ABSTRACT
Genetic optimization of Nucleic Acid immunogens is important for potentially improving their immune potency. A COVID-19 DNA vaccine is in phase III clinical trial which is based on a promising highly developable technology platform. Here, we show optimization in mice generating a pGX-9501 DNA vaccine encoding full-length spike protein, which results in induction of potent humoral and cellular immune responses, including neutralizing antibodies, that block hACE2-RBD binding of live CoV2 virus in vitro. Optimization resulted in improved induction of cellular immunity by pGX-9501 as demonstrated by increased IFN-γ expression in both CD8+ and CD4 + T cells and this was associated with more robust antiviral CTL responses compared to unoptimized constructs. Vaccination with pGX-9501 induced subsequent protection against virus challenge in a rigorous hACE2 transgenic mouse model. Overall, pGX-9501 is a promising optimized COVID-19 DNA vaccine candidate inducing humoral and cellular immunity contributing to the vaccine’s protective effects.
Introduction
SARS-CoV-2 is a single-strand positive-sense RNA virus that encodes multiple structural antigens including the entry relevant spike antigen, nucleocapsid, membrane protein, and E proteinCitation1, as well as multiple non-structural antigens. The spike proteins prime function is related to attaching itself to the host target cell receptor facilitating cell entry to initiate infection. Spike is composed of two distinct subunits, namely S1 which includes the receptor-binding domain (RBD) and the S2 entry domain composed of the transmembrane region of the Spike Ag. The two subunits provide the entry functions of the virus including receptor binding the angiotensin-converting enzyme 2 (hACE2) receptor for cell entry and infection.Citation2 Therefore, the spike protein represents an ideal immunogen candidate. Neutralizing antibodies that blocks the binding of RBD to hACE2 inhibit virus infection,Citation3 amino acid mutations within spike protein can affect the infectivity and stability of virusCitation4 as has been observed in the variants of concern (VOC).
Technologies for developing vaccines against the COVID-19 include a spectrum of inactivated virus, subunit proteins, mRNA, or recombinant viral vector-based approaches as well as DNA vaccine approaches. DNA vaccine represents an important platform that is highly scalable, cost-effective, thermally stable, and simple to administer.Citation5–7 Important studies demonstrated that optimized DNA vaccines induce both binding and neutralizing antibody responses against recent EID viruses including SARS, Ebola, Middle East respiratory virus (MERS), Zika virus, and SARS-CoV2Citation8–11 along with T cell responses. Most recently, ZyCoV-D of India advanced a three dose approaches that demonstrated that this COVID-19 DNA vaccine was safe, well tolerated, and immunogenic in Phase 1/2 trials and achieved a 67% efficacy in Phase III against COVID-19 cases caused mainly by the delta-variant SARS-CoV-2 circulating in India at the time of the trials, a very important outcome.Citation12 Further advancement of DNA vaccine technology by more efficient delivery as well as through genetic optimization is important to allow for lower dosing and faster immunization schedules. Here, we describe studies of pGX-9501 optimized DNA vaccine compared to a genetically non optimized construct efficiently delivered by the well-tolerated Cellectra system and show that pGX-9501 induce robust humoral and T cell immunity in a short two dose formats. These studies also show robust protection from challenge in a rigorous ACE2+ mouse lethal challenge model. These data are highly supportive of the continued advanced development of this important vaccine candidate for SARS-CoV2.
Methods and materials
Animal experiments
Female, C57BL/6 mice (6–8 weeks of age) and BALB/c mice (6–8 weeks of age) were purchased from Beijing Vital Laboratory Animal Technology Co., Ltd. (Beijing, China) and Shanghai Jiesjie Laboratory Animal Co., Ltd. (Shanghai, China), which were kept in SPF condition. hACE2 transgene BALB/c mice were from the Institute of Laboratory Animal Sciences, CAMS&PUMC. The Experimental Animals Committee of SHMC approved all animal experiments.
The mice were injected twice via the intramuscular route (i.m.) with 25 μg plasmid, and electroporation was followed at intervals of two weeks. Serum was collected 14 days after the second immunization.
Plasmid preparation
Plasmids of pGX-9501, pVAX1-S-WT, and pVAX1 were transformed into DH5a E. Coli, respectively. A single colony was undergone expansion in a one-liter flask for culturing in LB broth. Plasmids were extracted, purified by MaxPure Plasmid EF Giga Kit (Magen, China), dissolved in saline buffer at a final concentration at 1 mg/ml. The purity was measured by an agarose gel electrophoresis and a UV detector at a range of 1.8–2.0 OD260nm/280 nm. Endotoxin in those plasmids was below 30 EU/mg by LAL test.
Rare codon analysis
The sequence of wild type and the sequence optimized pGX-9501 was submitted to the GenScript Rare Codon Analysis Tool (https://www.genscript.com/tools/rare-codon-analysis). From origin organism to expression host, the tool compared the difference between these sequences by analyzing the DNA sequence features including cis-regulatory and negative repeat elements that could influence transcription and translation efficiencies in vivo.
