ABSTRACT
The currently used Japanese Oka and Korean MAV/06-attenuated varicella vaccine strains belong to clade 2 genotype varicella–zoster viruses (VZV). More than seven clades of VZV exist worldwide. In this study, we investigated the cross-reactivity of antibodies induced by clade 2 genotype vaccines against VZV strains belonging to clades 1, 2, 3, and 5 using a fluorescent antibody to membrane antigen (FAMA) test. Among 59 donors, 29 were vaccinated with the MAV/06 strain MG1111 (GC Biopharma, South Korea) and the other 30 were vaccinated with the Oka strain VARIVAX (Merck, USA). The sera were titrated using FAMA tests prepared with six different VZV strains (two vaccine strains, one wild-type clade 2 strain, and one each of clade 1, 3, and 5 strains). The ranges of geometric mean titers (GMTs) of FAMA against six different strains were 158.7–206.5 and 157.6–238.9 in MG1111 and VARIVAX groups, respectively. GMTs of the MG1111 group against all six strains were similar; however, GMTs of the VARIVAX group showed differences of approximately 1.5-fold depending on the strains. Nevertheless, the GMTs of the two vaccinated groups for the same strain were not significantly different. These results suggest that both MG1111 and VARIVAX vaccinations induce cross-reactive humoral immunity against other clades of VZV.
Introduction
Varicella–zoster virus (VZV) belongs to the genus Varicellovirus, which is a member of α-herpesvirinae of Herpesviridae. VZV is a highly contagious virus, and varicella vaccines have been developed using live-attenuated strains, derived from vOka and MAV/06. The vOka and MAV/06 strains originated from clinical isolates of varicella-infected children in Japan and Korea, respectively.Citation1–3 Attenuation was performed by serial passages in several cell lines at different temperatures.Citation4 According to whole-genome sequencing data, 72 passages of VZV in cell culture lead to 28 substitutions.Citation5 Compared to the wild-type parental Oka (pOka) strain, 42 base substitutions that lead to 20 amino acid changes and differences in the length of tandem repeat regions and in the origin of DNA replication were found in the attenuated vOka strain.Citation6 Among them, 15 substitutions leading to 8 amino acid changes occurred in the immediate-early gene 62 (IE62), a strong trans-activator that interferes with virus growth and spread activity.Citation6 Analysis of single-nucleotide polymorphisms (SNPs) suggests 24 vaccine-specific sites in the VZV genome that differentiate vaccine strains from wild-type strains, and 18 vaccine-specific sites have both wild-type and vaccine sequences.Citation7 It is well known that live-attenuated varicella vaccine induces tenfold less immunity than the wild-type virus.Citation8,Citation9 However, it is unclear whether vaccine immunogenicity is influenced by amino acid substitutions in vaccine-specific sites.
There are at least nine glycoproteins in the VZV envelope, among which glycoprotein E (gE), gB, and gH are immunodominant glycoproteins.Citation10–12 After varicella vaccination, gE and gB antibodies were predominantly observed. However, gH-specific antibodies showed the most potent neutralizing activity.Citation13 The fluorescent antibody to membrane antigen (FAMA) test detects antibodies against VZV glycoproteins on the cell membrane. The FAMA test is considered to be a more reliable protective immunogenicity test than the neutralization test because of the cell-associated nature of VZV.Citation9,Citation14
VZV genotype has proposed seven distinct major phylogenetic clades (1–6, 9) and one tentative clade (VIII) based on single-nucleotide polymorphisms in the whole genome of VZV.Citation15,Citation16 There were at least 30 SNPs in 18 open reading frames (ORFs) to distinguish VZV clades.Citation15,Citation16 Clade 1 and 3 strains are prevalent mainly in Europe, Americas, and Oceania, whereas clade 2 strains are dominant in Asia and clade 5 strains are dominant in Africa.Citation17 Both MAV/06- and vOka-derived vaccine strains belong to clade 2.Citation18 A previous study showed that vOka-derived Varilrix vaccine-induced antibody pooled from seven individuals had cross-reactivity against clade 1, 3, and 5 strains.Citation19
A new live-attenuated varicella vaccine (MG1111) was recently launched in Korea using the MAV/06 strain propagated in the Medical Research Council-5 (MRC-5) cell line.Citation20 To investigate whether MAV/06-based vaccine can protect against wild-type viruses and other clade viruses, we evaluated the cross-reactivity of the sera from MG1111- and VARIVAX-vaccinated individuals using six different VZV strains as FAMA antigens.
