ABSTRACT
Humoral immunity is crucial for an efficient host immune response against rabies virus (RABV) infection. But the B cell receptor (BCR) repertoire in human after RABV vaccine immunization remained unclear. To study the BCR repertoires in peripheral blood mononuclear cells (PBMCs) of human immunized with rabies virus vaccine. In this study, we conducted BCR complementarity determining region 3 (CDR3) repertoires in 4 healthy volunteers before and after immunization with RABV vaccine by high-throughput sequencing. The bioinformatics analysis process was performed. The results showed that RABV vaccination changed the BCR diversity and the usage of V/J gene segments, as well as V-J pairing. B cell clone expansion was induced by the vaccination and sequences of high expand CDR3 aa clones were identified. To the best of our knowledge, we firstly quantitative characterized B cell receptor repertoire of human immunized with c rabies virus vaccine. It might provide us with new insights into B cell receptor condition after RABV vaccination.
Introduction
Rabies is an infectious disease caused by rabies virus (RABV) infection in the mammalian central nervous system (CNS). Rabies is a zoonotic disease with a high mortality, once clinical symptoms occur, the mortality rate is as high as 100%. According to statistics from the World Health Organization, more than 50000 people die from rabies each year worldwide, but the vast majority of cases occur in developing countries such as Asia and Africa.Citation1,Citation2 RABV infects nearly all warm-blooded animals, dogs and cats are their main storage hosts, and humans and various mammals are hosts of transmission. The infected animals transmit RABV by scratching, and then infect the host’s CNS.Citation3 RABV is usually transmitted through saliva and blood, almost without gastrointestinal tract, respiratory tract and mother-to-child transmission. Most RABV infections start from a dermal or muscular wound. RABV replicate locally in muscle tissue, and then enter a neuron and spread to motor neurons through synapses between muscles and motor neurons. The RABV transports to CNS by retrograde axonal transporting. Once RABV reaches the CNS, the clinical symptoms of rabies appeared, where RABV elicited neuronal dysfunction and led to death.Citation4,Citation5 Although much advances have been achieved in prevention of RABV, the mechanism by which RABV causes fatal disease remains unclear.
If the body had been infected, it needs to have a post-exposure prophylaxis (PEP), which consists of administration of inactivated RABV vaccine together with passive antibody therapy with human rabies immunoglobulin (HRIG) or equine rabies immunoglobulin (ERIG).Citation6–8 Whether rabies is prevented, it depends on PEP provided promptly and correctly. The selection of rabies virus vaccine is particularly important. The traditional rabies virus vaccine is mainly inactivated vaccine. However, the current vaccine not only needs multiple injections, but also costs much time and money, what’s more, studies had reported some rabies-exposed patients were reported vaccination failures because of the failure of timely co-administer RIG.
Due to the repeated injection and the vaccine cost, developing a low-cost, highly effective vaccine is particularly urgent. Now some vaccine to prevent the rabies had been development.Citation9 However, for the pathogenesis of RABV infection, there are relatively few discoveries, which limits the design and improvement of rabies virus vaccine. In the current study, we firstly quantitative characterized B cell receptor (BCR) repertoire of human immunized with rabies virus vaccine. It might provide us with new insights into the biological role of B cells in protection of host against RABV infection and understanding the immune senescence to improve vaccine responses.
Materials and methods
Experimental design
Four healthy volunteers (V1: Male, 31 years old; V2: Female, 38 years old; V3: Male, 34 years old; V4: Female, 23 years old), with no rabies virus vaccine history, were recruited to receive three doses of Rabies Vaccine (Vero Cell) for Human Use, Freeze-dried (Cat # 201703027–1; Changchun Institute of Biological Products Co. Ltd), the active ingredient is inactivated fixed rabies virus strain aG. The vaccination was followed the pre-exposure procedure according to the manufacturer’s instructions. The immunization procedure contained 3 injections: 1 dose was injected on 0 days, 7 days, 21 days. And 10 ml peripheral blood samples were obtained from the volunteers at 0day and 28 days, the peripheral blood mononuclear cells (PBMCs) had been separated using Ficoll-Paque (GE Healthcare) gradient centrifugation for further BCR repertoire sequence. This study was conducted according to protocols approved by the Research Ethics Committee of Yuebei People’s Hospital, Shantou University Medical College, Shaoguan, China. Written informed consents were obtained from all participants.