Antigen-specific humoral immune responses
Enzyme-linked immunosorbent assay (ELISA) was utilized to measure the antigen-specific antibody responses induced in the animals by the DNA vaccinations against SARS-CoV2 antigens as previously described. Briefly, 96-well plates were, respectively, coated with 0.5μg/ml of pre-S1 (Sino biological, 40591-V05H1), 0.5 μg/ml of pre-S2 (Sino biological, 40590-V08B), and 0.17 μg/ml of RBD (Sino biological, 40592-V08B) protein (50 mM carbonate-bicarbonate buffer, pH 9.6) at 4°C overnight and blocked with 5% BSA in PBST (0.05% Tween 20 in PBS) at 37°C for 1 hour. The plates were incubated with diluted serum from different immunization groups for 1 hour at 37°C. Antibodies were detected with HRP-conjugated goat anti-mouse IgG (Southern Biotech, Birmingham, AL) after the enzymatic reaction was developed, the OD values were read at 450/620 nm by an ELISA plate reader (Bio-Rad, Hercules, CA).
hACE2-RBD blocking assay
The blockade of hACE2 binding to SARS-CoV-2 RBD in ELISA as previously reported.Citation3 Briefly, 96-well plates were coated with 0.34 μg/ml of RBD protein (50 mM carbonate-bicarbonate buffer, pH 9.6) at 4°C overnight and blocked with 5% BSA in PBST (0.05% Tween 20 in PBS) at 37°C for 1 hour. Plates were incubated with diluted serum samples from different immunization groups for 1 hour at 37°C. Then, 0.12 μg/ml of hACE2 (Sino Biological, 10108-H08H) was added and reacted for 1 hour at 37°C. The Goat anti-Rabbit IgG-HRP (Invitrogen, 32460) and Rabbit anti-hACE2 antibody (Sino Biological, 10108-RP01) were used to detect the concentration of hACE2 binding with the RBD. The following formula calculated the blocking ratio: Blocking Ratio = (1-(experimental group/control group)) *100%.
Live virus neutralization assays
Neutralization assays were performed at the Institute of Laboratory Animal Sciences, CAMS&PUMC of China. Seed SARS-CoV-2 (SARS-CoV-2/WH-09/human/2020) stocks and virus isolation studies were performed in Vero E6 cells, which are maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS), 100 IU/mL penicillin, and 100 µg /mL streptomycin, and incubated at 36.5°C, 5% CO2. Virus titers were determined using a standard 50% tissue culture infection dose (TCID50) assay. Serum samples collected from immunized animals were inactivated at 56°C for 30 min and serially diluted with a cell culture medium in two-fold steps. The diluted samples were mixed with a virus suspension of 100 TCID50 in 96-well plates at a ratio of 1:1, followed by 2 h incubation at 36.5°C in a 5% CO2 incubator. 1*104 Vero cells were then added to the serum-virus mixture, and the plates were incubated for 3–5 days at 36.5°C in a 5% CO2 incubator. Cytopathic effect (CPE) of each well was recorded under microscopes, and the neutralizing titer was calculated by the dilution number of 50% protective condition.
Flow cytometry
Single suspension cells from spleens and lymph nodes collected 14 days after the second immunization were prepared and stimulated with 10 μg/ml peptides at 37°C for 5 h. Cells were stained with viability dye eflour780 in PBS for 15 minutes on ice followed by washes twice with PBS supplemented with 2% FBS. To detect cell surface antigens, cells were stained with fluorochrome-tagged antibodies, as shown in the following table for 15 minutes on ice. To detect intracellular cytokines or intranuclear transcription factors, cells were fixed and permeabilized using an intercellular cytokine staining kit (Biolegend) or a commercial transcription factor staining kit (eBioscience). All stained samples were run on LSRFortessa (Biolegend) and analyzed by FlowJo (TreeStar).
SARS-CoV-2 spike protein peptide pool
COVID-19 spike RBD peptide pools (SARS-CoV-2 Spike protein 258–518aa) published previouslyCitation13 was used for the study.
Cytotoxic lymphocyte (CTL) killing ability
Single suspensions of splenocytes from naïve syngeneic mice were diluted to 1.5*108/ml by RPMI1640 with 10% FBS and 2% Penicillin and Streptomycin, respectively, pulsed with or without 5 μg/ml peptides as mentioned above at 37°C. After 4 hours, a higher concentration of eflour450 (eBioscience, 65–0842-85) at 5 mM was used to label pulsed peptide cells. Cells without peptide-pulsed were labeled with a low concentration of eflour450 at 0.5 mM at room temperature in the dark. After being rinsed by PBS three times, 4*106 of labeled and peptides-pulsed cells and another equal number of labeled cells without peptide-pulsed were adoptive transferred by tail vein injections into mice previously immunized with different vaccines, respectively. Six hours later, the percentage of labeled cells was detected with LSRFortessa flow cytometry (BD) and analyzed by FlowJo (TreeStar). The following formula calculated the specific cell lysis: Specific cell lysis ability = (1-(percentage of cells incubated with peptide/percentage of cells incubated without peptide)) *100%.
SARS-CoV-2 challenge study
SARS-CoV-2 (SARS-CoV-2/WH-09/human/2020/CHN) was isolated by the Institute of Laboratory Animal Sciences, CAMS&PUMC. Immunized hACE2 transgenic miceCitation14 were infected with SARS-CoV-2 (106TCID50) via the nasal route in a volume of 100 μl 7 days after the final immunization. Five days after infection, the lung was harvested for measuring virus loads by qPCR and H&E staining. Before the challenge, serum was collected for ELISA to evaluate the neutralizing antibody levels. After the challenge, daily weight loss was monitored until the animal was sacrificed at day 5 post-infection (5 dpi).