Materials and methods
Sera from vaccinees
Sera were collected from vaccinees who participated in the phase 3 clinical trial of MG1111 which had been performed in Thailand and South Korea (protocol No. NCT03375502).Citation20 During enrollment, children who had been exposed to varicella or previously vaccinated were excluded. Yeungnam University Medical Center IRB approved for the use of those samples for this study (YUMC 2019-09-081). To minimize the variability between two vaccine groups, we selected samples with the same FAMA titer as 128 at post-vaccination when analyzed with MAV/06 strain as FAMA antigen during the phase 3 clinical trial. To exclude the preexisting VZV antibody positive sample before vaccination, we selected FAMA-negative samples (FAMA titer <4) at pre-vaccination (Supplemental Figure S1, Supplemental Table S1). For final statistical analysis, 29 samples from the MG1111 (GC Biopharma, Yong-in, South Korea)-vaccinated group and 30 from the VARIVAX (Merck, Kenilworth, NJ, USA)-vaccinated group were included. In the MG1111 group, 24 and 5 samples were selected from Thailand and South Korea, respectively. In the VARIVAX group, 25 and 5 samples were selected from Thailand and South Korea, respectively (Supplemental Figure S1). The mean age was 2 y in both MG1111 and VARIVAX groups.
Cell and viruses
MRC-5 cells (ECACC, UK) were cultured in Eagle’s minimal essential medium (EMEM; Lonza, Breda, Netherlands) supplemented with 10% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY, USA), 1% MEM non-essential amino acid (NEAA; Sigma-Aldrich, St. Louis, MO, USA), 1% l-glutamine (Gibco BRL), and 1× antibiotic antimycotic (Gibco BRL) at 37°C in a humidified atmosphere containing 5% CO2.
MRC-5 cell cultures at 70–80% confluence were individually infected with 6 VZV strains () at a ratio of one infected cell to 200 normal cells. VZV-infected cells (FAMA antigens) were harvested at 72 h post-infection and stored in liquid nitrogen until use. YC03 strain was isolated in our laboratory from 8-y-old girl with zoster in 2012 and confirmed as clade 2 wild-type virus.Citation21 VZV strains 432/2008 (clade 1), 2308/2003 (clade 3), and 446/2007 (clade 5) were kindly provided by Dr Andreas Sauerbrei (Jena University, Germany). VZV strains 432/2008, 2308/2003, and 446/2007 were isolated from a 57-y-old patient with zoster, a 5-y-old girl with varicella, and a 1-y-old boy with varicella in Germany, respectively.Citation22
Table 1. Varicella–zoster virus strains used as FAMA antigen.
Fluorescent antibody to membrane antigen (FAMA) test
Sera were twofold serially diluted with Dulbecco’s phosphate-buffered saline (DPBS, Lonza, Breda, Netherlands), and mouse monoclonal anti-VZV glycoprotein H (gH), gB, gI, and gE antibodies (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) were diluted 1:50 in DPBS. WHO international standard for VZV immunoglobulin (National Institute for Biological Standards and Control (NIBSC) code: W1044, UK) was used as a positive reference serum.