Multiplex PCR procedure
The multiplex PCR primers were according a published paperCitation10 to amplify the CDR3 regions of IGH V/J. The total PBMC RNA had been subjected to DNase I digestion (NEB) in order to remove the genomic DNA and terminated with addition of 1 mM EDTA. First-strand cDNA was synthesized using SuperScript I reverse transcriptase (Invitrogen). First-strand cDNA was used as a template for multiplex PCR with the primers pool for the BCR repertoires. The primer pool is available in the published paper.Citation10 The reaction conditions were follows the QIAGEN Multiplex PCR Kit with the IGH primers pool. PCR conditions were 95°C for 15 min, 25–30cycles of 94°C for 30 s, 60°C for 90 s and 72°C for 30 s, and 72°C for 5 minutes. The PCR product was gel-extracted and purified.
Sequencing and bioinformatics analysis process
The sequencing process was performed as the reference,Citation10 including the cluster generation, template hybridization, isothermal amplification, linearization, blocking and denaturation and hybridization. Paired-end sequencing of samples was carried out with a read length of 150bp using the Illumina HiSeqTM Xten (Novaseq) platform.
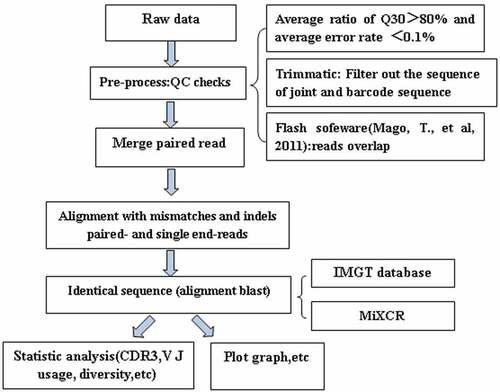
The bioinformatics analysis process was performed as followed: 1) The sequenced data are called raw data, and then the raw reads is to be quality control (QC) to determine whether the sequencing data is suitable for subsequent analysis, the average ratio of Q30 should be more than 80% and average error rate should less than 0.1%. 2) the joint in the sequence is intercepted, and the low-quality bases or sequences are removed; 3) the clean reads alignment after filtered clean was used to comparison with the reference sequence and do the next assembly to get specific functional areas, such as the CDR3 zone (clones) by the software of Trimmatic, Flash and MiXCR;Citation11,Citation12 4) the clone sequence which was qualified was used as the core clone for comparison and correction; 5) the clones obtained above are again compared to the V, D, J and C reference sequences in the IMGT database (http://www.imgt.org/) according to the reference,Citation13 and the final statistical documents include the sequence of cloning, the sequence of amino acid residues, the number of clones, the frequency of cloning, V/J gene combination and other information.Citation14 Follow-up can be done based on the information, such as clone distribution, gene recombination and diversity analysis.
Statistical analysis
The data were represented as the mean±SEM and comparisons between pre and post were performed using Student’s t test with GraphPad Prism 6 software. B cell receptor diversity index distribution calculated as Inverse Simpson coefficient.Citation15 The B cell receptor overlap was calculated according to the reference.Citation15,Citation16 *P < .05 were considered statistically significant. The CDR3 aa average hydrophilic grand average of hydropathicity (GRAVY) were calculated based on the Kyte and Doolittle method.Citation17
Result
Summary of the BCR CDR3 pre- and post-vaccination
Our study cohort consisted of four volunteers. The detail description of the sequence information is displayed in . The total number of clean reads was range from 7.1 × 106 to 1.8 × 107 before and after vaccination, the total clean reads achieved with 8.0 × 106 to 1.0 × 107. After bioinformatics analysis process (), the total sequence reads were 2.06E+07, with an average of 5.15E+06 reads for each sample before vaccination, and after vaccination the total sequence reads was 1.89E+07, with an average of 4.72E+06 reads. We identified a total of 37 distinct V gene segments and 7 distinct J gene segments among the participants. A detailed description of immune repertoire sequencing data including total clean reads, clones and other sequencing data are presented in .