Statistical analysis
The statistical analysis methods and sample sizes (n) are specified in the results section or figure legends for all quantitative data. All values are reported as means ± sem with the indicated sample size. No samples were excluded from the analysis. The difference among and between groups was determined by ordinary one-way ANOVA and Turkey test, respectively. P values less than 0.05 were considered statistically significant. All animal studies were performed after randomization. Statistics were performed using GraphPad Prism 7 software. In all data *p < .05, **p < .01, ***p < .001, and ****p < .0001.
Results
Comparison of expression of optimized vs unoptimized spike vaccine sequences
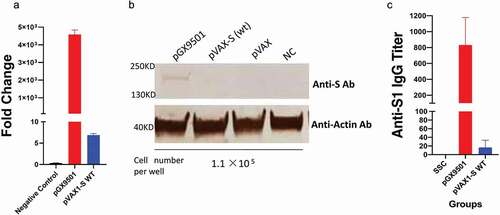
A DNA construct, pVAX1-S-wt, made from the wild-type sequence of the full-length spike protein of the SARS-CoV-2 was subcloned into the starting pVAX1 vector. As previously described, the sequence of the spike region was optimized via SynCon® technology, synthesized, and cloned into pVAX1 and utilized as pGX-950114. The two constructs were transfected separately into two batches of 293T cells in parallel under the same condition and after 24 hours were subjected to a qRT-PCR and Western blotting Analysis to compare their expression levels. As depicted in ), the level of mRNA and protein expression of the optimized pGX-9501 vs the wild type design was 100x that of the wild-type pVAX1-S-wt. We further identified that among the important differences between the two sequences were the increased content of of negative Cis-elements in the absence of optimization with five observed the wild-type construct and just one in the optimized pGX-9501 () vaccine.
Table 1. Sequence optimization score of the optimized and wild-type sequences
Figure 1. Comparison of expression and antibody levels of optimized versus non-optimized spike sequences. 293 T cells were transfected by pGX-9501, pVAX1-S-WT, and pVAX1 for 48hrs and lysed for RT-PCR analysis (a) and Western blotting (b), respectively. (c) BALB/c mice were immunized with either construct at 25 μg dose by using the 3P EP by the IM route. Immune analysis was performed at 2 weeks in an S1Elisa assay.
Effects of antibody production and functional assays induced by the vaccines
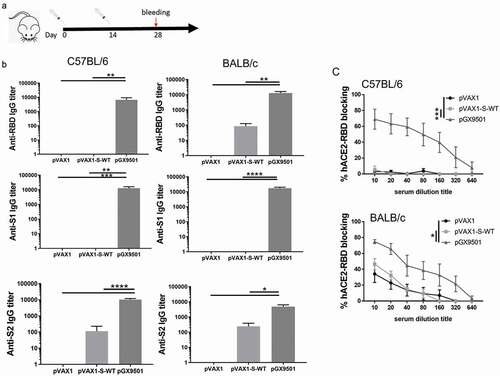
To evaluate the effects of pVAX1-S-wt and pGX-9501 DNA vaccines on the abilities to induce specific antibodies, mice were injected twice using 25 μg DNA vaccine each time via intramuscularly (i.m) at biweekly intervals and followed by electroporation by a Cellectra2000 device (). Serum IgG samples were serially diluted and tested for binding titers against recombinant spike proteins covering RBD, S1 region, and S2 region in the extracellular domain (ECD). Levels of antibodies taken from pGX-9501 immunized BALB/c, or the C57/BL6 animals, were a thousand times higher than those induced in the pVAX1-S-WT immunized (). In addition, these sera were also used to examine the inhibition experiment of RBD to hACE2 binding. We observed that a higher level of inhibition was achieved from sera immunized with pGX-9501 than pVAX1-S-WT (). We therefore selected pGX-9501 for further study for its ability to protect animals in a lethal challenge model. In these challenge models, a minimum of two doses of vaccines are necessary to see robust protection in most challenged animalsCitation14.
Figure 2. Effects of antibody production and functional assay. (a), The scheme of mice immunizations. (b), C57BL/6, or BALB/c mice (N = 6 per group) were either immunized with pVAX1 (blue circle) or vaccinated with pVAX1-S-WT (red square) and pGX-9501 (green triangle) intramuscularly, following by electroporation. Serum IgG binding titers (mean ± SEM) to SARS-CoV-2 pre-S1, S2, and RBD were measured on day 28. (c), Blocking abilities of RBD binding to the hACE2 with serum samples at serial dilutions on day 28. Data shown represent mean blocking efficiency (mean± SEM) for the five mice. Please add in the Single immunization group to the chart as well. Including bleeding.