Diluted sera (100 µL) or monoclonal antibodies were incubated with 2 × 105 FAMA antigen cells for 30 min at room temperature (RT) and then washed twice with DPBS. Goat anti-human IgG-Alexa 488 or goat anti-mouse IgG-Alexa 488 antibody (Invitrogen, Eugene, OR, USA) was used as the secondary antibody. After the secondary antibody reaction for 30 min at RT, FAMA antigen cells were washed three times with DPBS. FAMA antigen cells were loaded at 7 µL per well in a 14-well slide (Cel-Line®, Thermo Scientific, MA, USA) and then dried. Slides were mounted with VectaShield mounting medium containing DAPI (Vector Laboratories, Burlingame, CA, USA) and observed using an Axioscope fluorescence microscope equipped with an HBO 50 mercury lamp (Carl Zeiss, Jena, Germany).
Phylogenetic analysis of seven VZV strains
Whole nucleotide sequences of seven VZV strains, including the attenuated clade 2 MAV/06 (GenBank Accession No. JF306641) and VARIVAX-Oka (DQ008355), wild-type clade 2 YC03 (KJ808816) and parental Oka (AB097933), clade 1 wild-type 432/2008 (JN704695), clade 3 wild-type 2308/2003 (JN704699), and clade 5 wild-type 446/2007 (JN704707), were downloaded from GenBank. Among them, ten ORFs such as ORF1 (membrane protein), 5 (gK), 9A (gN), 14 (gC), 31 (gB), 37 (gH), 50 (gM), 60 (gL), 67 (gI), and 68 (gE) were analyzed. The evolution history was inferred using the Maximum Likelihood method and the Tamura–Nei model.Citation23 The tree with the highest log-likelihood is shown. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura–Nei model and then selecting the topology with a superior log-likelihood value. The tree was drawn to scale, with branch lengths indicating the number of substitutions per site. Evolutionary analyses were conducted using MEGA X.Citation24
Statistical analyses
Statistical analyses were performed using GraphPad Prism 9.5.0 (GraphPad Software, San Diego, CA, USA). Two-tailed non-parametric Mann–Whitney U tests were performed to compare geometric mean titers (GMTs) between the two vaccine groups for the same FAMA antigen strains (significance level: p < .05). To compare GMTs between six different FAMA antigen strains within the same vaccine group, repeated-measures two-way ANOVA with Tukey’s multiple comparisons test was performed (significance level: p < .05).
Results
Comparison of antigenicities of six different VZV strains against reference serum and glycoprotein-specific monoclonal antibodies
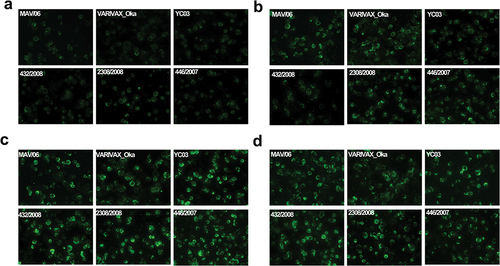
Before antibody titrations, antigenicities of FAMA antigens prepared with six strains (MAV/06, VARIVAX-Oka, YC03, 432/2008, 2308/2003, or 446/2007) were evaluated using positive reference serum (31.25 mIU/mL). All six strains showed 2+ grade fluorescent intensity relative to the positive reference serum (). After verifying the equivalent antigenicity of FAMA antigens prepared with the six strains, we used these six FAMA antigens to analyze sera from the MG1111 or VARIVAX groups.
Figure 1. Image of FAMA test with positive reference serum using six different VZV strains as FAMA antigens (400× magnification).
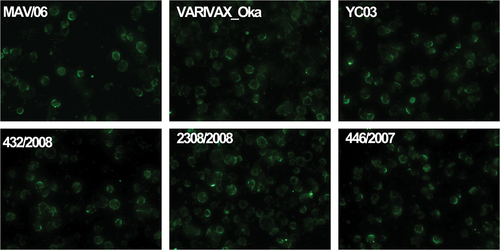
The antigenicities of highly immunogenic glycoproteins, such as gH, gB, gI, and gE, of the six different VZV strains were also evaluated using anti-VZV glycoprotein-specific monoclonal antibodies (). Anti-gH showed the weakest fluorescence intensity (), whereas anti-gI showed the strongest response compared to the other glycoprotein antibodies (). Anti-gH, anti-gI, and anti-gE showed similar fluorescence intensities against all six VZV strains; however, anti-gB showed weaker fluorescence intensity against the 432/2008 strain as compared to intensity against the other five strains ().
