consists of a complex set of continuous and discontinuous epitopes defined by disulfide bridging. The common “a determinant region” of MHR classically includes amino acids 124–147 of the HBsAg and is shared by all HBV genotypes and serotypes (adw, ady, ayw, ayr).Citation4,Citation26 It contains a major conformation-dependent neutralizing epitope of HBV which is the principal binding site of anti-HBs following natural infection, after immunization with Hepatitis B vaccine, and during HBIG prophylaxis.Citation4,Citation23,Citation27 Recent reports describe monoclonal antibodies that target this region of HBV.Citation11
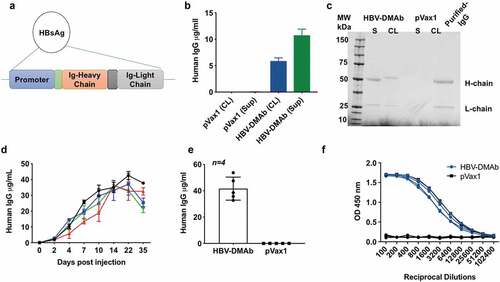
We systematically converted an anti-HBV MAb sequence into a single-plasmid antibody-encoding DNA cassette for insertion into a DNA plasmid and optimized it with the aim of increasing HBV-specific human IgG production from the construct. We optimized the leader sequences from the Ig and performed RNA and codon optimization as we have described.Citation28–Citation30 The synthetic DNA encoding the human H and L chains were synthesized independently and cloned into a mammalian expression plasmid pVax1 (). We then studied in vitro expression in cell lines for the HBV-DNA construct by quantitative enzyme-linked immunosorbent assay (ELISA) of supernatant and cell lysates of 293 T cells transfected with HBV-DMAb. These analyses confirmed intracellular expression as well as potent secretion of human IgG (). Moreover, Western blot analyses demonstrated the presence of IgG heavy and light chains in the supernatant and cell lysate of HBV-DMAb transfected cells (). To then study if the HBV-DMAb was expressed in vivo, athymic nude CAnN.Cg-Foxn1nu/Crl mice were administered HBV-IgG plasmid DNA at dose of 100 μg or 400 μg utilizing Cellectra 3P electroporation. Notably, significant human IgG expression persisted for at least 4–5-weeks post a single injection () with 100 μg dose. Mean expression levels in mice sera immunized with HBV-DMAb were 41.6 μg/ml (). The DMAb-expressed antibody exhibited substantial reactivity to HBV viral antigen in ELISA assay (). These results demonstrate that our synthetic DMAb plasmid is capable of inducing HBV-specific antibody expression in vitro and in vivo.
Figure 2. HBV-DMAb expression in vitro and in vivo.
Functional binding activity of in vivo expressed DMAb
The in vitro binding activity of the human IgG in sera collected from nude mice immunized with HBV-DMAb plasmid was next determined as previously described.Citation13,Citation14,Citation22,Citation28 The sera from the mice was found to bind plasma purified native HBsAg. The binding specificity of HBV-DMAb was also tested using HepG2.2.15 cells by IFA to determine reactivity against HBV-viral produced antigen. Sera from mice immunized with HBV-DMAb was found to bind to HBV produced by HepG2.2.15 cells ().
Figure 3. HBV-DMAb binds a specific epitope of HBsAg.
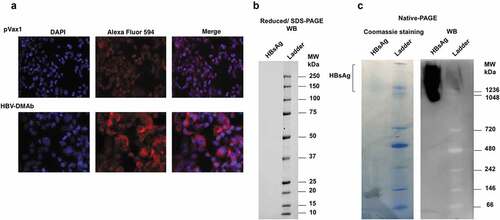
HBV-DMAb recognition of conformational antigenic epitope
The HBV-DMAb was developed from an antibody sequence reported to recognize a conformational epitope in the common “a determinant region” of s-HBsAg. To test whether the DMAb produced in vivo identified a conformational epitope, plasma-purified HBsAg, cells were separated under denaturing (reducing) and native conditions by polyacrylamide gel electrophoresis. Western blot analysis of denatured and native HBsAg using sera from nude mice immunized with HBV-DMAb indicated that the sera did not bind denatured HBsAg () but only to HBsAg in its native conformation (). These results indicated that the DMAb bound to a conformational epitope of s-HBsAg (). The functionality of this approach was next studied.
