ABSTRACT
Influenza is a major public health concern causing millions of hospitalizations every year. The current vaccines need annual updating based on prediction of likely strains in the upcoming season. However, mismatches between vaccines and the actual circulating viruses can occur, reducing vaccine effectiveness significantly because of the remarkably high rate of mutation in the viral glycoprotein, hemagglutinin (HA). Clearly, it would be of great interest to determine the potential role of universally conserved epitopes in inducing protective immunity. Here, an antibody against the 14-aa fusion peptide sequence at the N-terminus of the HA2 subunit (Uni-1) was investigated for its ability to elicit antibody-dependent cellular cytotoxicity (ADCC) in vitro and cross-protection against lethal infection in animals. Uni-1, known to neutralize influenza type A (IAV) in vitro, was found to induce strong ADCC against diverse influenza viruses, including human and avian IAVs and both lineages of type B (IBV). The ADCC effects against human IAVs by Uni-1 was comparable to ADCC induced by well-characterized antibodies, F10 and FI6V3. Importantly, mice treated with Uni-1 were protected against lethal challenge of IAV and IBV. These results revealed the versatile effector functions of this universal antibody against markedly diverse strains of both IAV and IBV.
Highlights
The fusion peptide is the only universally conserved epitope in both IAV and IBV
Mono-specific universal antibody induces strong ADCC against human and avian IAV
Mono-specific universal antibody induces strong ADCC against IBV from both genetic lineages of IBV
The antibody has bi-functional effector functions against several influenza viruses
Introduction
Influenza viruses cause 3 to 5 million hospitalizations and 290,000 to 650,000 deaths annually.Citation1 Although there is currently an annual vaccination strategy in place to protect against the circulating strains, influenza continues to be a major public health burden. Influenza viruses belong to the family Orthomyxoviridae and are divided into four types, A, B, C, and D.Citation2 Type A (IAV) and B (IBV) cause the greatest mortality with seasonal and pandemic outbreaks, while type C (ICV) can infect children with mild respiratory symptoms. The effect of type D (IDV) on human health is yet to be understood.Citation3,Citation4 While IBV and ICV are mainly restricted to humans, IAVs infect many animal species including pigs, dogs, cats, horses, sea mammals, and birds along with humans.Citation5,Citation6 In addition, IAVs are categorized into different subtypes based on the hemagglutinin (HA) and neuraminidase (NA) glycoproteins. There are at least 18 HA (H1-H18) and 11 NA (N1-N11) subtypes among IAV with many variant strains of each subtype being species-specific.Citation7,Citation8 Furthermore, the different HA subtypes fall into two different phylogenetic groups: group 1 and 2.Citation9 On the other hand, IBV is classified into two antigenically distinct lineages, Victoria and Yamagata, which co-circulate within the human population.Citation10–13
Influenza viruses encode their own RNA-dependent RNA polymerase that results in approximately one error per replicated genome.Citation2,Citation14,Citation15 This leads to antigenic drift where errors accumulate over time allowing the virus to escape from existing immunity.Citation16,Citation17 In addition, novel IAV strains or subtypes to which there is no existing immunity in the human population could emerge through antigenic shift where the genome is reassortedCitation18or transmitted to humans through direct jump from other species.Citation19–21 This can potentially lead to major pandemics such as the ones in 1918, 1957, 1968 and 2009.Citation22–24
The HA is one of the major surface glycoproteins of influenza and is divided into two subunits, HA1 and HA2. HA1 has the receptor binding site that binds the sialic acid on a cell after which the virus is endocytosed, initiating viral replication. The fusion peptide in the HA2 subunit is then exposed mediating viral and cell membrane fusion, and uncoating of viral genome.Citation2 Since the HA is constantly under immune pressure, it is prone to high mutation rate. These mutations usually occur in HA1 of both IAV and IBV. The HA2 subunit, shielded from immune pressures, is highly conserved in both types.Citation25 Specifically, the fusion peptide at the N-terminus of HA2 that facilitates viral and cell membrane fusion is universally conserved among these viruses.Citation26
IAV and IBV co-circulate within the human population. Either type can be dominant in a given season, causing disease with comparable severity.Citation27 Current quadrivalent vaccines provide strain-specific protection against two IAV and two IBV strains; the vaccines mainly elicit neutralizing antibodies (nAbs) targeting the variable HA1 subunit. As the strains are selected several months ahead of the influenza season, mismatch between the vaccine seeds and the actual circulating viruses could happen, resulting in significant reduction of vaccine effectiveness.Citation12,Citation28,Citation29 In a pandemic situation, such delays in generating strain-specific reagents for vaccine quantitation and production could further delay the deployment of vaccines.
Given the antigenic variability of HA1, HA2 could be explored as a potential target for a universal vaccine. However, as it is shielded by HA1, HA2 is less immunogenic.Citation30 While low levels of HA2-specific B cells are detected in influenza patients, circulating antibodies against the stalk are usually not detectable following natural infection in humans.Citation31 Nevertheless, antibodies with broad reactivities, i.e. broadly nAbs (BnAbs), targeting highly conserved epitopes, mostly in the HA2 domain of diverse influenza viruses from group 1 and/or group 2, could be isolated from B cells in the immune repertoire of humans and animals with prior exposure to influenza viruses and/or their vaccines.Citation32–34 Interestingly, most of these reported BnAbs shared the highly conserved fusion peptide at the N-terminus of HA2 subunit as part of their epitope.Citation34
We previously reported that animals immunized with the highly conserved fusion peptide could produce a mono-specific antibody capable of cross-neutralizing several subtypes of IAV in vitro.Citation26,Citation35 Yet, the neutralizing activity of this antibody was moderate compared to other reported antibodies; it remains to be seen if this antibody has other effector functions beyond neutralization, and, more importantly, whether it could protect animals from challenge with lethal doses of IAV and IBV. In this study, we reveal that this universal antibody has strong antibody-dependent cellular cytotoxicity (ADCC) against markedly diverse strains of IAV and IBV, and affords effective protection in mice against lethal challenge of IAV and IBV.
Materials and methods
Cells and viruses
Madin-Darby canine kidney (MDCK) cells were grown in DMEM (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS and penicillin-streptomycin. Influenza A/California/7/2009 (H1N1), A/Netherlands/602/09 (H1N1), A/Hong Kong/1/68 (H3N2), A/Equine/Prague/1/56 (H7N7), A/Duck/England/56 (H11N6), B/Victoria/2/87 (Victoria lineage), B/Brisbane/60/08 (Victoria lineage), B/Yamagata/16/88 (Yamagata lineage), and B/Florida/04/06 (Yamagata lineage) viruses were generated as follows. Virus was grown in 10-day-old embryonated chicken eggs (Canadian Food Inspection Agency, Ottawa, ON, Canada) for 3 days at 33°C Eggs were cooled down overnight to 4°C Allantoic fluid was harvested, centrifuged at 2,000 rpm for 10 min at 4°C and the supernatant was sucrose-purified. The purified virus was used for infections. The virus titer was determined on monolayers of MDCK cells.
Antibody production
Mono-specific universal antibody against the N-terminal 14-aa fusion peptide (GLFGAIAGFIEGGW), denoted Uni-1, was generated in rabbits (Covance, Princeton, NJ) as previously described.Citation26,Citation36 Uni-1 was then purified using affinity columns using the peptide as binding ligand as previously described.Citation26
Antibody-dependent cellular cytotoxicity (ADCC)
MDCK cells, seeded in a 96-well plate and 80–90% confluent, were infected at MOI of 5 for 20–24 h in media without TPCK-treated trypsin. The next day, purified Uni-1 antibody or purified rabbit IgG (Sigma-Aldrich, St. Louis, MO) were serially diluted and added to the infected cells. Mouse FcγRIV effector cells (Promega, Madison, WI) were also added to each well (100,000 cells/well). Following a 5-h incubation at 37℃ and 5% CO2, Bio-GloTM luciferase assay substrate (Promega, Madison, WI) was added. Luminescence values were read in relative luminescence units (RLU). ADCC activity was expressed as fold induction, a ratio of the RLU of a test sample to the RLU of the ‘rabbit IgG’ isotype control. ADCC assays with F10 and FI6V3 human monoclonal antibodies were done using Human FcγRIIIa effector cells (Promega, Madison, WI).
Mouse studies
All animal procedures were approved by the National Research Council of Canada Animal Care Committee and performed in accordance with institutional guidelines. Eight-week-old female BALB/c mice (Charles River, Saint Constant, QC) were intraperitoneally treated with 0.5 mL Uni-1 rabbit serum 2 h before infection, and 0.25 mL serum 1 day and 3 days post-infection. A control group received serum collected from the same rabbit prior to peptide immunization. Mice were intranasally challenged with 500 PFU of purified A/Netherlands/602/09 or 3.75x105PFU of purified B/Victoria/2/87 in 25 µL. All mice were monitored daily for weight loss and clinical score from the time of first treatment until 14 days post-challenge. A clinical score was assigned where: 0 means healthy; 1 means ruffled fur but lively; 2 means sick with ruffled fur and slowing activity; 3 means very sick with ruffled fur, hunched with very little activity, showing respiratory distress and eyes squeezed shut; 4 means moribund; and 5 means dead. Clinical score of 3 was set as the endpoint and mice were euthanized.
Statistical analysis
Statistical analysis was conducted using Mann–Whitney test when appropriate using GraphPad Prism 8 (San Diego, CA) software. Tests were performed at a 5% significance level. Area under the curve (AUC) in was calculated using GraphPad Prism 8 (San Diego, CA) software with a baseline of 1.
Figure 1. Uni-1 induces ADCC against several human and avian strains of influenza A. MDCK cells were infected at MOI of 5 for 20–24 hours in media without TPCK. Infected cells were then incubated with purified Uni-1 antibody or rabbit IgG along with Promega mFcγriv effector cells for 5 hours prior to detection with luciferase substrate. Fold induction of ADCC over isotype control is shown for human strains, A/California/7/2009 (H1N1) and A/Hong Kong/1/68 (H3N2), and avian strains, A/Equine/Prague/1/56 (H7N7) and A/Duck/England/56 (H11N6). Purified human monoclonal antibodies, F10 and FI6V3, were also tested along with Promega FcγRIIIa effector cells. N = 3 per virus per antibody treatment group; data shown is mean ± SEM representative of 3 independent experiments.
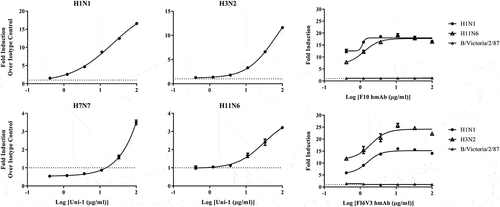
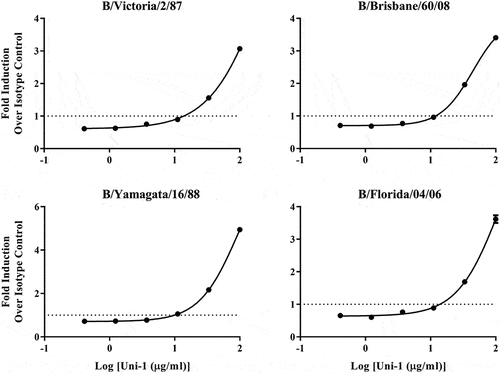
Figure 2. Uni-1 induces ADCC against several circulating strains of influenza B from both lineages. MDCK cells were infected at MOI of 5 for 20–24 hours in media without TPCK. Infected cells were then incubated with purified Uni-1 antibody or rabbit IgG along with Promega mFcγriv effector cells for 5 hours prior to detection with luciferase substrate. Fold induction of ADCC over isotype control is shown for strains from the Victoria lineage (top), B/Victoria/2/87 and B/Brisbane/60/08, and the Yamagata lineage (bottom), B/Yamagata/16/88 and B/Florida/04/06. N = 3 per virus per antibody treatment group; data shown is mean ± SEM representative of 3 independent experiments.
Results
The universal antibody demonstrates ADCC effects against diverse strains of IAV and IBV in vitro
The current study was done to investigate a universal antibody (designated as Uni-1) targeting the most conserved 14-aa region at the N-terminus of HA2, which is nearly 100% conserved. Indeed, Uni-1 is capable of recognizing HA from all IAV and IBV subtypes.Citation26,Citation36 Notably, while Uni-1 could neutralize diverse subtypes of IAV by inhibiting the pH-dependent fusion of viral and cellular membranes,Citation35 it was not clear if Uni-1 could mediate other effector functions or protect animals from lethal viral infection.
We sought to determine if Uni-1 could mediate ADCC as ADCC has been increasingly recognized in providing protective immunity against influenza infection.Citation37–42 For the target viruses in this study, we chose H1N1 and H3N2 as they are IAV subtypes circulating in recent years, while H7N7 and H11N6 are avian virus strains. For IBV, viruses from both genetic lineages were used. Specifically, MDCK cells were infected with several strains of IAV, including H1N1, H3N2, H7N7, and H11N6, and IBV, including B/Victoria (Victoria lineage), B/Brisbane (Victoria lineage), B/Yamagata (Yamagata lineage), and B/Florida (Yamagata lineage) before incubation with Uni-1 and FcγRIV effector cells.Citation27
As shown in , Uni-1 demonstrated strong ADCC activities against cells infected with all IAV strains tested (). Specifically, over 10-fold increase in ADCC activity was observed against the human H1N1 and H3N2 IAV strains with an AUC of 15.5 and 6.7, respectively, while at least 3- to 6-fold induction was detected against the two avian IAV strains with an AUC of 1.3 for H7N7 and 1.6 for H11N6 (). Furthermore, Uni-1 also induced ADCC against 4 strains of IBV derived from either Victoria or Yamagata lineages (). The AUC for B/Victoria, B/Brisbane, B/Yamagata, and B/Florida is 1.1, 1.4, 1.7, and 1.3, respectively (). While the different magnitude of ADCC amongst the strains tested is likely due to various factors including the differed susceptibility of the particular cell lines used in the study (see more in Discussion), these results revealed the marked breadth of ADCC activity elicited by Uni-1 against IAV and IBV.
We also tested well-studied antibodies, F10 and FI6V3, for their ADCC activity (). F10,Citation32 a human monoclonal antibody (mAb) that binds group 1 HA’s, elicited 15- to 18-fold induction of ADCC against H1N1 and H11N6 with an AUC of 38.2 and 34.4, respectively. FI6V3, another human mAb that binds both group 1 and 2 HA’s,Citation43 elicited 14- to 23-fold induction of ADCC against H1N1 and H3N2 with an AUC of 28.1 and 46.5, respectively. As expected, neither F10 nor FI6V3 induced any ADCC against B/Victoria since these antibodies have not been shown to bind IBVs. Overall, the fold induction of ADCC elicited by Uni-1 against some of the IAV strains tested, specifically H1N1 and H3N2, are comparable to the levels observed with F10 and FI6V3 (). However, it is important to note that since F10 and FI6V3 are human antibodies, human ADCC reporter cells were used whereas mouse reporter cells were used in the Uni-1 ADCC assays, which may explain the observed differences in the ADCC activities. Nonetheless, these results collectively suggest that universal antibodies, such as Uni-1, F10, and FI6V3, could inhibit influenza viruses through neutralization and Fc effector functions (ADCC).
Uni-1 effectively protects mice from lethal challenge of IAV and IBV
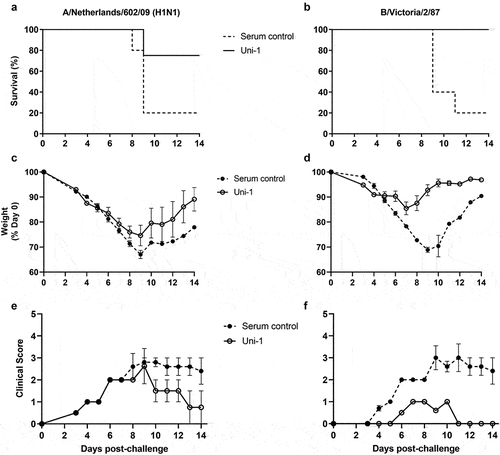
Having observed the broad functionality of Uni-1 in vitro, we sought to determine its protective ability in vivo. To this end, BALB/c mice were treated with Uni-1 once before and twice after viral challenge with lethal doses of IAV (500 PFU of A/Netherlands/602/09) or IBV (3.75x105PFU of B/Victoria/2/87). While 80% of mice in the control group succumbed to the IAV and IBV infections, 75% and 100% of the Uni-1 treated mice survived the IAV and IBV challenge, respectively ()). The survival data was in agreement with the changes in body weight during the course of infection. Specifically, although both the Uni-1 treated and control mice showed similar body weight loss in the first few days after the challenge, the Uni-1 treatment prevented the mice from further weight loss from Day 6 post-challenge with both IAV and IBV ()). The protective effects of Uni-1 were further confirmed with clinical scores ()), with Uni-1 treatment resulting in significantly reduced clinical presentations in mice challenged with either IAV (p = 0.0441) or IBV (p = 0.0002).
Figure 3. Treatment with Uni-1 provides effective cross-protection in mice against lethal challenge of influenza a and B viruses. BALB/c mice were intraperitoneally treated with Uni-1 serum once before and twice after an intranasal challenge with A/Netherlands/602/09 (A, C, and E) or B/Victoria/2/87 (B, D, and F). Mice were monitored for 14 days post-challenge. Survival (A and B), weight (C and D), and clinical score (E and F) are shown. Clinical scores were assigned daily based on the criteria outlined in the Materials and Methods section. Data shown is mean ± SEM; n = 5 per group.
Discussion
While heterosubtypic immunity against influenza is mainly mediated by cross-reactive cytotoxic T lymphocytes (CTLs),Citation44,Citation45 which target conserved epitopes in the viral internal proteins such as nucleoprotein and matrix protein,Citation46–48 cross-reactive antibodies against HA and BnAbs have also been long reported,Citation34,Citation49–52 with viral epitopes identified in either HA1 or HA2 or both.Citation32,Citation33,Citation53,Citation54 BnAbs targeting HA1 mainly inhibit the viral replication through disrupting the binding of the HA protein with sialic acids receptor of the cells, while HA2-targeting BnAbs prevent viral entry through either hindering the pH-dependent conformational change or stabilizing the prefusion conformation of the HA protein.Citation32–34,Citation46–52,Citation55–57 Given the much higher conservation rates in the HA2,Citation26,Citation58 BnAbs targeting the stem are more broadly reactive, with conformational epitopes found to involve at least three distinct parts of the HA protein, i.e. the HA1, HA2 and the fusion peptide at the N-terminus of the HA2 subunit.Citation32–34,Citation58–63
In this brief communication, we further characterized Uni-1, a mono-specific antibody targeting the N-terminal 14-aa of the fusion peptide, which is the only universally conserved sequence among all influenza viruses.Citation26 We previously reported that it could inhibit multiple strains of IAV at the fusion step of viral entry.Citation25,Citation35 One of novelties of this current work is that we unraveled a functional feature of Uni-1 in ADCC against influenza viruses from a wide range of subtypes or genetic lineages. The diversity of viral targets against which Uni-1 was able to induce ADCC effects is underscored by the wide range of viruses analyzed in the experiments, including human and avian IAV as well as two genetic lineages of IBV. This finding is in agreement with our recent observation where a HA2-based prototype universal vaccine with the N-terminal deleted resulted in a significant reduction in ADCC effector functions as well as in vivo protection.Citation27 Moreover, Uni-1 was not found to neutralize IBV in vitro but both groups of IAV.Citation35 Here, Uni-1 was found to induce significant ADCC response against IBV () and provide complete protection in mice against a lethal challenge (), suggesting effector functions of Uni-1 are sufficient to fully protect the mice from lethal challenge by IBV. However, Uni-1 appeared to inhibit IAV through both neutralization and effector functions, which could be the main mechanisms underlying the full protection of the animals against lethal IAV challenge ().
While the breadth of ADCC effects exerted by Uni-1 is clear in this study, we are not really surprised by the seemingly differed magnitudes of ADCC effects on different virus strains; indeed, as such phenomena regarding broadly reactive antibodies against influenza have been well documented, i.e., sequences outside the conserved region could contribute to the difference by affecting the accessibility of the antibodies to the ADCC epitopes.Citation32,Citation43,Citation64–74 While these previous observations, along with our results presented here, underline the potential challenge for the designing of universal vaccines in general,Citation66 epitopes targeted by non-neutralizing antibodies with anti-viral effector functions are more conserved, meaningfully informing the rationale designing of universal influenza vaccines.Citation75,Citation76
It should be mentioned that antisera, not purified Uni-1, was used in the in vivo challenge studies due to technical difficulties of purifying large quantities of Uni-1 antibodies using peptide-affinity columns, an observation likely resulting from the extreme hydrophobic nature of the peptide.Citation26 While higher yield antibodies could be generated using recombinant techniques to express the antibodies in cell cultures, the use of pre- and post-immunized sera, which has been well characterized for antigenic specificity,Citation26,Citation36 should be sufficient to determine the anti-viral effects of the antibodies.
In short, BnAbs targeting conformational or linear epitopes in the HA2 could also exert their inhibitory effects through effector mechanisms such as ADCC,Citation37,Citation42,Citation73 which is one of the key effector functions in the control of viral replication.Citation37–42,Citation77,Citation78 Here, we found that Uni-1, a mono-specific antibody targeting the universally conserved linear epitope, could induce strong ADCC and in vivo protection, a new observation consistent with our recent vaccine studies showing deletion of the fusion peptide could reduce the ADCC as well as weaken the vaccine efficacy induced by a HA2 vaccine.Citation27 The ADCC activity, along with neutralizing activities,Citation25 could collectively contribute to the effective protection of animals from lethal challenges of both IAV and IBV as presented in this short report.
Acknowledgements
We thank Bozena Jaentschke for technical assistance. We are grateful to Dr. Wayne Marasco of Dana Farber Cancer Institute for providing the F10 antibody and Dr. Jeffrey Boyington from NIH for providing the FI6V3 antibody.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- World Health Organization. Influenza update - 331. Geneva (Switzerland): WHO; 2018.
- Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26:1. [accessed 2021 Jun 25]. /pmc/articles/PMC3074182/.
- Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, Wang D, Nelson EA, Li F, Palese P. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the orthomyxoviridae family. Mbio. 2014;5(2). [accessed 2021 Jun 25]. /pmc/articles/PMC3958797/.
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9(2):e1003176. [accessed 2021 Jun 25]. /pmc/articles/PMC3567177/.
- Zambon MC. Epidemiology and pathogenesis of influenza. J Antimicrob Chemother. 1999;44(suppl_2):3–8. [accessed 2021 Jul 17]. doi:10.1093/jac/44.suppl_2.3.
- Capua I, Alexander DJ. Human health implications of avian influenza viruses and paramyxoviruses. Eur J Clin Microbiol Infect Dis. 2003;23(1):1–6. [accessed 2022 Feb 21]. doi:10.1007/s10096-003-1059-3.
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. New world bats harbor diverse influenza a viruses. PLoS Pathog. 2013;9(10):e1003657. [accessed 2021 Jun 25]. /pmc/articles/PMC3794996/.
- Gao R, Sheng Z, Sreenivasan CC, Wang D, Li F. Influenza a virus antibodies with antibody-dependent cellular cytotoxicity function. Viruses. 2020;12. [accessed 2021 Jun 25]. /pmc/articles/PMC7150983/.
- Russell RJ, Kerry PS, Stevens DJ, Steinhauer DA, Martin SR, Gamblin SJ, Skehel JJ. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S a. 2008;105(46):17736–17741. [accessed 2022 Feb 21]. doi:10.1073/pnas.0807142105.
- Koutsakos M, Nguyen TH, Barclay WS, Kedzierska K. Knowns and unknowns of influenza B viruses. Future Microbiol. 2016;11(1):119–135. [accessed 2021 Jun 25]. doi:10.2217/fmb.15.120.
- Jennings L, Huang QS, Barr I, Lee PI, Kim WJ, Buchy P, Sanicas M, Mungall BA, Chen J Literature review of the epidemiology of influenza B disease in 15 countries in the Asia-Pacific region. Influenza Other Respi Viruses. 2021;12(3):383–411. [accessed 2018 Jun 25]]. https://pubmed.ncbi.nlm.nih.gov/29127742/.
- De Sandt CE V, Bodewes R, Rimmelzwaan GF, De Vries RD. Influenza B viruses: not to be discounted. Future Microbiol. 2015;10:1447–1465. [accessed 2021 Jun 25]. https://pubmed.ncbi.nlm.nih.gov/26357957/.
- Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175(1):59–68. [accessed 2021 Jun 25]. doi:10.1016/0042-6822(90)90186-U.
- Boivin S, Cusack S, Ruigrok RWH, Hart DJ. Influenza a virus polymerase: structural insights into replication and host adaptation mechanisms. J Biol Chem. 2010;285:28411–28417. [accessed 2021 Jun 25]. /pmc/articles/PMC2937865/.
- Air GM. Influenza virus antigenicity and broadly neutralizing epitopes. Curr Opin Virol. 2015;11:113–121. [accessed 2021 Jun 25]. /pmc/articles/PMC4456283/.
- Huang KYA, Rijal P, Schimanski L, Powell TJ, Lin TY, McCauley JW, Daniels RS, Townsend AR. Focused antibody response to influenza linked to antigenic drift. J Clin Invest. 2015;125(7):2631–2645. [accessed 2021 Jun 25]. /pmc/articles/PMC4613558/.
- Schmidt AG, Do KT, McCarthy KR, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC. Immunogenic stimulus for germline precursors of antibodies that engage the influenza hemagglutinin receptor-binding site. Cell Rep. 2015;13(12):2842–2850. [accessed 2021 Jun 25]. /pmc/articles/PMC4698027/.
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza a viruses. Microbiol Rev. 1992;56(1):152–179. [accessed 2022 Feb 21]. doi:10.1128/mr.56.1.152-179.1992.
- Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, et al. Characterization of an avian influenza a (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279(5349):393–396. [accessed 2022 Feb 21]. doi:10.1126/science.279.5349.393.
- Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, et al. Avian-To-Human transmission of H9N2 subtype influenza a viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A. 2000;97(17):9654–9658. [accessed 2022 Feb 21]. doi:10.1073/pnas.160270697.
- Fouchier RAM, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SAG, Munstert V, Kuikent T, Rimmelzwaan GF, Schutten M, Van Doornum GJJ, et al. Avian influenza a virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101(5):1356–1361. [accessed 2022 Feb 21]. doi:10.1073/pnas.0308352100.
- Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006;12(1):9–14. [accessed 2021 Jun 25]. /pmc/articles/PMC3291411/.
- Von Holle TA, Anthony Moody M. Influenza and antibody-dependent cellular cytotoxicity. Front Immunol. 2019;10:1457. [accessed 2021 Jun 25]. /pmc/articles/PMC6611398/.
- Fowlkes AL, Arguin P, Biggerstaff MS, Gindler J, Blau D, Jain S, Dhara R, McLaughlin J, Turnipseed E, Meyer JJ, et al. Epidemiology of 2009 pandemic influenza a (H1N1) deaths in the United States, April-July 2009. Clin Infect Dis. 2011;52(Supplement 1):S60–S68. [accessed 2021 Jun 25]. doi:10.1093/cid/ciq022.
- M Hashem A, M Doyle T, Van Domselaar G, Farnsworth A, Li C, Wang J, He R, Li X. Recent developments in bioinformatics analyses of influenza a virus surface glycoproteins and their biological relevance. Curr Bioinform. 2011;6(4):415–426. doi:10.2174/157489311798073007.
- Chun S, Li C, Van Domselaar G, Wang J, Farnsworth A, Cui X, Rode H, Cyr TD, He R, Li X. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza a viral hemagglutinins. Vaccine. 2008;26(48):6068–6076. doi:10.1016/j.vaccine.2008.09.015.
- Gravel C, Muralidharan A, Duran A, Zetner A, Pfeifle A, Zhang W, Hashem A, Tamming L, Farnsworth A, Loemba H, et al. Synthetic vaccine affords full protection to mice against lethal challenge of influenza B virus of both genetic lineages. iScience. 2021;24(11):103328. [accessed 2022 Feb 21]. doi:10.1016/j.isci.2021.103328.
- Heikkinen T, Ikonen N, Ziegler T. Impact of influenza B lineage-level mismatch between trivalent seasonal influenza vaccines and circulating viruses, 1999–2012. Clin Infect Dis. 2014;59(11):1519–1524. [accessed 2021 Jun 25]. doi:10.1093/cid/ciu664.
- Tafalla M, Buijssen M, Geets R, Vonk Noordegraaf-Schouten M. A comprehensive review of the epidemiology and disease burden of influenza B in 9 European countries. Hum Vaccines Immunother. 2021;12(4):993–1002.[accessed 2016 Jun 25]. doi:10.1080/21645515.2015.1111494.
- Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Dunand CJH, Taylor WM, Lim S, Huang M, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7(316):316ra192. [accessed 2021 Jun 25]. /pmc/articles/PMC4770855/.
- Krammer F, Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol. 2013;3(5):521–530. [accessed 2021 Jun 25]. /pmc/articles/PMC3804342/.
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza a viruses. Nat Struct Mol Biol. 2009;16(3):265–273. [accessed 2021 Jun 25]. https://www.nature.com/articles/nsmb.1566.
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RHE, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–251. [accessed 2021 Jun 25]. doi:10.1126/science.1171491.
- Hashem AM Prospects of HA-based universal influenza vaccine. Biomed Res Int. 2015. [accessed 2022 Feb 21]. https://pubmed.ncbi.nlm.nih.gov/25785268/.
- Hashem AM, Van Domselaar G, Li C, Wang J, She YM, Cyr TD, Sui J, He R, Marasco WA, Li Xuguang X. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza a virus. Biochem Biophys Res Commun. 2010;403(2):247–251. [accessed 2021 Jun 25]. doi:10.1016/j.bbrc.2010.11.030.
- Li C, Jaentschke B, Song Y, Wang J, Cyr TD, Van Domselaar G, He R, Li X. A simple slot blot for the detection of virtually all subtypes of the influenza a viral hemagglutinins using universal antibodies targeting the fusion peptide. Nat Protoc. 2010;5(1):14–19. [accessed 2021 Jun 25]. doi:10.1038/nprot.2009.200.
- Dilillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk–specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med. 2014;20(2):143–151. [accessed 2022 Feb 21]. doi:10.1038/nm.3443.
- Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ Antibody-Dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. 2013 [accessed 2022 Feb 21]. http://jvi.asm.org/.
- Dunand CJH, Leon PE, Huang M, Palese P, Krammer F, Wilson PC. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. 2016. [accessed 2022 Feb 21]. doi:10.1016/j.chom.2016.05.014.
- Jegaskanda S, Laurie KL, Amarasena TH, Winnall WR, Kramski M, De Rose R, Barr IG, Brooks AG, Reading PC, Kent SJ. Age-Associated cross-reactive antibody-dependent cellular cytotoxicity toward 2009 pandemic influenza a virus subtype H1N1. 2013;208(7):1051–1061. [accessed 2022 Feb 21]. https://academic.oup.com/jid/article-abstract/208/7/1051/2192734.
- Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de RR, Winnall WR, Stratov I, Brooks AG, Reading PC, et al. Cross-Reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol. 2013;190(4):1837–1848. [accessed 2022 Mar 3]. https://www.jimmunol.org/content/190/4/1837.
- DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest. 2016;126(2):605–610. [accessed 2022 Feb 21]. doi:10.1172/JCI84428.
- Wu NC, Xie J, Zheng T, Nycholat CM, Grande G, Paulson JC, Lerner RA, Wilson IA. Diversity of functionally permissive sequences in the receptor-binding site of influenza hemagglutinin. Cell Host Microbe. 2017;21(6):742–753.e8. [accessed 2022 Feb 23]. doi:10.1016/j.chom.2017.05.011.
- Takada A, Matsushita S, Ninomiya A, Kawaoka Y, Kida H. Intranasal immunization with formalin-inactivated virus vaccine induces a broad spectrum of heterosubtypic immunity against influenza a virus infection in mice. Vaccine. 2003;21(23):3212–3218. doi:10.1016/S0264-410X(03)00234-2.
- Rimmelzwaan GF, Fouchier RA, Osterhaus AD. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr Opin Biotechnol. 2007;18(6):529–536. [accessed 2021 Jun 25]. doi:10.1016/j.copbio.2007.11.002.
- Gerhard W, Mozdzanowska K, Zharikova D. Prospects for universal influenza virus vaccine. Emerg Infect Dis. 2006;12(4):569–574. [accessed 2021 Jun 25]. doi:10.3201/eid1204.051020.
- Epstein SL, Kong WP, Misplon JA, Lo CY, Tumpey TM, Xu L, Nabel GJ. Protection against multiple influenza a subtypes by vaccination with highly conserved nucleoprotein. Vaccine. 2005;23(46–47):5404–5410. [accessed 2021 Jun 25]. doi:10.1016/j.vaccine.2005.04.047.
- Epstein SL. Prior H1N1 influenza infection and susceptibility of Cleveland family study participants during the H2N2 pandemic of 1957: an experiment of nature. J Infect Dis. 2006;193(1):49–53. [accessed 2021 Jun 25]. doi:10.1086/498980.
- Sánchez-Fauquier A, Villanueva N, Melero JA. Isolation of cross-reactive, subtype-specific monoclonal antibodies against influenza virus HA1 and HA2 hemagglutinin subunits. Arch Virol. 1987;97(3–4):251–265. [accessed 2021 Jun 25]. https://link.springer.com/article/10.1007/BF01314425.
- Tkáčová M, Varečková E, Baker IC, Love JM, Ziegler T. Evaluation of monoclonal antibodies for subtyping of currently circulating human type a influenza viruses. J Clin Microbiol. 1997;35(5):1196–1198. [accessed 2021 Jun 25]. /pmc/articles/PMC232728/?report=abstract.
- Sanchez-Fauquier A, Guillen M, Martin J, Kendal AP, Melero JA. Conservation of epitopes recognized by monoclonal antibodies against the separated subunits of influenza hemagglutinin among type a viruses of the same and different subtypes. Arch Virol. 1991;116(1–4):285–292. [accessed 2021 Jun 25]. doi:10.1007/BF01319250.
- Russ G, Polakova K, Kostolansky F, Styk B, Vancikova M. Monoclonal antibodies to glycopolypeptides HA1 and HA2 of influenza virus haemagglutinin. Acta Virol. 1987;31:374–386.
- Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, et al. First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes. Sci Transl Med. 2021;13(596). [accessed 2022 Feb 21]. doi:10.1126/scitranslmed.abg4535.
- Guthmiller JJ, Han J, Utset HA, Li L, Lan LYL, Henry C, Stamper CT, McMahon M, O’-Dell G, Fernández-Quintero ML, et al. Broadly neutralizing antibodies target a haemagglutinin anchor epitope. Nature. 2022;602(7896):314–320. [accessed 2022 Feb 21]. doi:10.1038/s41586-021-04356-8.
- Corti D, Suguitan AL, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120(5):1663–1673. [accessed 2022 Feb 21]. doi:10.1172/JCI41902.
- Ekiert DC, Friesen RHE, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJWM, Brandenburg B, et al. A highly conserved neutralizing epitope on group 2 influenza a viruses. Science. 2011;333(6044):843–850. [accessed 2022 Feb 21]. doi:10.1126/science.1204839.
- Friesen RHE, Lee PS, Stoop EJM, Hoffman RMB, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, et al. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S a. 2014;111(1):445–450. [accessed 2022 Feb 21]. doi:10.1073/pnas.1319058110.
- Bliss CM, Freyn AW, Caniels TG, Leyva-Grado VH, Nachbagauer R, Sun W, Tan GS, Gillespie VL, McMahon M, Krammer F, et al. A single-shot adenoviral vaccine provides hemagglutinin stalk-mediated protection against heterosubtypic influenza challenge in mice. Mol Ther. 2022 [accessed 2022 Feb 21]. https://pubmed.ncbi.nlm.nih.gov/34999208/
- Kirkpatrick E, Qiu X, Wilson PC, Bahl J, Krammer F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci Rep. 2018;8(1). [accessed 2022 Feb 21]. doi:10.1038/s41598-018-28706-1.
- Roubidoux EK, Carreño JM, McMahon M, Jiang K, van Bakel H, Wilson P, Krammer F, Schultz-Cherry S. Mutations in the hemagglutinin stalk domain do not permit escape from a protective, stalk-based vaccine-induced immune response in the mouse model. Mbio. 2021;12(1):1–14. [accessed 2022 Feb 21]. doi:10.1128/mBio.03617-20.
- Krammer F. The human antibody response to influenza a virus infection and vaccination. Nat Rev Immunol. 2019;19(6):383–397. [accessed 2022 Feb 21]. doi:10.1038/s41577-019-0143-6.
- Tan J, Asthagiri Arunkumar G, Krammer F. Universal influenza virus vaccines and therapeutics: where do we stand with influenza B virus? Curr Opin Immunol. 2018;53:45–50. [accessed 2022 Feb 21]. https://pubmed.ncbi.nlm.nih.gov/29677684/.
- Nachbagauer R, Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect. 2017;23(4):222–228. [accessed 2022 Feb 21]. doi:10.1016/j.cmi.2017.02.009.
- Van Baalen CA, Els C, Sprong L, Van Beek R, Van Der Vries E, Adme O, Rimmelzwaan GF, McAdam AJ. Detection of nonhemagglutinating influenza a(h3) viruses by enzyme-linked immunosorbent assay in quantitative influenza virus culture. J Clin Microbiol. 2014;52(5):1672–1677. [accessed 2022 Feb 21]. doi:10.1128/JCM.03575-13.
- Thyagarajan B, Bloom JD. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. Elifesciences Org Thyagarajan Bloom eLife. 2014;3:3300.
- Wu NC, Thompson AJ, Lee JM, Su W, Arlian BM, Xie J, Lerner RA, Yen HL, Bloom JD, Wilson IA. Different genetic barriers for resistance to HA stem antibodies in influenza H3 and H1 viruses. Science. 2020;368(6497):1335–1340. [accessed 2022 Feb 23]. doi:10.1126/science.aaz5143.
- Asaoka N, Tanaka Y, Sakai T, Fujii Y, Ohuchi R, Ohuchi M. Low growth ability of recent influenza clinical isolates in MDCK cells is due to their low receptor binding affinities. Microbes Infect. 2006;8(2):511–519. [accessed 2022 Feb 23]. doi:10.1016/j.micinf.2005.08.006.
- Dolskiy AA, Grishchenko IV, Yudkin DV. Cell cultures for virology: usability, advantages, and prospects. Int J Mol Sci. 2020;21(21):1–23. [accessed 2022 Feb 21]. doi:10.3390/ijms21217978.
- Lombardo T, Dotti S, Renzi S, Ferrari M. Susceptibility of different cell lines to avian and swine influenza viruses. J Virol Methods. 2012;185(1):82–88. [accessed 2022 Feb 21]. doi:10.1016/j.jviromet.2012.06.008.
- Ilyushina NA, Ikizler MR, Kawaoka Y, Rudenko LG, Treanor JJ, Subbarao K, Wright PF. Comparative study of influenza virus replication in MDCK cells and in primary cells derived from adenoids and airway epithelium. J Virol. 2012;86(21):11725. [accessed 2022 Feb 21]. /pmc/articles/PMC3486302/.
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun. 2014;5. [accessed 2022 Feb 21]. https://pubmed.ncbi.nlm.nih.gov/24717798/.
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. [accessed 2021 Jul 17]. https://www.science.org/doi/abs/10.1126/science.1222908.
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza a hemagglutinins. Science. 2011;333(6044):850–856. [accessed 2022 Feb 21]. doi:10.1126/science.1205669.
- Doud MB, Hensley SE, Bloom JD, Lauring AS. Complete mapping of viral escape from neutralizing antibodies. PLoS Pathog. 2017;13(3):e1006271. [accessed 2022 Feb 23]. doi:10.1371/journal.ppat.1006271.
- Jennewein MF, Mabuka J, Papia CL, Boudreau CM, Dong KL, Ackerman ME, Ndung’-U T, Alter G Tracking the trajectory of functional humoral immune responses following acute HIV infection. Front Immunol. 2020;11. [accessed 2022 Mar 5]. https://pubmed.ncbi.nlm.nih.gov/32849622/.
- Boudreau CM, Burke JS, Shuey KD, Wolf C, Katz J, Tielsch J, Khatry S, LeClerq SC, Englund JA, Chu HY, et al. Dissecting Fc signatures of protection in neonates following maternal influenza vaccination in a placebo-controlled trial. Cell Rep. 2022;38(6):110337. [accessed 2022 Mar 5]. doi:10.1016/j.celrep.2022.110337.
- Florek NW, Weinfurter JT, Jegaskanda S, Brewoo JN, Powell TD, Young GR, Das SC, Hatta M, Broman KW, Hungnes O, et al. Modified vaccinia virus ankara encoding influenza virus hemagglutinin induces heterosubtypic immunity in macaques. J Virol. 2014;88:13418–13428. [accessed 2021 Jun 25]. /pmc/articles/PMC4249095/.
- Vanderven HA, Liu L, Ana-Sosa-Batiz F, Nguyen THO, Wan Y, Wines B, Mark Hogarth P, Tilmanis D, Reynaldi A, Parsons MS, et al. Fc functional antibodies in humans with severe H7N9 and seasonal influenza. J Clin Invest. 2017;2. [accessed 2021 Jun 25]. /pmc/articles/PMC5499372/.