Geometric mean titers of vaccinated sera analyzed using six different VZV strains FAMA antigens
FAMA titers of 59 sera from the MG1111- and VARIVAX-vaccinated groups were analyzed using FAMA antigens prepared using six different VZV strains: clade 2 attenuated MAV/06 and VARIVAX-Oka, clade 2 wild-type YC03, clade 1 wild-type 432/2008, clade 3 wild-type 2308/2003, and clade 5 wild-type 446/2007 strains. GMTs of the MG1111 and VARIVAX groups were not significantly different when analyzed using the same FAMA antigen strain (p > .05, ). A repeated-measures two-way ANOVA followed by Tukey multiple comparisons test was performed to compare the GMTs of MG1111- and VARIVAX-vaccinated sera, which were analyzed using the six different VZV strains. There were no statistically significant differences between the FAMA titers of MG1111- and VARIVAX-vaccinated sera according to FAMA antigens (p = .361) (). GMTs analyzed using clade 2 VARIVAX-Oka strain were significantly different from those analyzed using clade 5 446/2007 strain in both MG1111 and VARIVAX groups (p = .015 and p = .007, respectively) (). In the VARIVAX group, the GMT analyzed using the MAV/06 strain was significantly different even from that analyzed using the other clade 2 strains, VARIVAX-Oka and YC03 (p = .029 and p = .006, respectively) (). Especially, the GMT analyzed using clade 5 446/2007 strain was different from that analyzed using all the other strains, except MAV/06 in the VARIVAX group (). However, the differences of GMTs were within 1.5-fold.
Figure 3. Geometric mean FAMA titer for six different VZV strains in MG1111-vaccinated sera and VARIVAX-vaccinated sera (a). Statistical differences in MG1111-vaccinated sera (b) and VARIVAX-vaccinated sera (c). Repeated-measures two-way ANOVA by Tukey multiple comparisons test at a significance level of *p < .05, **p < .01.
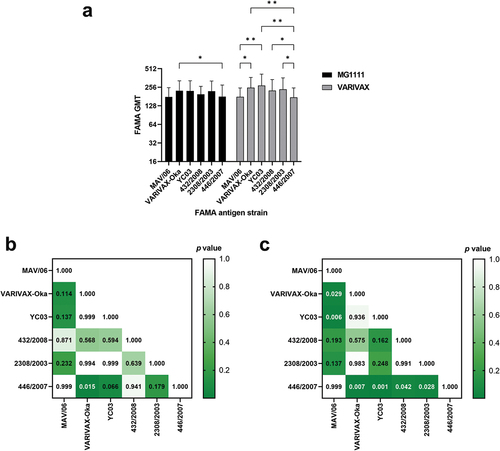
Table 2. Geometric mean titers of MG1111- and VARIVAX-vaccinated sera against six different VZV strain FAMA antigens.