Efficacy of protective response against SARS-CoV-2 challenge in hACE2 transgenic mice
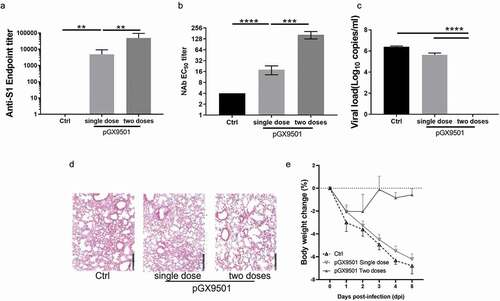
The goal of a vaccine is to protect the host against symptoms of infection and protect the host from disease and their consequences including death by infection of the virus. We examined the protective efficacy of these DNA vaccine immunization(s) in hACE2 transgenic animals in a SARS-CoV-2 challenge model (). Following 1 or 2 immunizations of human ACE2 transgenic mice, which are susceptible to infection, disease and SARS-CoV2 morbidity, the animals were then challenged 7 days after the last immunization. Challenge was by the intranasal route using a potent 1*106 TCID50 of SARS-CoV-2 (SARS-CoV-2/WH-09/human/2020/CHN). A virus neutralizing assay was performed using serum samples from the pGX-9501 immunized animals and the samples from pVAX1 as the control. The neutralizing level from pGX-9501 immunized once was around 1:18 but increased significantly after the two immunizations at 1:166, compared with the control (). The animals were sacrificed 5 days after the challenge for analysis of viral loads and pathological changes in the lungs. We observed almost 6log10 reduction of viral loads in lungs from mice immunized twice with the pGX-9501, showing robust and nearly complete protection against SARS-CoV-2 infection, whereas the 1–2 log10 reductions were reached in animals immunized once with pGX-9501 (). Pathological analysis of H&E stained lung tissue sections was performed. Compared to control mice the mice immunized 2x showed close to normal lung histology with only occasional slight interstitial pneumonia changes, and only 2/6 animals showing even slight infiltration around some blood vessels observed. In contrast, the single-dose and control groups showed moderate interstitial pneumonia with the marked widening of the alveolar septum and a slight infiltration of inflammatory cells around the vessels ( & ). After the SARS-CoV-2 challenge, the control mice demonstrated marked weight loss. The weight loss was dramatically observed from 1 dpi and continued till 5 dpi with approximately 5% of their body weight lost. Similar weight loss was observed in the single-dose immunization group. On the contrary, we observed that less than 2% of weight loss occurred at 1 dpi and rapidly recovered after 2 dpi in the two doses of pGX-9501 immunization group ().
Table 2. Analysis of H&E staining of lung and weight loss of mice challenged with SARS-CoV-2
Figure 3. pGX-9501 protects against challenges with SARS-CoV-2 in BALB/c mice. Mice treated with the vaccine were challenged by SARS-CoV-2 (105TCID50) in a volume of 100 μl 7 days after the second immunization (single dose group was challenged by virus 14 days after immunization). Five days after the challenge, Serum was collected for anti-s1 ELISA(a), and lung was harvested for measuring virus load by qRT-PCR (b). (c), Mice post vaccination were challenged by SARS-CoV-2 (105TCID50) in a volume of 100 μl 7 days after the second immunization (single dose group was challenged by virus 14 days after immunization). Serum was collected for ELISA to evaluate the Neutralizing antibody. (d), The histochemistry analysis of lung after H&E staining. €, Daily weight loss were monitored as shown.
Effects of Cell-Mediated Immunity (CMI)
This optimized DNA vaccine has the potential induce CMI due to its consistent post-inoculation expression in host cells in skin and muscle on multiple cell types allowing for presentation of the vaccine S Ag via both MHC-I and -II antigens. To assess the CMI, mice immunized with either pGX-9501 or pVAX1-S-WT in BALB/c and C57/BL6 mice on day 14 after the second immunization were used to isolate T cells from spleens or Lymph nodes for analyzing intracellular cytokine expressions by flow cytometry. As depicted in , percentages of IFN-γ-expressing CD4 T cells, representing Th1 response, were higher in both C57/BL6 and BALB/c mice immunized with pGX-9501 than that of the other two groups ( & b). However, expression of IL-5 and IL-13 in CD4 T cells, representing Th2 response, did not show increases among the groups ( & d). Levels of IFN-γ and TNF-α from CD8 T cells were dominant with close to 1% of CD8 stimulated cells expressing IFN-γ observed in the spleen and LN in the group immunized with pGX-9501 in both C57/BL6 and BALB/c mice ( & b). Expression of Granzyme B (Gz-B) in CD8 T cells was observed as robust as obtained as the IFN-γ (Figure S1A&S1B). The data support that this is a very Th1 immune potent vaccine with high protective value.
Figure 4. pGX-9501 promoted a biased CD8 T cell-based Th1-type cytokine phenotype and did not induce a TH2-associated phenotype. Single suspensions of splenocytes and lymphoid cells of lymph nodes harvested from C57BL/6 (a) or BALB/c (b) mice immunized were stimulated with 10 mg/mL SARS-CoV-2 peptide pools in vitro for 4 to 6 hours, and IFN-γ production from CD4+ T cells was analyzed by flow cytometry in both the C57BL/6 and BALB/c mice strains. Cytokine expression was studied using the SARS-CoV-2 peptide pool for immune stimulation.