Table 1. The detail description of the sequence information
A reduced BCR CDR3 diversity pre- and post-vaccination
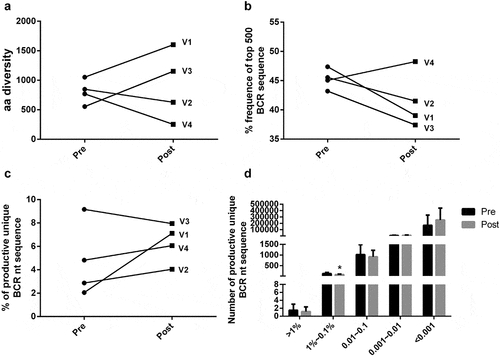
In the diversity of the BCR CDR3, we analysis the diversity index of amino acid (aa) sequence and found that there was no difference whether the volunteers had the vaccination or not (). But, the male-volunteers had higher aa diversity than female, and both the female had a decreased diversity. What’s more, the ratio of the productive unique BCR sequence to the number of productive sequence (namely total clones) provides a general assessment of sample diversity. The ratio of pre-vaccination was 4.72%±1.58%, after vaccination, it reached 6.29%±0.84%, while there was no significant difference vaccination or not (). The expansion level of each unique clone is another major measurement for immune diversity. Clonal expansion was further assessed by calculating the cumulative percentage of the repertoire that was constituted by the top 500 BCR nucleotide sequences. It was shown that the top 500 BCR nucleotide sequences consisted almost half the total BCR repertoire. The average fraction of the top 500 BCR sequences was 45.29% pre-vaccination, 41.56% after vaccination, suggesting the vaccination can decrease the clonally expanded BCR nucleotide sequences (). The BCR nucleotide sequences were similarly diverse before and after vaccination, but only significantly different in copy numbers lower than 1% to 0.1% ().
Figure 2. Clonal distribution of B cells receptor in pre and post vaccination. (A) Data show the distribution of B-cell clonal frequencies through the measurement of amino acid diversity. Each line represented a volunteer’s diversity changes. (B) Percentage frequency of top 500 BCR nucleotide sequence in pre and post vaccination. The line represents the percentage of top 500 BCR sequence in the total repertoire of each volunteer. (C) Percentage of productive unique BCR nt sequence in volunteers. The line represents the percentage of unique sequence in the total productive B-cell receptor repertoire before and after vaccination. (D) Frequency distribution of BCR nucleotide sequence from volunteers. Data are represented as mean±SEM of distribution of volunteers. The differences between groups were compared using one-way ANOVA. *P < .05. Abbreviation: nt, nucleotide; aa, amino acid
The usage patterns of V, J gene and V-J pairing
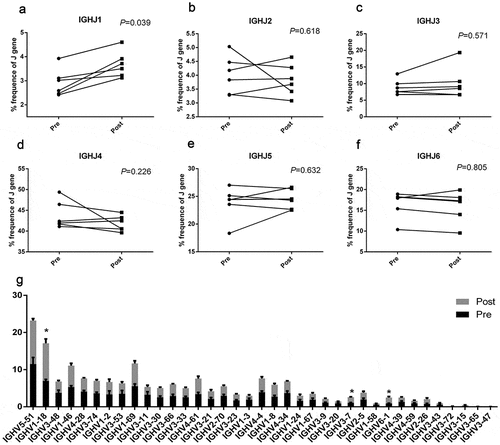
Because of the different genetic background or the viral infected, et al., different people had different repertoires of the B cell receptor. Here we analysis the distribution of the V and J gene before and after vaccination and found these differences. The usage of J gene among the different volunteers was list as followed (pre vs post, %):IGHJ1: 2.84 ± 0.36% vs 3.83 ± 0.30%; IGHJ2: 4.15 ± 0.38% vs 3.81 ± 0.18%; IGHJ3: 9.29 ± 1.38% vs 10.83 ± 2.98%; IGHJ4: 44.66 ± 1.97% vs 41.3 ± 1.08%; IGHJ5: 23.55 ± 1.84% vs 25.03 ± 1.95%; IGHJ6: 15.49 ± 1.82% vs 15.19 ± 2.23%; (). In the usage patterns of V gene segments, we identified total 37 distinct V gene segments, and found only IGV3-7 and IGV2-26 had difference between pre and post vaccination. The present data thus demonstrate a preferential usage of V and J genes from B-cell clones in the volunteers.