Phylogenetic analysis of ten ORFs in seven different VZV strains
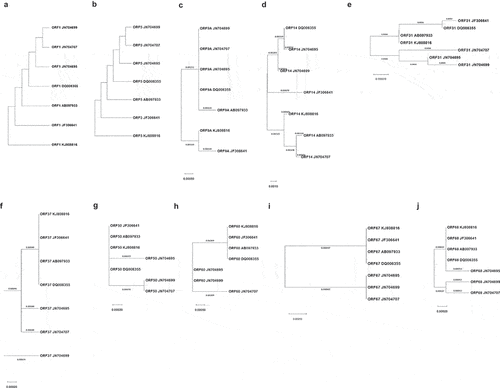
Phylogenetic trees of ten ORFs in seven different VZV strains that were used for preparing FAMA antigens, including the parental Oka (pOka) strain, were generated (). Membrane protein (ORF1) and gK (ORF5) had the same sequences in all seven strains (). All clade 2 VZV strains had the same ancestors for gH (ORF37), gM (ORF50), gL (ORF60), gI (ORF67), and gE (ORF68), and there was no divergence, regardless of attenuation. Therefore, two attenuated vaccine strains, MAV/06 and VARIVAX-Oka, shared the same sequences for seven ORFs among ten ORFs, including highly immunogenic gE and gH. However, three ORFs, gN (ORF9A), gC (ORF14), and gB (ORF31), were different between two attenuated vaccine strains, and gC was the most divergent. Strains 432/2008 (clade 1), 2308/2003 (clade 3), and 446/2007 (clade 5) showed different patterns for gC, gB, gH, gM, gL, and gE. For gE, 432/2008 strain branched from the same ancestor of clade 2 strains, but 2308/2003 and 446/2007 strains did not. For gB, strains 432/2008, 2308/2003, and 446/2007 originated from ancestors distinct from clade 2 strains, while strains 432/2008 and 2308/2003 had the same ancestor. For gH, 2308/2003 showed distinct clustering from clade 2 strains. However, sequence divergences were between 0.11 and 0.24 bases for every 100 bases for all of the above three glycoproteins.
Figure 4. NJ phylogenetic tree of ten ORF sequences of 7 VZV strains. Each tree was constructed using the Neighbor-Join method and BioNJ algorithms. Branch lengths are proportional to the divergence between the tax. (a) ORF1, (b) ORF5, (c) ORF9A, (d) ORF14, (e) ORF31, (f) ORF37, (g) ORF50, (h) ORF60, (i) ORF67, (j) ORF68.
Discussion
Two live-attenuated clade 2 genotype VZV strains, vOka and MAV/06, are used in VZV vaccines worldwide. vOka-derived strains are used in several vaccines, including the Biken varicella vaccine (Biken, Osaka, Japan), VARIVAX (Merck & Co., Inc., Kenilworth, NJ, USA), Varilrix (GSK, Rixensart, Belgium), Vari-L (Changchun Institute of Biological Products, Changchun-si, China), Sky Varicella (SK Bioscience, Seongnam, Korea), Zostavax (Merck & Co., Inc., Kenilworth, NJ, USA), and Sky Zoster (SK Bioscience, Seongnam, Korea). The MAV/06 strain developed by GC BioPharma (Yongin, Korea) has been used for Suduvax grown in the LuMA cell line since 1994. Recently, MG1111, a new varicella vaccine made with the MAV/06 strain grown in the MRC-5 cell line, was approved by the Ministry of Food and Drug Safety (MFDS) of Korea in March 2020 and named BARYCELA (GC Biopharma, Yongin, Korea).
In this study, we assessed whether sera from clade 2 live-attenuated VZV vaccine-immunized individuals cross reacted with world-wide prevalent clades of varicella strains, including the wild-type clade 2 VZV. Sera from MG1111-vaccinated (n = 29) and VARIVAXTM-vaccinated individuals (n = 30) were analyzed using the FAMA test prepared with six different VZV strains, namely MAV/06 (live-attenuated clade 2), VARIVAX-Oka (live-attenuated clade 2), YC03 (wild-type clade 2), 432/2008 (clade 1), 2308/2003 (clade 3), and 446/2007 (clade 5) as FAMA antigens.