HBV-DMAb neutralizes hepatitis B virus
The neutralizing potential of HBV-DMAb produced IgG was tested using differentiated HepaRG cells. Cells were infected at a multiplicity of infection of 500 HBV-DNA copies/cell in the presence or absence of HBV-DMAb containing sera (10 µg). Nabi-HB-(Hepatitis B Immune Globulin in a sterile solution of immunoglobulin containing antibodies to hepatitis B surface antigen; 1.25 mg) served as a positive control for the neutralization experiment. Analysis of infection in HepaRG cells was performed using culture supernatants and cells collected 8 days post infection as indicated in . Analysis of viral load in the culture supernatant was by quantitative PCR. Analysis of viral load in the culture supernatant demonstrated that there was a significant reduction in the level of HBV-DNA in culture supernatant of cells treated with HBV-DMAb containing sera as compared to untreated cells (p = .0001) (). Quantification of total HBV-RNA and 3.5kb pre-genomic RNA in HepaRG cells was performed by Real-Time Quantitative Reverse Transcription PCR (qRT-PCR). The expression of total HBV-RNA in DMAb-containing sera treated cells was over 80-fold lower compared to untreated cells (p = .0001) () and was further reduced compared to Nabi-HB treated samples.
Figure 4. HBV-DMAb neutralizes HBV and blocks infection of HepaRG cells.
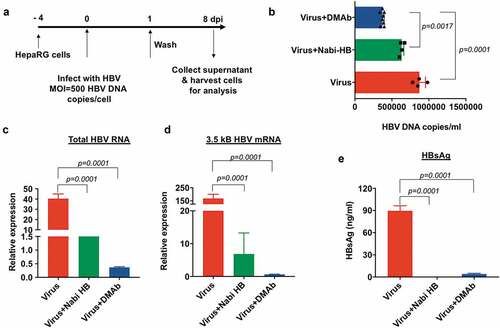
A similar trend in the expression level of 3.5kB HBV mRNA was observed for HepaRG cells treated with DMAb-containing sera where a more than 150-fold reduction in virus was observed (). ELISA-based detection of HBsAg secreted into the culture supernatants demonstrated close to complete suppression of viral antigen (HBsAg) as compared to untreated cells (p = .0001) (). HBV-DMAb performed exceedingly well in neutralizing HBV as compared to Nabi-HB treated, as just 10 μg HBV-DMAb containing sera administered was compared to 1.25 mg Nabi-HB in these neutralization experiments demonstrating the high potency of DMAb-containing sera for prevention of infection in this important assay system.
Discussion
Hepatitis B virus infection is an important cause of acute and chronic liver disease in the United States and globally.Citation15,Citation18,Citation31,Citation32 The total costs for this infection due to hospitalizations in the United States in 2006 were estimated to be 1.3 USD billion. The expenditure for prophylaxis for patients receiving liver transplantation (LT) is extremely high ranging between 2,000 USD and 10,000 USD per month in various countries for an undefined period and presumably for life.Citation1,Citation10 As a consequence, there is a need for additional non-blood based and more cost-efficient modes of therapy to treat HBV infection.
In the present study, we report the construction and in vivo delivery of an optimized plasmid DNA which was designed to express an anti-HBV human monoclonal antibody which targets a functional region of the HBsAg. The human DMAb is directed against the common “a determinant region” of HBsAg which carries a major HBV neutralizing epitope for anti-HBs that is highly conserved.Citation11,Citation20 We observed that the DMAb reacted well with the native antigen as well as HepG2.2.15 cells but reacted poorly with the denatured antigen, supporting the three-dimensional nature of the epitope targeted. Advantages of the DMAb platform include the ease and cost-effectiveness of producing DNA plasmids compared to protein therapies and the ease of delivery of plasmid DNA as well as its ability to generate sustained in vivo monoclonal antibody expression which in the present study lasted at least one-month post administration. Moreover, the epitope specificity of the monoclonal antibody was known and is well characterized. For HBIG, the preparation method is based on the physical characteristic of immunoglobulins and not on the specificity of the antibodies, the precise antigen specificity of most HBV immunoglobulin preparations is not known and thus far, are only determined to be reactive with HBV surface antigen proteins in general. This provides a source of variability in these preparations, that can facilitate poor control or viral escape as well as having the complication of being a blood-based product. The DMAb technology might have advantages in this regard as a stand-alone additional tool or to be used in combination with current HBV immunotherapies.
The ability of HBV-DMAb to neutralize HBV is relevant. Using the Nabi-HB as a positive control for viral neutralization we observed that the concentration of HBV DMAb required to neutralize HBV was 100x less than the required dose of Nabi-HB (10 µg DMAb vs 1.25 mg of Nabi-HB) supporting the potency of this platform. This was illustrative in the drastic reduction of HBV 3.5kb pre-genomic RNA and total HBV-specific RNAs detected in cells treated with HBV DMAb as compared to untreated cells and the reduction in the level of secreted HBsAg from HBV-infected HepaRG cells which was studied in the treatment groups.