Figure 3. V gene and J gene usage of clonotypes in the volunteers. Data show the percentage frequency of J gene (IGHJ1, IGHJ2, IGHJ3, IGHJ4, IGHJ5 and IGHJ6) and V gene usage by clonotypes in volunteers. Data show mean ±SEM frequency of each individual. Each dot represents each individual volunteer’s information. Data were compared to pre-vaccine using paired t tests. **P < .01 and *P < .05
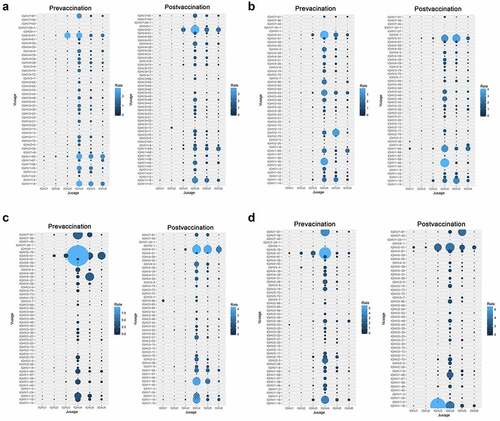
shows all the 4 volunteers’ V-J pairing frequencies, there are 243 possible V-J combinations for BCR repertoires. In our dataset, we successfully captured the majority of V-J combinations from BCR almost 90%) repertoires. Due to the unequal usage of V and J genes in BCR repertoires, we found that the usage of V-J pairing in BCR repertoire were also non-uniform (). In the BCR repertoire, we found that only three V-J combinations have statistical significance, as follows: IGHV3-33/IGHJ2 (0.056% vs 0.20%), IGHV6-1/IGHJ5 (0.20% vs 0.59%) and IGHV4-28/IGHJ2 (0.22% vs 0.12%) ().
Figure 4. The usage frequencies of all V-J pairing in the B cell receptor repertoires A-D represent the individual volunteer’s (V1-V4) V-J pairing frequencies. The x axis represents the J genes and the y axis represents the V genes. The area of the circle is proportional to the frequency of each V-J pairing
CDR3 overlapping sequences pre- and post-vaccination with rabies virus vaccine
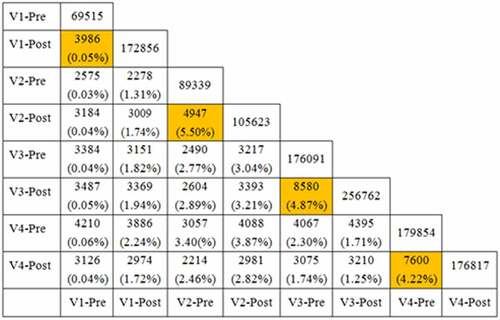
In , it demonstrated the results of overlapping CDR3 repertoires in the 4 healthy volunteers pre- and post-vaccination with rabies virus vaccine. The proportions of the common sequence in different samples were rather low. The CDR3 sequence was not identical in the 4 healthy volunteers. Furthermore, we obtained a high overlapping proportion CDR3 repertoires in the same individual and at the same time (all above 1% and up to 5.50%). A small number of public CDR3 sequences were found in the 4 healthy volunteers before and after immunizations, so as among each other. However, no identical IgG H chain CDR3 sequence was found among the 4 healthy volunteers.