GMTs of MG1111- and VARIVAX-vaccinated sera were not significantly different when analyzed using the same FAMA antigen strains (). It is highly likely that MAV/06 and VARIVAX-Oka strains share the identical sequences for most glycoproteins. In particular, both strains share the same sequences for membrane proteins and six glycoproteins, including highly immunogenic gE and gH (). The sequence of highly immunogenic gB differs by only 0.02/100 base pairs between two vaccine strains. GMTs of MG1111-vaccinated sera were not significantly different when analyzed using clades 1, 2, and 3 VZV FAMA antigens, regardless of the virus being attenuated or wild-type. These results were similar to recent data obtained using MAV/06 vaccine-immunized guinea pig sera.Citation25 MAV/06-vaccinated guinea pig sera cross-reacted not only with eight clade 2 wild-type YC strains but also with clade 1, 3, and 5 strains.Citation25 GMTs of VARIVAX-vaccinated sera were also not significantly different when analyzed using VARIVAX-Oka, YC03, clade 1, and clade 3 strain FAMA antigens. These results are similar to previous data showing that Varilrix®-vaccinated human sera had protective immunity against not only clade 2 but alsoclades 1, 3, and 5.Citation19 By 2021, varicella vaccine has been used as an universal vaccine in 44 countries, even though vaccination schedule, doses, and ages are vary depending on countries.Citation26 In particular, the most prevalent varicella strain in the United States is found to be clade 1, while varicella-associated disease declined > 97% in all age groups and 99% in <20 y of age at 25 y after the introduction of varicella vaccination program.Citation27 In Europe, Canada, and Australia, where clades 1 and 3 are predominant area, hospitalization rate was significantly reduced in children aged 1–4 y after introduction of varicella vaccine.Citation28 Cade 2 is the most prevalent clade in East Asia such as Korea, Japan, China, Taiwan. In Korea, varicella vaccine was introduced in 1988 and included in National Immunization Program (NIP) since 2005. The vaccine coverage rate was over 97%. However, there were no reliable epidemiologic data for varicella incidence or hospitalization before inclusion in NIP, it was very hard to interpret vaccine effectiveness. However, according to the Health Insurance Review and Assessment Service data, the number of varicella diseases decreased gradually.Citation29 Japan introduced varicella vaccine as universal vaccine in 2014 with two dose schedules, the effectiveness of varicella vaccine in children younger than 15 y was between 76% and 94% depending on doses.Citation30 In Taiwan, the incidence of varicella and hospitalization were reduced 82% and 85%, respectively, after the introduction of universal varicella vaccination program.Citation31 The clinical follow-up study of MG1111 had been conducted until 6 months after vaccination in Thailand and Korea. Among 814 children who enrolled in phase 2/3, 17 subjects who had at least one varicella-like rash were reported. VZV genotyping was conducted in 15 samples. The VZV genotyping results showed that four out of seven subjects in the VARIVAX group were confirmed as a vOka vaccine strain and eight out of eight subjects in MG1111 group and three out of seven subjects in VARIVAX group were confirmed as not detectable results (unpublished data). Therefore, there were no evidence of other clades or wild-type clade 2 VZV infection until 6 months after MG1111 vaccination. Therefore, the effectiveness of clade 2 varicella vaccine has been clinically validated to reduce disease burden in worldwide.
In this study, GMTs of the sera of VARIVAX-vaccinated individuals were significantly different even for clade 2 FAMA antigen strains such as VARIVAX-Oka vs. MAV/06 (p = .029) and YC03 vs. MAV/06 (p = .006). Vaccine strains MAV/06 (Suduvax, MG1111) and vOka (VARIVAX, Varilrix) have 24 vaccine-specific sequence sites compared to wild-type YC03 strain.Citation7 However, 18 of the vaccine-specific sequence sites are heterogeneous, having both wild-type and vaccine-specific sequences. The average frequencies of wild-type sequences at the 24 vaccine-specific sites were 36.26%, 12.22%, and 99.83% in VARIVAX, MAV/06, and YC03, respectively.Citation7 Therefore, VARIVAX strain had more wild-type sequences than MAV/06 strain. This might explain higher response of VARIVAX-vaccinated sera to VARIVAX-Oka or YC03 FAMA antigens compared with that to MAV/06 FAMA antigens. Most sequence differences between wild-type and attenuated vaccine strains are in ORF62 (immediate early 62 protein).Citation7 Thus, the attenuation might not alter the major immunogenicity induced by glycoproteins. GMTs of MG1111-vaccinated sera were different when analyzed using clade 5 (446/2007 strain) and clade 2 VARIVAX-Oka strain FAMA antigens. GMTs of VARIVAX-vaccinated sera analyzed using clade 5 446/2007 was significantly different from GMTs analyzed by all the other strains (p < .05) except clade 2 MAV/06 (p = .999) (). This result might be due to the higher divergence of clade 5 446/2007 strain compared to other clade strains (). Nevertheless, the highest GMT differences between 446/2007 and other strains were only 1.3- and 1.5-fold in MG1111 and VARIVAX groups, respectively. Unfortunately, varicella vaccine is not introduced as a universal vaccination program in Africa and Bangladesh, where clade 5 strain is predominant, there are no available clinical data of varicella vaccine effectiveness on clade 5 yet. Therefore, it will be necessary to further investigate the clinical effectiveness of varicella vaccines in clade 5 prevalent area.
This study has some limitations. First, the sample size of each vaccine group was small. Second, we used only one strain each from clades 1, 3, and 5 of VZV. Further evaluation of additional strains and clades as FAMA antigens will be interesting to investigate in the future.
In conclusion, MAV/06-based vaccine demonstrates comparable immunogenicity to clade 2 VZV strains and clades 1, 3, and 5 VZV strains. In addition, when evaluated using the same strain FAMA antigens, there were no variations in GMTs between MG1111- and VARIVAXTM-vaccinated sera. Therefore, MG1111 could provide effective measures to reduce disease burden of varicella in worldwide.
Supplemental Material
Download PDF (173 KB)Acknowledgments
The authors thank Dr Andreas Sauerbrei (Jena University, Germany) for providing VZV strains used in this study.
Disclosure statement
Younchul Shin is an employee of GC BioPharma. Involvement of GC Biopharma employee did not compromise the scientific integrity of this work. The other authors declare no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website at https://doi.org/10.1080/21645515.2023.2210961.
Additional information
Funding
References
- Takahashi M, Otsuka T, Okuno Y, Asano Y, Yazaki T. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet. 1974;2(7892):1–8. doi:10.1016/s0140-6736(74)90144-5. PMID: 4139526.
- Hwang KK, Park SY, Kim SJ, Ryu YW, Kim KH. Restriction fragment length polymorphism analysis of varicella-zoster virus isolated in Korea. J Korean Soc Virol. 1991;21:201–10.
- Park SY, Hwang KK, Choi MK, Ryu YW, Paik SB, Kim KH. Propagation of varicella-zoster virus isolated in Korea. J Korean Soc Virol. 1991;21:1–9.
- Hwang KK, Chun BH, Park HS, Park SY, Kim KH, Moon HM. Marker test for attenuation of varicella-zoster viruses isolated in Korea. J Korean Soc Virol. 1992;22:105–9.
- Tyler SD, Peters GA, Grose C, Severini A, Gray MJ, Upton C, Tipples GA. Genomic cartography of varicella–zoster virus: a complete genome-based analysis of strain variability with implications for attenuation and phenotypic differences. Virology. 2007;359(2):447–58. doi:10.1016/j.virol.2006.09.037. PMID: 17069870.
- Gomi Y, Sunamachi H, Mori Y, Nagaike K, Takahashi M, Yamanishi K. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J Virol. 2002;76(22):11447–59. doi:10.1128/jvi.76.22.11447-11459.2002. PMID: 12388706.
- Jeon JS, Won YH, Kim IK, Ahn JH, Shin OS, Kim JH, Lee CH. Analysis of single nucleotide polymorphism among varicella-zoster virus and identification of vaccine-specific sites. Virology. 2016;496:277–86. doi:10.1016/j.virol.2016.06.017. PMID: 27376245.
- Ndumbe PM, Cradock-Watson J, Levinsky RJ. Natural and artificial immunity to varicella zoster virus. J Med Virol. 1988;25(2):171–8. doi:10.1002/jmv.1890250207. PMID: 2839610.