Delivery of DMAbs is a novel approach, which only recently has been developed.Citation13,Citation22,Citation29,Citation33,Citation34 Utilizing this approach, we can bypass protein IgG delivery, and provide a potential alternative technology, which may address the limitations of conventional delivery of protein-based IgG. DMAbs have the potential to provide rapid protection against infectious diseases such as HBV through in vivo antibody production. Conceptually, this technology provides the simplicity of immunization with the power of specific mAb delivery. Advantages of this approach include rapid protection or treatment of affected populations, including health care workers, travelers, and immune-compromised personnel. Furthermore, this technology has the possibility to enable routine mAb delivery to treat other circulating and emerging infectious diseases. Immune escape by hepatitis B viruses is a challenge for all form of antiviral therapeutics, including DMAbs; however, through the use of combination therapeutics such as co-administration of broadly neutralizing DMAbs with small molecule antivirals or HBIG, potentially superior control over the HBV infection could be achieved. Also, it is necessary to determine that with each DMAb target and strategy that a sufficient level of mAb is produced to mediate the desired biological effect in terms of quantity and duration compared to conventional delivery of protein-based mAbs.
In conclusion, for the first time, we demonstrate the expression of a human monoclonal antibody from a DMAb platform with a potent HBV neutralizing ability which may be used for immunoprophylaxis of HBV infection.
Author contributions
U.Z. and K.M. designed, analyzed the data analysis and prepared the manuscript. U.Z., S.K., M.K., A.P.P., H.C., M.H., F.Z., performed experiments and analyzed the data. K.U., aided in resources, and manuscript preparation. D.B.W and K.M.; review & editing and oversaw final manuscript preparation.
Disclosure of potential conflicts of interest
KM and DBW note funding by Inovio Pharmaceuticals, PA, USA. KM discloses grant funding, industry collaborations, speaking honoraria, and fees for consulting from Inovio Pharmaceuticals related to DNA and DMAb vaccine development. He has a patent application for DNA vaccine development and delivery of DNA-encoded monoclonal antibodies pending to Inovio Pharmaceuticals. Remuneration includes direct payments. DBW discloses grant funding, SAB and Board service, industry collaborations, speaking honoraria, and fees for consulting. His service includes serving on scientific review committees and advisory boards. Remuneration includes direct payments and/or stock or stock options. He notes potential conflicts associated with this work with Pfizer, Bristol Myers Squibb, Inovio Pharmaceuticals, Merck, VGXI, Geneos, AstraZeneca, and potentially others. Licensing of technology from this laboratory has created over 150 jobs in the biotech/pharma industry. J.J.K is employer of Inovio Pharmaceuticals and as such receive salary and benefits, including ownership of stock and stock options. The other authors declare no competing financial interests. The other authors declare no competing financial interests. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No writing assistance was utilized in the production of this manuscript. No writing assistance was utilized in the production of this manuscript.
Acknowledgments
The authors would like to acknowledge other members of the Weiner laboratory for their significant contributions and/or critical reading and editing of the paper. We would like to thank the Wistar Flow Cytometry Facility and Animal Facility for their technical assistance.
Additional information
Funding
References
- Hofmeister MG, Foster MA, Teshale EH. Epidemiology and transmission of Hepatitis A virus and Hepatitis E virus infections in the United States. Cold Spring Harb Perspect Med. 2019;9(4):a033431. doi:10.1101/cshperspect.a033431.
- Nelson KE. The changing epidemiology of hepatitis A virus infections in the United States. J Infect Dis. 2015;212(2):171–72. doi:10.1093/infdis/jiu835.
- Dosik H, Jhaveri R. Prevention of neonatal hepatitis B infection by high-dose hepatitis B immune globulin. N Engl J Med. 1978;298(11):602–03. doi:10.1056/NEJM197803162981105.
- Coleman PF. Detecting hepatitis B surface antigen mutants. Emerg Infect Dis. 2006;12:198–203. doi:10.3201/eid1203.050038.
- Chirikov VV, Marx SE, Manthena SR, Strezewski JP, Saab S. Development of a comprehensive dataset of Hepatitis C patients and examination of disease epidemiology in the United States, 2013–2016. Adv Ther. 2018;35:1087–102. doi:10.1007/s12325-018-0721-1.