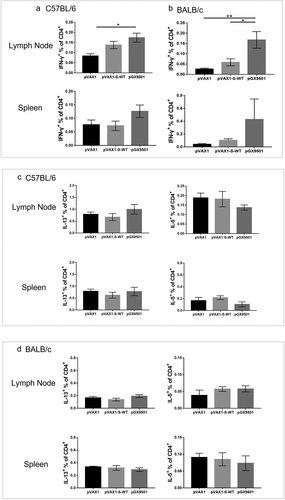
Figure 5. pGX-9501 induces effective specific cytotoxic lymphocyte(CTL) killing activity in vivo with enhanced IFN-g dominated cytokine expression in specific CD8 + T cells. Single suspension lymphocytes of spleens or lymph nodes from immunized C57BL/6 (a) and BALB/c (b) mice were stimulated with 10 mg/mL SARS-CoV-2 peptide pools in vitro for 4 to 6 hours. Levels of IFN-γ and TNF-α production in CD8 + T cells were measured by flow cytometry. C, Antigen-specific cytotoxic lymphocyte driven (CTL) killing ability was evaluated using an in vivo CTL assay. Target cells at 4*106/ml from naïve mice labeled with eFlour450 were incubated with 10 mg/mL SARS-CoV-2 peptide pools in vitro for 4–6 h before transferring into immunized mice by the intravenous route. The intensity of eFlour450 peptide labeled target cells were compared with the non-peptide labeled negative control cells after 5 hrs by flow cytometry to demonstrate in vivo killing. In vivo killing is only observed in the optimized pGX-9501 vaccinated animals.
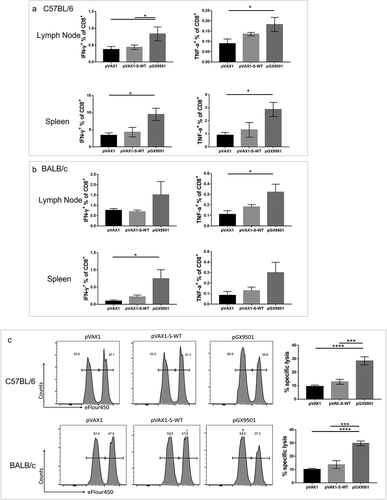
Functional CD8 T cells serve as cytolytic killer T cells to destroy virally infected host cells, resulting in a sterilizing immunity. Since its function can be tested in the in vivo CTL assay, we examined the level of specific CTL killing activities in these immunized groups. We observed that both BALB/c mice and C57/BL6 mice immunized with pGX-9501 obtained more robust CTL killing activity against SARS-CoV2 level than the other two groups (). In this assay, the wild-type unoptimized construct elicited much more minimal activity, just above the negative control group.
Discussion
COVID-19 DNA vaccines can drive immune responses in the clinic and result in protection from infection,Citation12,Citation13 as was recently reportedCitation12 using a modest delivery system and a three dose vaccine regime. Studies to generate additionally advanced DNA vaccines are important for protection of global populations and to focus on two dose regimes. In this regard COVID-19 DNA vaccine (pGX-9501) has been found to induce a significantly high level of immunogenicity in animal studies and human trialsCitation13 in a two dose regime. Here, we examined details of the optimization of pGX-9501 in a robust mouse challenge system. Prior to advancing candidate candidates for development of pGX-9501 we observed that designing viral vaccines using wild-type sequence of SARS-CoV-2 Spike as a DNA vaccine, pVAX-S-wt, induced less potent vaccine candidates. We compared the vaccine versions head-to-head in this study and observed that the expression level of mRNA of the pGX-9501 transfected into culture cells was over a hundred times higher than that of the pVAX-S-wt (). Such a superior level was also shown from direct studies corresponding to the expressed spike protein (). The higher expression level translates into a more robust vaccine resulting in an increased level of anti-spike antibodies, blocking RBD to hACE2 binding and neutralizing activities against live viruses infecting host cells. It was surprising to observe the low antibody-induced binding level from pVAX-S-wt-immunized BALB/c mice to RBD antigen and almost no antibody binding against the S1 protein. This inability to induce anti-spike antibodies of pVAX-S-wt was not due to a lower amount of plasmid DNA used to immunize since both DNA vaccines were delivered using the same dose for each immunization. The difference is likely related to a low expression level of pVAX-S-wt as shown in . Furthermore, not only was the induction of antibodies greatly enhanced but also the induction of T cells, including CD4 and CD8 T cells producing IFN-γ and TNF-α, and killing activities were also augmented. Notably, the level of cytolytic CD8 cells was increased significantly, demonstrated by the in vivo CTL assay, supporting a broader ability for clearance of viral infection. As T cells are less restricted than neutralizing antibodies, this greater T cell activity would likely be an additional advantage for the vaccine to protect against the emergence of new VOC. Most impactful, the codon-optimized pGX-9501 vaccine was highly protective in a lethal ACE-2 transgenic mouse model showing protection of the lungs with reduced lung pathogenesis in a dose-dependent manner and reduced time challenge model.
As the immunogenicity of pGX-9501 has been previously investigated and published,Citation13 the comparison study has not performed. Although optimization of codons in a foreign gene could significantly enhance its expression in vivo, in general, has been reported, the over hundreds if not thousand fold increases in expression from the pGX-9501 over the pVAX-S-wt is very impressive and is among the more dramatic descriptions of the impact of such optimization, suggesting its importance for SARS-CoV2 DNA vaccines. Rare codon analysis observed that five negative Cis-regulatory elements were situated within the coding region of wild-type spike versus only one with the pGX-9501Citation13(). Previous studies reported that gene expression can be inhibited by these negative elements. We reasoned that the decreasing number of negative CIS-regulatory elements in the optimized pGX-9501 could account for the higher mRNA and protein expression over that of wild type in addition to advantages over rare codon usage. These data support that wild-type sequence of spike antigen of SARS-CoV-2 and its variants would not be as efficient for nucleic acid or genetic-based vaccine development against COVID-19.