High-frequency CDR3 amino acid sequence analysis of BCR repertoires after vaccination with rabies virus vaccine
As shown in , in the CDR3 repertoires, the proportion of sequence of CDR3 frequency > 0.4% was relatively high and consistent among the 4 healthy volunteers after immunization with rabies virus vaccine. In the same volunteer, most of the frequence of the high cloned compared sequence pre- to post-vaccination had change, and the sequence were different among volunteers. But in the volunteer 4, there are 2 same CDR3 aa exhibit different frequence, i.e. CARTPLFGVVYYFDSW (1.93% vs 0.50%) and CARMRTSQARRFRAEYIQHW (0.48% vs 0.81%). In the high frequency expressed CDR3 sequence, the BCR library of the 4 healthy volunteers after immunization with rabies vaccine demonstrated that the CDR3 region had high frequency of mutations. In the CDR3 aa, we also analysis properties of the peptide, like the isoelectric point (pI) and GRAVY. And we found that the high cloned CDR3 sequence distributions were relatively wide, most of GRAVY of the peptides were negative and exhibited a hydrophilic interaction between the CDR3 and the ligands.
Table 2. Characteristics of high frequency CDR3 sequences (>0.4%) in the B cell receptor repertoires after immunization with rabies virus vaccines in 4 volunteers
Conclusions
In conclusion, with the IR-SEQ, it can help us to compare naïve and immune repertoires after the rabies vaccination and obtain a deeper understanding of the humoral immune system, which could aid in designing vaccination strategies. Here we first investigated the immune system change with the rabies virus vaccine-induced. We believe the comprehensive BCR repertoires of human will be of substantial interest and help to the broad scientific communities. In the further study, we would investigate the construction of precise lineage trees in mutation analysis and CDR region during affinity maturation after vaccination.
Discussion
RABV is one of the most fatal CNS zoonosis cases. It is still a serious threat to human public health. Although many efforts have been made in the study of rabies, there is still no effective treatment. Now the only way to prevent the outbreak of rabies is the rapid immunization after exposure. Therefore, though effective vaccine is available, rabies still has about 70 thousand deaths per year.Citation18 The incubation period of canine disease is different, rang from a few days to more than ten years or even decades. The rabies was almost 100% deaths, and there is still no special therapeutic drug. When the infected person is unable to receive the correct treatment in time, the rabies virus enters the central nervous system and causes the infective restlessness, attack, progressive paralysis, and final death.Citation19 Vaccination can prevent rabies. The selection of rabies virus vaccine is particularly important. The traditional rabies virus vaccine is mainly based on inactivated vaccine. However, the current vaccine not only needs multiple injections, but also costs much time and money.Citation19
The natural infection of rabies virus is mainly characterized by neurotropism. The replication of virus is almost limited in neurons.Citation20 When the virus first entered the body’s wound, rabies virus was not detected in blood. RABV did not enter the blood circulation but replicated in the bitten muscle tissue and then intruded into the peripheral nervous system by neurons and axons.Citation21–24 If the patient has neurological symptoms, it will cause ineffective immunity and lead to death. The human and other mammalian immune systems are unable to cope with RABV without active therapy. By using modern vaccines both before and after the infection, it can prevent the fatal cases from RABV infection. What’s more, it also recommended to use RIG with the vaccine to enhance the efficiency of anti-rabies prophylaxis. In addition to ERIG, HRIG is also used for passive immunization. However, the HRIG is high cost and an insufficient number of donors.Citation25 So, the rabies virus vaccine is the most important way to against rabies infection, especially the PEP.
It is critical to complete the regimen of vaccination for the subjects who are potentially exposed to rabies. But the protective effects raised by the vaccines differ significantly among vaccinees. In the current study, we for the first time compared the profiles of B cell receptor repertoires in PBMCs of human pre- or post-vaccination with commercial rabies virus vaccine.
A comprehensive analysis including V, J and V-J pairing usage patterns, CDR3 length distribution, the composition of amino acids and the high frequencies of the CDR3 sequence were precisely performed. Immune repertoire sequencing technology is an important tool for genetic repertoire studies and/or antibody therapy.Citation26 Due to the different immune effect, humoral immune response to the rabies virus vaccine provides complementary information on the condition of the immune system of vaccinated individuals. By defining the antibody repertoire elicited by rabies virus antigens, IR-SEQ help us to established to examine BCR repertoires and diversifications.Citation14 After antibody exposure to rabies virus vaccine, an affinity maturation process generates diversity from which antibodies with higher affinity are selected, as the antigen concentration decreases during the secondary immune response.Citation27 The IGHV gene family was selected from the group consisting of IGHV1, IGHV2, IGHV3, IGHV4, IGHV5 and IGHV6. Our data showed that the V and J gene family after IR-SEQ consisted 37 distinct V gene segments and 7 distinct J gene segments among the volunteers, which was accord with previous study.Citation27,Citation28 Some studies had shown that V gene had involved in the viral infection,Citation29,Citation30 IGHV1-69 polymorphism modulates anti-influenza antibody repertoires and make broadly neutralizing antibodies to the influenza A.Citation31 Though here we had found IGV3-7 and IGV2-26 had difference between pre and post vaccination, whether both critical for anti-RABV in humoral immunity is still need further investigation.
Limitations associated with the present study warrant mention. First, because the number of samples tested was small and we were unable to compare RABV vaccine responders with non-RABV vaccine responders. thus, it is not possible to determine if these changes are RABV-specific. Second, B-cell interactions with the RABV vaccine were not clarified in this study. Furthermore, some reports suggest that there is a relationship between age and repertoire diversity. However, the participants in the present study were all the same young age, so we were unable to evaluate the age-related response to the RABV vaccine or the relationship between age and repertoire diversity during RABV vaccination. Despite these limitations, our results suggest that B cell clone expansion was induced by the vaccination and sequences of high expand CDR3 aa clones were identified. It could potentially serve as a surrogate prediction marker for a better response to the RABV vaccine. Further studies with additional subjects are needed to confirm this possibility. In addition, while the sample size of the present work is too small for detecting correlations between anti-RABV responses and specific factors, our data may be useful for future metadata reviews.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Organization WH. WHO expert consultation on rabies. World Health Organ Tech Rep. 2005;931:1–88.
- Hampson K, Dobson A, Kaare M, Dushoff J, Magoto M, Sindoya E, Cleaveland S. Rabies exposures, post-exposure prophylaxis and deaths in a region of endemic canine rabies. Plos Negl Trop Dis. 2008;2(11):e339. doi:10.1371/journal.pntd.0000339.
- Mcgettigan JP. Experimental rabies vaccines for humans. Expert Rev Vaccines. 2010;9(10):1177. doi:10.1586/erv.10.105.
- Kelly RM, Strick PL. Rabies as a transneuronal tracer of circuits in the central nervous system. J Neurosci Methods. 2000;103(1):63–71. doi:10.1016/S0165-0270(00)00296-X.
- Lafon M. Evasive strategies in rabies virus infection. Adv Virus Res. 2011;79:33–53.
- Both L, Banyard AC, Van DC, Horton DL, Ma JK, Fooks AR. Passive immunity in the prevention of rabies. Lancet Infect Dis. 2012;12(5):397–407. doi:10.1016/S1473-3099(11)70340-1.
- Ertl HC. Novel vaccines to human rabies. PLoS Negl Trop Dis. 2009;3(9):e515. doi:10.1371/journal.pntd.0000515.
- Warrell M. Rabies and African bat lyssavirus encephalitis and its prevention. Int J Antimicrob Agents. 2010;36(1):S47. doi:10.1016/j.ijantimicag.2010.06.021.
- Zhu S, Guo C. Rabies control and treatment: from prophylaxis to strategies with curative potential. Viruses. 2016;8(11):279. doi:10.3390/v8110279.
- Zhang W, Du Y, Su Z, Wang C, Zeng X, Zhang R, Hong X, Nie C, Wu J, Cao H. IMonitor: a robust pipeline for TCR and BCR repertoire analysis. Genetics. 2015;201(2):459–72. doi:10.1534/genetics.115.176735.
- Magoä T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–63. doi:10.1093/bioinformatics/btr507.
- Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12(5):380. doi:10.1038/nmeth.3364.
- Giudicelli V, Mp L. IMGT/junctionanalysis: IMGT standardized analysis of the V-J and V-D-J junctions of the rearranged immunoglobulins (IG) and T cell receptors (TR). Cold Spring Harb Protoc. 2011;2011:716.
- Yaari G, Kleinstein SH. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med. 2015 Nov 20;7(1):1–14. doi:10.1186/s13073-015-0243-2.
- Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods. 2007;321(1):182–95. doi:10.1016/j.jim.2007.01.019.
- Ma L, Wang X, Bi X, Yang J, Shi B, He X, Ma R, Ma Q, Yao X. Characteristics peripheral blood IgG and IgM heavy chain complementarity determining region 3 repertoire before and after immunization with recombinant HBV vaccine. PLoS One. 2017;12(1):e0170479. doi:10.1371/journal.pone.0170479.
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–32. doi:10.1016/0022-2836(82)90515-0.
- Knobel DL, Cleaveland S, Coleman PG, Fèvre EM, Meltzer MI, Miranda ME, Shaw A, Zinsstag J, Meslin FX. Re-evaluating the burden of rabies in Africa and Asia. Bull World Health Organ. 2005;83(5):360.
- Schnell M, Mcgettigan J, Wirblich C, Papaneri A. The cell biology of rabies virus: using stealth to reach the brain. Nat Rev Microbiol. 2010;8(1):51. doi:10.1038/nrmicro2260.
- Lafon M. Rabies virus receptors. J Neurovirol. 2005;11(1):82–87. doi:10.1080/13550280590900427.
- Macedo CI, Carnieli JP, Brandão PE, Es TDR, Oliveira RN, Castilho JG, Medeiros R, Machado RR, Oliveira RC, Carrieri ML. Diagnosis of human rabies cases by polymerase chain reaction of neck-skin samples. Braz J Infect Dis. 2006;10(5):341. doi:10.1590/S1413-86702006000500008.
- Ugolini G. Rabies virus as a transneuronal tracer of neuronal connections. Adv Virus Res. 2011;79:165–202.
- Jackson AC, Ye H. Extraneural organ involvement in human rabies. Lab Invest. 1999;79:945–51.
- Jogai S, Radotra BD, Banerjee AK. Rabies viral antigen in extracranial organs: a post-mortem study. Neuropathol Appl Neurobiol. 2002;28(4):334–38. doi:10.1046/j.1365-2990.2002.00400.x.
- Ilina EN, Larina MV, Aliev TK, Dolgikh DA, Kirpichnikov MP. Recombinant monoclonal antibodies for rabies post-exposure prophylaxis. Biochemistry. 2018;83(1):1–12. doi:10.1134/S0006297918010017.
- Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GMR. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106(48):20216–21. doi:10.1073/pnas.0909775106.
- Kramer RA, Marissen WE, Goudsmit J, Visser TJ, Clijsters‐Van Der Horst M, Bakker AQ, De Jong M, Jongeneelen M, Thijsse S, Backus HH, et al. The human antibody repertoire specific for rabies virus glycoprotein as selected from immune libraries. Eur J Immunol. 2005;35(7):2131–45. doi:10.1002/eji.200526134.
- Wildt RMTD, Hoet RMA, Venrooij WJV, Tomlinson IM, Winter G. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol. 2012;285(3):895–901. doi:10.1006/jmbi.1998.2396.
- Kostareli E, Hadzidimitriou A, Stavroyianni N, Darzentas N, Athanasiadou A, Gounari M, Bikos V, Agathagelidis A, Touloumenidou T, Zorbas I. Molecular evidence for EBV and CMV persistence in a subset of patients with chronic lymphocytic leukemia expressing stereotyped IGHV4-34 B-cell receptors. Leukemia. 2009;23(5):919–24. doi:10.1038/leu.2008.379.
- Mehdi H. The analysis of VH and VL genes repertoires of Fab library built from peripheral B cells of human rabies virus vaccinated donors. Hum Immunol. 2014;75(8):745–55. doi:10.1016/j.humimm.2014.05.005.
- Avnir Y, Watson CT, Glanville J, Peterson EC, Tallarico AS, Bennett AS, Qin K, Fu Y, Huang CY, Beigel JH. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci Rep. 2016;6(20842).