- Park JH, Cho EW, Lee YJ, Shin SY, Kim KL. Determination of the protective effects of neutralizing anti-hepatitis B virus (HBV) immunoglobulins by epitope mapping with recombinant HBV surface-antigen proteins. Microbiol Immunol. 2000;44:703–10. doi:10.1111/j.1348-0421.2000.tb02552.x.
- Brown SE, Howard CR, Zuckerman AJ, Steward MW. Affinity of antibody responses in man to hepatitis B vaccine determined with synthetic peptides. Lancet. 1984;2:184–87. doi:10.1016/S0140-6736(84)90479-3.
- Said ZN, Abdelwahab KS. Induced immunity against hepatitis B virus. World J Hepatol. 2015;7:1660–70. doi:10.4254/wjh.v7.i12.1660.
- Zuckerman JN. Review: hepatitis B immune globulin for prevention of hepatitis B infection. J Med Virol. 2007;79:919–21. doi:10.1002/jmv.20816.
- Hyun Kim B, Ray Kim W. Epidemiology of hepatitis B virus infection in the United States. Clin Liver Dis (Hoboken). 2018;12:1–4. doi:10.1002/cld.732.
- Cerino A, Bremer CM, Glebe D, Mondelli MU. A human monoclonal antibody against hepatitis B surface antigen with potent neutralizing activity. PLoS One. 2015;10:e0125704. doi:10.1371/journal.pone.0125704.
- Tan W, Meng Y, Li H, Chen Y, Han S, Zeng J, Huang A, Li B, Zhang Y, Guo Y, et al. A bispecific antibody against two different epitopes on hepatitis B surface antigen has potent hepatitis B virus neutralizing activity. MAbs. 2013;5(6):946–55. doi:10.4161/mabs.26390.
- Perales-Puchalt A, Duperret EK, Yang X, Hernandez P, Wojtak K, Zhu X, et al. DNA-encoded bispecific T cell engagers and antibodies present long-term antitumor activity. JCI Insight. 2019 Apr 18;4(8). pii: 126086. doi:10.1172/jci.insight.126086. eCollection 2019 Apr 18.
- Patel A, DiGiandomenico A, Keller AE, Smith TRF, Park DH, Ramos S, Schultheis K, Elliott STC, Mendoza J, Broderick KE, et al. An engineered bispecific DNA-encoded IgG antibody protects against Pseudomonas aeruginosa in a pneumonia challenge model. Nat Commun. 2017 Sep 21;8(1):637. doi:10.1038/s41467-017-00576-7.
- Michailidis E, Pabon J, Xiang K, Park P, Ramanan V, Hoffmann -H-H, Schneider WM, Bhatia SN, de Jong YP, Shlomai A, et al. A robust cell culture system supporting the complete life cycle of hepatitis B virus. Sci Rep. 2017;7(1):16616. doi:10.1038/s41598-017-16882-5.
- Muthumani K, Marnin L, Kudchodkar SB, Perales-Puchalt A, Choi H, Agarwal S, Scott VL, Reuschel EL, Zaidi FI, Duperret EK, et al. Novel prostate cancer immunotherapy with a DNA-encoded anti-prostate-specific membrane antigen monoclonal antibody. Cancer Immunol Immunother. 2017;66(12):1577–88. doi:10.1007/s00262-017-2042-7.
- Duperret EK, Trautz A, Stoltz R, Patel A, Wise MC, Perales-Puchalt A, Smith T, Broderick KE, Masteller E, Kim JJ, et al. Synthetic DNA-encoded monoclonal antibody delivery of anti-CTLA-4 antibodies induces tumor shrinkage in vivo. Cancer Res. 2018;78(22):6363–70. doi:10.1158/0008-5472.CAN-18-1429.
- Scheel TK, Luna JM, Liniger M, Nishiuchi E, Rozen-Gagnon K, Shlomai A, Auray G, Gerber M, Fak J, Keller I, et al. A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe. 2016;19(3):409–23. doi:10.1016/j.chom.2016.02.007.
- Choi H, Kudchodkar SB, Reuschel EL, Asija K, Borole P, Ho M, Wojtak K, Reed C, Ramos S, Bopp NE, et al. Protective immunity by an engineered DNA vaccine for Mayaro virus. PLoS Negl Trop Dis. 2019;13(2):e0007042. doi:10.1371/journal.pntd.0007042.
- Shlomai A, Schwartz RE, Ramanan V, Bhatta A, de Jong YP, Bhatia SN, Rice CM. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A. 2014;111(33):12193–98. doi:10.1073/pnas.1412631111.