- Kim Y, Hwang JY, Lee KM, Lee E, Park H. Cross-sectional study of varicella zoster virus immunity in healthy Korean children assessed by glycoprotein enzyme-linked immunosorbent assay and fluorescent antibody to membrane antigen test. Vaccines (Basel). 2021;9(5):492. doi:10.3390/vaccines9050492. PMID: 34065863.
- Brunell PA, Novelli VM, Keller PM, Ellis RW. Antibodies to the three major glycoproteins of varicella-zoster virus: search for the relevant host immune response. J Infect Dis. 1987;156(3):430–5. doi:10.1093/infdis/156.3.430. PMID: 3039010.
- Giller RH, Winistorfer S, Grose C. Immunogenicity of purified varicella zoster virus glycoproteins. Pediatr Res. 1987;21(4):312. doi:10.1203/00006450-198704010-00868.
- Haumont M, Jurdan M, Kangro H, Jacquet A, Massaer M, Deleersnyder V, Garcia L, Bosseloir A, Bruck C, Bollen A, et al. Neutralizing antibody responses induced by varicella‐zoster virus gE and gB glycoproteins following infection, reactivation or immunization. J Med Virol. 1997;53:63–8. doi:10.1002/(sici)1096-9071(199709)53:1<63:aid-jmv11>3.0.co;2-y. PMID: 9298734.
- Sullivan NL, Reuter-Monslow MA, Sei J, Durr E, Davis CW, Chang C, McCausland M, Wieland A, Krah D, Rouphael N, et al. Breadth and functionality of varicella-zoster virus glycoprotein-specific antibodies identified after Zostavax vaccination in humans. J Virol. 2018;92(14):e00269–18. doi:10.1128/JVI.00269-18. PMID: 29743372.
- Breuer J, Schmid DS, Gershon AA. Use and limitations of varicella-zoster virus-specific serological testing to evaluate breakthrough disease in vaccinees and to screen for susceptibility to varicella. J Infect Dis. 2008;197(Suppl 2):S147–51. doi:10.1086/529448. PMID: 18419389 .
- Breuer J, Grose C, Norberg P, Tipples G, Schmid DS. A proposal for a common nomenclature for viral clades that form the species varicella-zoster virus: summary of VZV Nomenclature Meeting 2008, Barts and the London School of Medicine and Dentistry, 24–25 July 2008. J Gen Virol. 2010;91(4):821–8. doi:10.1099/vir.0.017814-0. PMID: 20071486.
- Jensen NJ, Rivailler P, Tseng HF, Quinlivan ML, Radford K, Folster J, Harpaz R, LaRussa P, Jacobsen S, Scott Schmid D. Revisiting the genotyping scheme for varicella-zoster viruses based on whole-genome comparisons. J Gen Virol. 2017;98(6):1434–8. doi:10.1099/jgv.0.000772. PMID: 28613146.
- Schmidt-Chanasit J, Sauerbrei A. Evolution and world-wide distribution of varicella–zoster virus clades. Infect Genet Evol. 2011;11(1):1–0. doi:10.1016/j.meegid.2010.08.014. PMID: 20817040.
- Kim JI, Jung GS, Kim YY, Ji GY, Kim HS, Wang WD, Park HS, Park SY, Kim GH, Kwon SN, et al. Sequencing and characterization of Varicella-zoster virus vaccine strain SuduVax. Virol J. 2011;8(1):547. doi:10.1186/1743-422X-8-547. PMID: 22176950.
- Sauerbrei A, Stefanski J, Gruhn B, Wutzler P. Immune response of varicella vaccinees to different varicella-zoster virus genotypes. Vaccine. 2011;29(22):3873–7. doi:10.1016/j.vaccine.2011.03.054. PMID: 21459174.
- Choi UY, Kim KH, Lee J, Eun BW, Kim DH, Ma SH, Kim CS, Lapphra K, