ABSTRACT
The rapid replacement of Omicron BA.1 by BA.2 sublineage is very alarming, raising the question of whether BA.2 can escape the immunity acquired after BA.1 infection. We compared the neutralizing activity toward the Omicron BA.1 and BA.2 sub-lineages in five groups: COVID-19 patients; subjects who had received two doses of mRNA vaccine; subjects naturally infected with SARS-CoV-2 who had received two doses of mRNA; and subjects who had received three doses of homologous or heterologous vaccine. The results obtained highlight the importance of vaccine boosters in eliciting neutralizing antibody responses against Omicron sub-lineages, and suggest that the adenovirus vectored vaccine elicits a lower response against BA.1 than against BA.2 sub-lineage.
Brief report
Despite the efficacy of spike-based vaccines and monoclonal antibodies, the SARS-CoV-2 pandemic continues more than two and a half years after its onset. Relentless investigation has outlined a causal dynamic between host-derived antibodies and reciprocal viral subversion. Integrating this paradigm into the architecture of next-generation antiviral strategies will be critical for successfully fighting the emerging SARS-CoV-2 variants.Citation1
Since the first isolation of SARS-CoV-2 in January 2020 in China, several viral variants have been detected.
The Omicron B.1.1.529 variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported to the World Health Organization (WHO) in November 2021.Citation2 Compared to the ancestral virus isolated in Wuhan in 2019, the Omicron BA.1 sub-lineage bears a total of 60 mutations on the non-structural proteins, spike (S) protein, envelope protein, membrane protein, and nucleocapsid (NP) protein. Of these mutations, 32 are on the S protein, 15 of which affect the receptor-binding domain (RBD). The BA.2 sub-lineage shares 32 mutations with BA.1 but has 28 distinct ones.Citation3
The S protein plays an essential role in SARS-CoV-2 infection and constitutes the main target of neutralizing antibodies.Citation4 The current vaccine formulations are designed to target the S protein of the wild-type (wt) virus, derived from the original Wuhan strain, and have proven to offer a high degree of protection. Currently, six vaccines have been authorized in EuropeCitation5: the BNT162b2 (Pfizer-BioNTech) and mRNA-1273 (Moderna) vaccines were developed by using the mRNA vaccine platform, Ad26.COV2.S (Janssen/Johnson & Johnson) and ChAdOx1-S (AstraZeneca) are adenovirus vectored vaccines, NVX-CoV2373 (Novavax) is a recombinant SARS-CoV-2 nanoparticle vaccine and VLA2001 (Valneva) is an inactivated virus vaccine.
The very rapid replacement of Omicron BA.1 by BA.2 during the period February–March 2022 suggests that Omicron BA.2 is inherently more transmissible than BA.1 and capable of vaccine breakthrough.Citation6,Citation7
Furthermore, approved antibodies with neutralizing activity against Omicron BA.1 are ineffective against a subset of Omicron subvariants containing a R346K substitution, BA.1.1 and BA.2, demonstrating the continued need to discover and characterize candidate therapeutic antibodies with the breadth and potency of neutralizing activity required to treat future SARS-CoV-2 variants.Citation8
The purpose of this study was to compare the neutralizing antibody response toward the two Omicron sub-lineages BA.1 and BA.2 by analyzing COVID-19 patients, subjects who had received two doses of mRNA vaccine, subjects naturally infected with SARS-CoV-2 who had received two doses of mRNA, who had received three doses of mRNA vaccine, and who had received two doses of an adenovirus vectored vaccine plus a booster dose of mRNA vaccine. Neutralizing titers were determined by live virus microneutralization (MN) assay.
Serum samples were grouped into five different cohorts.
Thirty-seven (37) serum samples from COVID-19 patients hospitalized at Humanitas Gavazzeni (Italy) during the first pandemic wave (March–May 2020) were included in the present study. Subjects’ characteristics and study procedures have been described in detail elsewhere.Citation9 This study was approved by the Ethics Committee of the University of Siena (approval number 17373) and by the Ethics Committee of Humanitas Gavazzeni (approval number 236).
Fifty (50) serum samples were collected at the Bari correctional facility (Italy) approximately 20 days after the second vaccination with mRNA vaccine (mRNA-273 and BNT162b2). These samples were negative to antibodies against the NP protein (IDScreenSARS-CoV-2 Double Antigen Multi-species ELISA, ID.vet, France).
Twenty-three (23) serum samples were collected at the Bari correctional facility (Italy) a mean of 20 days after the second dose of mRNA vaccine (mRNA-1273 and BNT162b2). These samples were positive to antibodies against the NP protein (as described above).
Forty-four (44) serum samples were collected at the Bari correctional facility (Italy), a mean of 21 days after the third mRNA vaccine dose (mRNA-1273 and BNT162b2).
Thirty-five (35) serum samples were collected from employees of the University of Bari, a mean of 42 days after the third dose of mRNA vaccine. These subjects received two doses of adenovirus vectored vaccine and a booster dose with mRNA vaccine (mRNA-1273 or BNT162b2).
The research protocol was approved by the Ethics Committee of the University Hospital of Bari (n.6955, prot.N.0067544-02082021) and subjects provided informed consent to participate in the study.
African green monkey kidney Vero E6 cells ([ATCC] #CRL-1586/Vero C1008) were cultivated in Dulbecco’s Modified Eagle’s Medium (DMEM) high Glucose with 2 mM L-Glutamine, 100 U/mL of penicillin−100 µg/mL streptomycin (complete DMEM), and 10% Fetal Bovine Serum (FBS). Cells were maintained at 37°C in a humidified 5%CO2 environment and passaged every 3–4 days; 18–24 hours before execution of the microneutralization (MN) assay, plates were seeded with 100 μL/well of cells (1.5 × 105cell/mL) diluted in complete DMEM with 2% FBS and incubated at 37°C, 5%CO2, until use.
Wuhan SARS CoV-2 2019 (2019-nCoV strain 2019-nCov/Italy-INMI1) virus was purchased from EVAg. The live-virus Omicron BA.1 SARS-CoV-2 and the Omicron BA.2 SARS-CoV-2 sub-lineages were provided by Piet Maes from Rega Institute (Belgium). Viral propagation was performed in 175 cm2 tissue-culture flasks pre-seeded with 50 mL of Vero E6 cells (1 × 106 cells/mL) diluted in DMEM 2% FBS. After a 18–20-hour incubation at 37°C, 5%CO2, flasks were washed twice with sterile Dulbecco’s phosphate buffered saline (DPBS) and inoculated with SARS-CoV-2 virus at a multiplicity of infection (MOI) of 0.001. The sub-confluent cell monolayer was incubated with the virus for 1 hour at 37°C, 5%CO2, and then flasks were filled with 50 mL of DMEM 2% FBS and incubated at 37°C, 5%CO2. Cells were monitored daily until they manifested 80–90% cytopathic effect (CPE). Supernatants of the infected culture were harvested, centrifuged at 469 g for 5 minutes at 4°C to remove cell debris, aliquoted, and stored at −80°C. The propagated viral stocks were titrated in 96-well plates. The viral titer was calculated by using the 50% tissue culture infectious dose per mL (TCID50/mL) as endpoint and defined as the reciprocal of the highest virus dilution yielding at least 50% CPE in the inoculated wells, according to the Reed and Muench formula.Citation10
For the MN assay, 12 twofold serial dilutions of the samples (starting dilution 1:20) were prepared in duplicate in DMEM 2% FBS and added to two different 96-well plates (final volume in the first dilution well: 120 μl). Plates were incubated for 1 hour at 37°C with a concentration of live virus (100TCID50/50 µl, sample–virus ratio 1:1). Following incubation, the virus–sample mixture was added to sub-confluent Vero E6 cells, to allow their infection by the residual virus. After 72 hours (Wuhan) or 96 hours (Omicron sub-lineages), cells were inspected for signs of CPE. The highest sample dilution able to inhibit at least 50% of viral growth, in terms of CPE, was regarded as the neutralization titer.
A cell-only and a virus-only control were added to each row of each plate in order to monitor the status of the cell monolayer and the virus itself within each plate. A negative control sample (plasma cod. 20/142 from NIBSC panel 20/268) and a positive control sample (high positive of anti-SARS-CoV-2 immunoglobulins, cod. 20/150 from NIBSC panel 20/268) were added in one separate plate, in duplicates, as control of the assay session. Parallel titrations of the viruses were performed in 96-well plates containing sub-confluent Vero E6 cells as previously described to monitor the viral titer.
A descriptive analysis of antibody titers was made via a box plot. Differences in titration of viral strains were tested by “Per-Term Linear Model,” a nonparametric analysis of variance on models with numeric response. For each group analyzed, an aligned rank transformation of the factorial model was made. The response variable of the model and the predictor variable were represented as the logarithm (base2) of the geometric mean titers and viral strains, respectively. A linear model for aligned rank transformation data was applied, and a post-hoc analysis using the estimated marginal means (EMMs) was conducted. The family-wise significance level was set at p < .05. Differences between viral strains were recorded, according to Bonferroni’s adjusted p-value. Applying the Bonferroni correction, the new significance level was set at p < .0167. All statistical analyses were performed by means of R software.
Neutralizing antibodies against Wuhan strain, Omicron BA.1 and BA.2 sub-lineages in the five cohorts of subjects are reported in .
Figure 1. Distribution of logarithm of titers geometric mean to base 2 (log2(gm)) for each group of subjects and for each viral strain (Wuhan, BA.1, BA.2). Patients: COVID-19 patients; T2: subjects receiving two doses of mRNA vaccine; Naturally_infected_T2: naturally infected subjects receiving two doses of mRNA vaccine; T3: individuals administered three doses of homologous vaccine (mRNA), T3_HETERO: individuals administered two doses of mRNA and one dose of adenoviral vaccine.
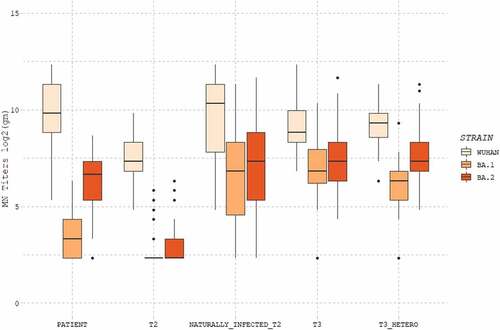
Looking at the five groups analyzed, Wuhan strain turns out to have the highest values of neutralizing titers (median of neutralizing titers being 9.82 in patients, 7.32 after two doses, 10.32 after natural infection plus two doses, 8.82 after three homologous doses, 9.32 after three heterologous doses). The BA.1 sub-lineage shows the lowest values, particularly in patients (3.32) and subjects with two doses of vaccine (2,32). Natural exposure to SARS-CoV-2 coupled with two mRNA vaccine doses resulted in a substantial increase in the response against BA.1 (6.82), reaching a similar GMT to those elicited by administration of three doses of homologous (6.82), or heterologous (6.32) vaccine. Higher titers were elicited against the BA.2 sub-lineage compared to the BA.1 in all groups (patients: 6.65, naturally infected and two doses 7.32, three doses homologous 7.32 and heterologous 7.32), except the one receiving two vaccine doses (2.32).
To assess whether the differences among the neutralizing titers obtained for Wuhan strain, BA.1 and BA.2 sub-lineages were statistically significant, the post-hoc analysis using the EMMs test was performed ().
Figure 2. Post-Hoc analysis results for each group (Patients: COVID-19 patients; T2: subjects receiving two doses of mRNA vaccine; Naturally_infected_T2: naturally infected subjects receiving two doses of mRNA vaccine; T3: individuals administered three doses of homologous (mRNA) vaccine; T3_HETERO: individuals administered two doses of mRNA and one dose of adenoviral vaccine) for each viral strain (WT, BA.1, BA.2). Differences between viral strains are shown per each group.
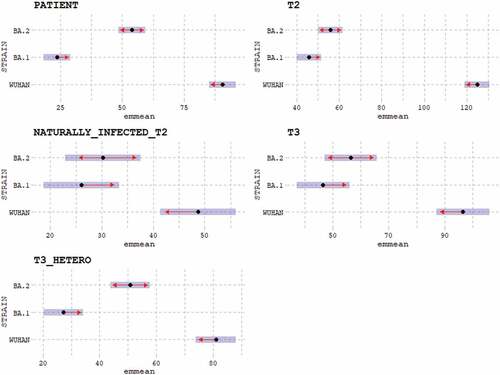
Comparison of neutralizing titers obtained for BA.1 and BA.2 sub-lineages per each cohort shows no significant differences in subjects who received two doses of vaccine (p value .0480), naturally infected receiving two vaccine doses (p value 1.000), and subjects receiving three doses of homologous vaccines (0.4297). In contrast, statistically significant differences between the two sub-lineages were observed when antibody responses elicited in COVID-19 patients (p value <.0001) and subjects who received three doses of heterologous vaccines (p value <.0001) were examined.
In this study, we compared the neutralizing antibody response toward Omicron BA.1 and BA.2 sub-lineages in serum samples collected from different cohorts of subjects: COVID-19 patients, subjects who received two doses of mRNA vaccine, SARS-CoV-2 naturally infected subjects who received two doses of mRNA vaccine, individuals administered three doses of homologous vaccine, and individuals receiving two doses of an adenovirus-based vaccine followed by a booster dose of mRNA vaccine. The neutralizing activity against the three strains analyzed (Wuhan, Omicron BA.1 and BA.2) shows a similar trend for all five groups examined, as also reported by Evans et al.Citation11 The neutralizing immune response against the Wuhan strain resulted to be the highest, whereas titers raised against the Omicron BA.1 sub-lineage were substantially lower. Although inferior to that elicited against the ancestral virus, the BA.2 cross-reactive response was higher than BA.1 in all subjects analyzed, particularly when looking at patients and subjects vaccinated with 3 heterologous doses.
In our study, natural SARS-CoV-2 infection induces an almost absent neutralizing response against Omicron BA.1, whereas BA.2 titers markedly rise; the functional antibody response toward the Omicron BA.2 sub-lineage shows similarity to that raised against the Wuhan strain, as natural infection alone triggers high neutralizing titers in both cases. Conversely, Omicron BA.1 cross-neutralizing titers in infected unvaccinated SARS-CoV-2 patients are remarkably low.
Two (2) vaccine doses are not sufficient to give a good antibody coverage against the two Omicron sub-lineages, inducing low-to-negligible neutralizing activity versus BA.2 and BA.1, respectively. These results are in agreement with data published by Ai et al.Citation12 that underline that two doses of vaccine are insufficient to provide complete protection against the Omicron sub-lineages.
In summary, our data show a marked improvement in serum neutralization in subjects who received a booster dose compared to those who did not and highlight the importance of vaccine boosters in eliciting neutralizing antibody responses against Omicron sub-lineages. Most vaccine formulations based on protein antigens require indeed repeated vaccinations in order to generate immunological memory and to maintain antibody responses above protective levels. The level of antigen–antibody binding avidity, a qualitative response index, can also correlate with protection and can potentially be enhanced by repeated immunization or natural infection. Conversely, inadequate levels of avidity maturation can heighten susceptibility to viral infection.Citation13
In our work, previous SARS-CoV-2 exposure coupled with administration of two doses of vaccine resulted in a substantial increase in BA.2 and BA.1 neutralizing responses, reaching a titer level similar to that triggered by a triple homologous or heterologous vaccination regimen. This strengthens the importance of hybrid immunity and could be associated with the antibody avidity index of the different groups analyzed: it is plausible that natural infection plus two doses of vaccine, as well as the administration of the third dose (despite containing a heterologous spike compared with BA.1 and BA.2) similarly increase antigen–antibody binding avidity.Citation14,Citation15
Vaccination or natural infection seems to play a major role in proper avidity maturation by prolonging the availability of antigens. As the post-vaccination antibody concentration wanes over time, higher avidity may sustain immunity and maintain the ability to fight future viral infection at reduced antibody levels.
This study has some limitations. Serum samples from COVID-19 patients have been collected during the first pandemic wave and may not be completely representative of the currently infected population. The number of enrolled subjects is relatively small (n = 189); however, all the considered cohorts reflect the common immunological situation in the general population. The timing of post-vaccination blood draws was not perfectly matched between subjects receiving the heterologous vaccination and the other cohorts. Furthermore, we did not evaluate other branches of immunity such as T cell responses that could contribute to protection even in the absence of or reduced neutralizing antibodies. Lastly, we did evaluate the neutralizing antibody response toward SARS-CoV-2 variants BA.1 and BA.2, which are no longer dominant.
Although this report analyzes SARS-CoV-2 variants that are no longer circulating, the results may be of relevance to the current circulating VOC. From the analyses performed in this study, it emerges that it is extremely important to keep administering vaccine doses in order to increase antibody coverage, which, despite the vaccine being formed from the “ancestral” protein, is maintained (although reduced) against BA.1 and BA.2 variants. BA.1 and BA.2 are found to be antigenically distinct,Citation16 but our results show that both are neutralized to some extent by antibodies yielded by pre-VOC SARS-CoV-2 or by repeated exposure with the ancestral spike. It is hence speculated that prior encounter of a different pre-Omicron variant (as in the case of COVID-19 patients), or repeated immunization with the ancestral spike (as in the case of vaccinees) could boost immunity toward conserved spike epitopes shared by different Omicron sub-lineages, potentially also those of newly emerged VOC such as BA.4 and BA.5 (which share most of the BA.2 spike sequence).Citation17 Indeed, circulating VOC BA.4 and BA.5 have shown to have additional antibody-escaping mutations (e.g. F486 V and L452 R) compared to BA.2.Citation17,Citation18 Variants-adapted vaccines are currently being developed and will likely be administered in the context of either homologous or heterologous vaccination regimens, which might help overcome this issue.
Overall, the aim of this brief report was to compare different groups of infected and vaccinated subjects with different regimens (representative of the general population), in order to verify the neutralizing response elicited toward the two sub-lineages BA.1 and BA.2 as compared to the wt virus. Although this should be further investigated, results of our study led us to conclude that the repeated exposure to the antigen, whether by natural infection or booster dose, can boost immunity toward more conserved antigens, generating cross-neutralizing antibodies that might retain their neutralizing power also against distinct (future) Omicron sub-lineages; and that this increased exposure allows maturity of antibody avidity and thus a prolonged antigen availability, supporting the idea that antibodies with high avidity and high neutralizing potential can increase cross-protection against variants that carry several mutations on the RBD.
Acknowledgement
The authors thank Ida Paciello and Giulio Pierleoni for their technical assistance.
Disclosure statement
FD, ML, CS, PP, GP, LB, E. Molesti and A.M. are employed by VisMederi srl. M.L. and L.B. are employed by VisMederi Research srl. R.R. is an employee of the GSK group of companies. E.A. and R.R. are listed as inventors of full-length human monoclonal antibodies described in Italian patent applications n. 102020000015754 filed on June 30th 2020, 102020000018955 filed on August 3rd 2020 and 102020000029969 filed on 4th of December 2020, and the international patent system number PCT/IB2021/055755 filed on the 28th of June 2021. All patents were submitted by Fondazione Toscana Life Sciences, Siena, Italy. CMT is an external consultant of VisMederi Research srl. E. Montomoli is founder and Chief Scientific Officer of VisMederi srl and VisMederi Research srl.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Errico JM, Adams LJ, Fremont DH. Antibody-mediated immunity to SARS-CoV-2 spike. Adv Immunol. 2022;154:1–5.
- Chen J, Wei GW. Omicron BA.2 (B.1.1.529.2): high potential to becoming the next dominating variant. J Phys Chem Lett. 2022;13(17):3840–49.
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180(2):281–92. doi:10.1016/j.cell.2020.02.058.
- Martinez-Flores D, Zepeda-Cervantes J, Cruz-Reséndiz A, Aguirre-Sampieri S, Sampieri A, Vaca L. SARS-CoV-2 vaccines based on the spike glycoprotein and implications of new viral variants. Front Immunol. 2021;12:701501.
- European Commission. Safe COVID-19 vaccines for Europeans; European Commission. 2022.
- Elliott P, Eales O, Steyn N, Tang D, Bodinier B, Wang H, Elliott J, Whitaker M, Atchison C, Diggle PJ, et al. Twin peaks: the Omicron SARS-CoV-2 BA.1 and BA.2 epidemics in England. Science. 2022;376(6600). doi:10.1126/science.abq4411.
- Duty JA, Kraus T, Zhou H, Zhang Y, Shaabani N, Yildiz S, Du N, Singh A, Miorin L, Li D, et al. Discovery and intranasal administration of a SARS-CoV-2 broadly acting neutralizing antibody with activity against multiple Omicron subvariants. Med (NY),3;1–17. 2022; S2666-6340(22):00321-X.
- Marchi S, Viviani S, Remarque EJ, Ruello A, Bombardieri E, Bollati V, Milani GP, Manenti A, Lapini G, Rebuffat A, et al. Characterization of antibody response in asymptomatic and symptomatic SARS-CoV-2 infection. PLoS One. 2021;16(7):e0253977. doi:10.1371/journal.pone.0253977.
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;27(3):493‐497. doi:10.1093/oxfordjournals.aje.a118408.
- Lyngse FP, Kirkeby CT, Denwood M, Christiansen LE, Mølbak K, Møller CH, Skov RL, Krause TG., Rasmussen M, Sieber RN, et al. Transmission of SARS-CoV-2. 1. Omicron VOC subvariants BA.1 and BA.2: 2: evidence from Danish households. MedRxiv. 2022.
- Evans JP, Zeng C, Qu P, Faraone J, Zheng Y-M, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, et al. Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microb. 2022;30:1093–1102.
- Ai J, Wang X, He X, Zhao X, Zhang Y, Jiang Y, Li M, Cui Y, Chen Y, Qiao R, et al. Antibody evasion of SARS-CoV-2 Omicron. BA.1, BA.1.1, BA.2, and BA.3 sub-lineages. Cell Host Microbe. 2022;30(8):1–7. doi:10.1016/j.chom.2022.05.001.
- Dapporto F, Marchi S, Leonardi M, Piu P, Lovreglio P, Decaro N, Buonvino N, Stufano A, Lorusso E, Bombardieri E, et al. Antibody avidity and neutralizing response against SARS-CoV-2 Omicron variant after infection or vaccination. J Immunol Res. 2022;2022:4813199. doi:10.1155/2022/4813199.
- Pratesi F, Caruso T, Testa D, Tarpanelli T, Gentili A, Gioè D, Migliorini P. Bnt162b2 mRNA SARS-CoV-2 vaccine elicits high avidity and neutralizing antibodies in healthcare workers. Vaccines (Basel). 2021;9(6). doi:10.3390/vaccines9060672.
- Richterman A, Scott J, Cevik M. Covid-19 vaccines, immunity, and boosters. BMJ. 2021;375:n3105.
- Rössler A, Netzl A, Knabl L, Schäfer H, Wilks SH, Bante D, Falkensammer B, Borena W, von Laer D, Smith D, et al. BA.2 omicron differs immunologically from both BA.1 omicron and pre-omicron variants [Preprint]. doi:10.1101/2022.05.10.22274906
- Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, Subramoney K, Makatini Z, Moyo S, Amoako DG, et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med. 2022;28(9):1785–90. doi:https://doi.org/10.1038/s41591-022-01911-2.
- Wang Q, Guo Y, Iketani S, Nair MS, Li Z, Mohri H, Wang M, Yu J, Bowen AD, Chang JY, et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608(7923):603–08. doi:10.1038/s41586-022-05053-w.