- Khoshnejad M, Patel A, Wojtak K, Kudchodkar SB, Humeau L, Lyssenko NN, Rader DJ, Muthumani K, Weiner DB. Development of novel DNA-Encoded PCSK9 monoclonal antibodies as lipid-lowering therapeutics. Mol Ther. 2019;27(1):188–99. doi:10.1016/j.ymthe.2018.10.016.
- Esquivel RN, Patel A, Kudchodkar SB, Park DH, Stettler K, Beltramello M, Allen JW, Mendoza J, Ramos S, Choi H, et al. In vivo delivery of a DNA-encoded monoclonal antibody protects non-human primates against Zika virus. Mol Ther. 2019;27(5):974–85. doi:10.1016/j.ymthe.2019.03.005.
- Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64(12):1972–84. doi:10.1136/gutjnl-2015-309809.
- Ditah I, Ditah F, Devaki P, Ditah C, Kamath PS, Charlton M. Current epidemiology of hepatitis E virus infection in the United States: low seroprevalence in the National Health and Nutrition Evaluation Survey. Hepatology. 2014;60(3):815–22. doi:10.1002/hep.27219.
- Suehiro T, Shimada M, Kishikawa K, Shimura T, Soejima Y, Yoshizumi T, Hashimoto K, Mochida Y, Maehara Y, Kuwano H, et al. Prevention of hepatitis B virus infection from hepatitis B core antibody-positive donor graft using hepatitis B immune globulin and lamivudine in living donor liver transplantation. Liver Int. 2005;25(6):1169–74. doi:10.1111/j.1478-3231.2005.01165.x.
- Ditah I, Ditah F, Devaki P, Ewelukwa O, Ditah C, Njei B, Luma HN, Charlton M. The changing epidemiology of hepatitis C virus infection in the United States: National Health and Nutrition Examination Survey 2001 through 2010. J Hepatol. 2014;60(4):691–98. doi:10.1016/j.jhep.2013.11.014.
- Shields PL, Owsianka A, Carman WF, Boxall E, Hubscher SG, Shaw J, O’Donnell K, Elias E, Mutimer DJ. Selection of hepatitis B surface “escape” mutants during passive immune prophylaxis following liver transplantation: potential impact of genetic changes on polymerase protein function. Gut. 1999;45(2):306–09. doi:10.1136/gut.45.2.306.
- Elliott STC, Kallewaard NL, Benjamin E, Wachter-Rosati L, McAuliffe JM, Patel A, Smith TRF, Schultheis K, Park DH, Flingai S, et al. DMAb inoculation of synthetic cross reactive antibodies protects against lethal influenza A and B infections. NPJ Vaccines. 2017;2(1):18. doi:10.1038/s41541-017-0020-x.
- Flingai S, Plummer EM, Patel A, Shresta S, Mendoza JM, Broderick KE, Sardesai NY, Muthumani K, Weiner DB. Protection against dengue disease by synthetic nucleic acid antibody prophylaxis/immunotherapy. Sci Rep. 2015;5(1):12616. doi:10.1038/srep12616.
- Muthumani K, Falzarano D, Reuschel EL, Tingey C, Flingai S, Villarreal DO, Wise M, Patel A, Izmirly A, Aljuaid A, et al. A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Transl Med. 2015;7:301ra132. doi:10.1126/scitranslmed.aac7462.
- Kourtis AP, Bulterys M, Hu DJ, Jamieson DJ. HIV-HBV coinfection–a global challenge. N Engl J Med. 2012;366:1749–52. doi:10.1056/NEJMp1201796.
- Jourdain G, Ngo-Giang-Huong N, Harrison L, Decker L, Khamduang W, Tierney C, Salvadori N, Cressey TR, Sirirungsi W, Achalapong J, et al. Tenofovir versus Placebo to prevent perinatal transmission of hepatitis B. N Engl J Med. 2018;378(10):911–23. doi:10.1056/NEJMoa1708131.
- Muthumani K, Block P, Flingai S, Muruganantham N, Chaaithanya IK, Tingey C, Wise M, Reuschel EL, Chung C, Muthumani A, et al. Rapid and long-term immunity elicited by DNA-encoded antibody prophylaxis and DNA vaccination against chikungunya virus. J Infect Dis. 2016;214(3):369–78. doi:10.1093/infdis/jiw111.
- Patel A, Park DH, Davis CW, Smith TRF, Leung A, Tierney K, Bryan A, Davidson E, Yu X, Racine T, et al. In vivo delivery of synthetic human DNA-encoded monoclonal antibodies protect against ebolavirus infection in a mouse model. Cell Rep. 2018;25:1982–93 e4. doi:10.1016/j.celrep.2018.10.062.