In SARS-CoV-2 infection, neutralizing antibodies are important for preventing viral entry and correlates analysis has identified that antibodies and T cells are important in the protection from disease and progression.Citation15 Antibodies to spike protein are believed to be important early in infection in part for controlling progression of SARS-CoV-2.Citation16,Citation17 More than 11 anti-spike monoclonal antibodiesCitation18 including LY-CoV555,Citation19 414–1,Citation18 and CB6Citation20with high-affinity binding to the RBD and hACE2 binding inhibitions are being used to develop therapies for SARS-CoV2 patients focusing to their neutralizing potency. We evaluated the specific anti-Spike binding antibody and RBD-hACE2 binding inhibition ability, of the optimized pGX-9501 that demonstrated a significantly higher potency than the native sequence wild-type vaccine. The higher levels of expression of the encoded mRNA and protein expression driven by the optimized pGX-9501 provides a rationale for the potent antibody induction compared to the wild-type constructs.
T cell immunity is indispensable for viral clearance as demonstrated in animal models infected with viruses like JEV, DENV, and recently Zika among others.Citation21–26 In a SARS-CoV infection model, enhanced CD8 + T cell results in earlier virus clearance and increased survival.Citation27 Clinical data in SARS infected patients show better recovery is frequently associated with the development of T cell immunity.Citation28 A similarity has been observed in SARS-CoV-2 infected patients in that COVID-19 patients with low- to mild-symptoms showed significantly higher levels of anti-SARS-CoV-2 CD4 and CD8 T cells versus the lower levels of T cell responses commonly found in severe patients including those who progress to fatal disease,Citation29,Citation30 and lymphopenia can be accentuated in symptomatic SARS-CoV-2 patients with pneumonia compared with those without pneumonia again indicating that T cell immunity plays an essential role in protective immunity against disease in SARS-CoV-2.Citation29Citation31–33
Activation of Ag-specific T cells was correlated with protective efficacy induced by some recently developed mRNA vaccines (BNT162b2 and mRNA-1273)Citation34 as well as the adenoviral vector vaccines including AZD1222.Citation35 In a recent study of AD26.COV2.S vaccine, CD8 T cell immunity is responsible for providing a broad spectrum of protection controlling SARS-CoV-2 variants.Citation36 Several prior studies have reported that DNA vaccines can induce high T cell-mediated responses in the clinic. Directly relevant, pGX-9501 induced T cell responses in mice, monkeys, and humans recently in phase I & II studies prior to moving into Phase III trials. Here, we observed that mice immunized with the pGX-9501 produced a strong induction of T cell immunity framed by induction of a Th1 type response including IFN-γ and a vigorous CTL activity, and was associated with robust protection of mice from SARS-CoV-2. However, there were some limitations of this study. The SARS-CoV-2 challenge was only performed at Day 14 after the final immunization, which was rapid compared to some other studies with other platforms studying SARS-CoV-2 or SARS-CoV vaccine development. For further study, the interval between challenge and immunizations could be extended allowing for the development of more robust memory responses with analysis of B and T cell responses performed at that time. Our study found that the pGX-9501 immunized twice showed a better virus clearance and Lung protection than the pGX-9501 immunized once. Similar results have been reported for other vaccine platforms.Citation37 This was also observed in clinical studies of both mRNA as well as Adenoviral vaccines both of which are much more effective after subsequent boosting.Citation38–41 In the twice immunized group, more robust neutralizing antibodies were induced and likely are important for the observed protection.Citation16 Also mice immunized twice showed a more robust IFN-γ and TNF-α induction in their T cell response, again which is important in viral clearance.Citation23,Citation26,Citation31 In conclusion, the wild-type coding sequence is a much poorer immunogen and behaves as a much weaker vaccine. Genetically optimized pGX-9501 appears to generate a much more robust immune profile and appears to be a highly promising DNA candidate vaccine for prevention of SARS-CoV-2. Additional clinical development remains important.
Supplemental Material
Download MS Power Point (305 KB)Supplemental Material
Download MS Word (32.9 KB)Acknowledgments
David Weiner was supported in part by the WW.Smith Trust and the Wistar Corona Virus Discovery Fund. We wish to thank Dr. David Hokey for his proofreading and editing.
Disclosure statement
D.B.W. has received grant funding, participates in industry collaborations, has received speaking honoraria, and received fees for consulting, including serving on scientific review committees, SAB, and BOD functions. Remuneration received by D.B.W. includes direct payments, stock, or stock options, and in the interest of disclosure, D.B.W. discloses the following paid associations with commercial partners: Pfizer (Advisory Board), Geneos (Advisory, SRA), Advaccine (Advisory) Astrazeneca (Advisory, Speaker), Inovio (BOD, SRA, Stock ownership), Sanofi (Advisory Board), BBI (Advisory Board, SRA).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website at https://doi.org/10.1080/21645515.2021.2016201
Additional information
Funding
References
- Amanat F, Krammer F. SARS-CoV-2 Vaccines: status report. Immunity. 2020;52(4):583. doi:10.1016/j.immuni.2020.03.007.
- Sakamoto A, Kawakami R, Kawai K, Gianatti A, Pellegrini D, Kutys R, Guo L, Mori M, Cornelissen A, Sato Y, et al. ACE2 (Angiotensin-Converting Enzyme 2) and TMPRSS2 (Transmembrane Serine Protease 2) expression and localization of SARS-CoV-2 infection in the human heart. Arterioscler Thromb Vasc Biol. 2020. doi:10.1161/ATVBAHA.120.315229.
- Huang WC, Zhou S, He X, Chiem K, Mabrouk MT, Nissly RH, Bird IM, Strauss M, Sambhara S, Ortega J, et al. SARS‐CoV‐2 RBD neutralizing antibody induction is enhanced by particulate vaccination. Adv Mater. 2020;32:2005637. doi:10.1002/adma.202005637.
- Berger I, Schaffitzel C. The SARS-CoV-2 spike protein: balancing stability and infectivity. Cell Res. 2020;30:1059. doi:10.1038/s41422-020-00430-4.
- Alarcon JB, Waine GW, McManus DP. DNA vaccines: technology and application as anti-parasite and anti-microbial agents. Adv Parasitol. 1999;42:343. doi:10.1016/s0065-308x(08)60152-9.
- Rice J, Ottensmeier CH, Stevenson FK. DNA vaccines: precision tools for activating effective immunity against cancer. Nat Rev Cancer. 2008;8:108. doi:10.1038/nrc2326.
- Robinson HL, Pertmer TM. DNA vaccines for viral infections: basic studies and applications. Adv Virus Res. 2000;55:1. doi:10.1016/s0065-3527(00)55001-5.
- Tebas P, Roberts CC, Muthumani K, Reuschel EL, Kudchodkar SB, Zaidi FI, White S, Khan AS, Racine T, Choi H, et al. Safety and immunogenicity of an Anti-Zika virus DNA vaccine - preliminary report. N Engl J Med. 2017. doi:10.1056/NEJMoa1708120.
- Modjarrad K, Roberts CC, Mills KT, Castellano AR, Paolino K, Muthumani K, Reuschel EL, Robb ML, Racine T, Oh M-d, et al. Safety and immunogenicity of an anti-middle east respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect Dis. 2019;19:1013. doi:10.1016/S1473-3099(19)30266-X.
- Muthumani K, Falzarano D, Reuschel EL, Tingey C, Flingai S, Villarreal DO, Wise M, Patel A, Izmirly A, Aljuaid A, et al. A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against middle east respiratory syndrome coronavirus in nonhuman primates. Sci Transl Med. 2015;7(301):132. doi:10.1126/scitranslmed.aac7462.
- Yang Z-Y, Kong W-P, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561. doi:10.1038/nature02463.
- Momin T, Kansagra K, Patel H, Sharma S, Sharma B, Patel J, Mittal R, Sanmukhani J, Maithal K, Dey A, et al. Safety and Immunogenicity of a DNA SARS-CoV-2 vaccine (ZyCoV-D): results of an open-label, non-randomized phase I part of phase I/II clinical study by intradermal route in healthy subjects in India. EClinicalMedicine. 2021;38:101020. doi:10.1016/j.eclinm.2021.101020.
- Smith T, Patel A, Ramos S, Elwood D, Zhu X, Yan J, Gary EN, Walker SN, Schultheis K, Purwar M, et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. 2020;11(1):2601. doi:10.1038/s41467-020-16505-0.
- Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830. doi:10.1038/s41586-020-2312-y.
- Rydyznski MC, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996. doi:10.1016/j.cell.2020.09.038.
- Dispinseri S, Secchi M, Pirillo MF, TMSolazzi M, Borghi M, Brigatti C, De Angelis ML, Baratella M, Bazzigaluppi E, Venturi G, et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun. 2021;12(1):2670. doi:10.1038/s41467-021-22958-8.
- Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang M-L, Jerome KR, Bloom JD, Greninger AL. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J Clin Microbiol. 2020;58(11). doi:10.1128/JCM.02107-20.
- Wan J, Xing S, Ding L, Wang Y, Gu C, Wu Y, Rong B, Li C, Wang S, Chen K, et al. Human-IgG-neutralizing monoclonal antibodies block the SARS-CoV-2 infection. Cell Rep. 2020;32(3):107918. doi:10.1016/j.celrep.2020.107918.
- Jones BE, Brown-Augsburger PL, Corbett KS, Westendorf K, Davies J, Cujec TP, Wiethoff CM, Blackbourne JL, Heinz BA, Foster D, et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci Transl Med. 2021;13(593):f1906. doi:10.1126/scitranslmed.abf1906.
- Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature (London). 2020;584(7819):120. doi:10.1038/s41586-020-2381-y.
- Wen J, Elong Ngono A, Regla-Nava JA, Kim K, Gorman MJ, Diamond, Shresta S. Dengue virus-reactive CD8(+) T cells mediate cross-protection against subsequent Zika virus challenge. Nat Commun. 2017;8(1):1459. doi:10.1038/s41467-017-01669-z.
- Hassert M, Harris MG, Brien JD, Pinto AK. Identification of protective CD8 T cell responses in a mouse model of Zika virus infection. Front Immunol. 2019;10:1678. doi:10.3389/fimmu.2019.01678.
- Bollard CM, Heslop HE. T cells for viral infections after allogeneic hematopoietic stem cell transplant. Blood. 2016;127(26):3331. doi:10.1182/blood-2016-01-628982.
- Kolls JK. CD4+T-cell subsets and host defense in the lung. Immunol Rev. 2013;252(1):156. doi:10.1111/imr.12030.
- Larena M, Regner M, Lee E, Lobigs M. Pivotal role of antibody and subsidiary contribution of CD8+ T cells to recovery from infection in a murine model of Japanese Encephalitis. J Virol. 2011;85(11):5446. doi:10.1128/JVI.02611-10.
- Prestwood TR, Morar MM, Zellweger RM, Miller R, May MM, Yauch LE, Lada SM, Shresta S. Gamma interferon (IFN-gamma) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-alpha/beta receptor-deficient mice. J Virol. 2012;86(23):12561. doi:10.1128/JVI.06743-11.
- Zhao J, Zhao J, Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol. 2010;84(18):9318. doi:10.1128/JVI.01049-10.
- Liu WJ, Zhao M, Liu K, Xu K, Wong G, Tan W, Gao GF. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antivir Res. 2017;137:82. doi:10.1016/j.antiviral.2016.11.006.
- Bonifacius A, Tischer-Zimmermann S, Dragon AC, Gussarow D, Vogel A, Krettek U, Gödecke N, Yilmaz M, Kraft ARM, Hoeper MM, et al. COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses. Immunity. 2021;54(2):340. doi:10.1016/j.immuni.2021.01.008.
- Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate, Wu JE, Alanio C, Kuri-Cervantes L, Pampena MB, D’Andrea K, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508). doi:10.1126/science.abc8511.
- Zhang G, Zhang J, Wang B, Zhu X, Wang Q, Qiu S. Analysis of clinical characteristics and laboratory findings of 95 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a retrospective analysis. Resp Res. 2020;21(1):74. doi:10.1186/s12931-020-01338-8.
- Zhang X, Cai H, Hu J, Lian J, Gu J, Zhang S, Ye C, Lu, Jin C, Yu G, et al. Epidemiological, clinical characteristics of cases of SARS-CoV-2 infection with abnormal imaging findings. Int J Infect Dis. 2020;94:81. doi:10.1016/j.ijid.2020.03.040.
- Wang F, Nie J, Wang H, Zhao Q, Xiong Y, Deng L, Song S, Ma Z, Mo P, Zhang Y, et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221(11):1762. doi:10.1093/infdis/jiaa150.
- Thompson MG, Burgess JL, Naleway AL, Tyner H, Yoon SK, Meece J, Olsho LEW, Caban-Martinez AJ, Fowlkes AL, Lutrick K, et al. Prevention and attenuation of Covid-19 with the BNT162b2 and mRNA-1273 vaccines. N Engl J Med. 2021;385(4):320–11. doi:10.1056/NEJMoa2107058.
- Ewer KJ, Barrett JR, Belij-Rammerstorfer S, Sharpe H, Makinson R, Morter R, Flaxman A, Wright D, Bellamy D, Bittaye M, et al. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat Med. 2020;27:270–78. doi:10.1038/s41591-020-01194-5
- Alter G, Yu J, Liu J, Chandrashekar A, Borducchi EN, Tostanoski LH, McMahan K, Jacob-Dolan C, Martinez DR, Chang A, et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature (London). 2021;596(7871):268–72. doi:10.1038/s41586-021-03681-2.
- Corbett KS, Gagne M, Wagner DA, O' Connell S, Narpala SR, Flebbe DR, Andrew SF, Davis RL, Flynn B, Johnston TS, et al. Protection against SARS-CoV-2 beta variant in mRNA-1273 vaccine-boosted nonhuman primates. Science. 202174(6573):1343–53. doi: 10.1126/science.abl8912.
- Sanchez-Felipe L,Vercruysse T, Sharma S, Ma J, Lemmens V, Van Looveren D, Arkalagud Javarappa MP, Boudewijns R, Malengier-Devlies B, Liesenborghs L, et al. A single-dose live-attenuated YF17D-vectored SARS-CoV-2 vaccine candidate. Nature. 2020;590(7845):320–25. doi:10.1038/s41586-020-3035-9.
- Sun W, McCroskery S, Liu W-C, Leist S, Liu Y, Albrecht R, Slamanig S, Oliva J, Amanat F, Schaefer A, et al. A Newcastle disease virus (NDV) expressing membrane-anchored spike as a cost-effective inactivated SARS-CoV-2 vaccine. bioRxiv. 2020. doi:10.1101/2020.07.30.229120.
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403. doi:10.1056/NEJMoa2035389.
- Lederer K, Castaño D, Gómez Atria D, Oguin TH, Wang S, Manzoni TB, Muramatsu H, Hogan MJ, Amanat F, Cherubin P, et al. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity. 2020;53(6):1281. doi:10.1016/j.immuni.2020.11.009.
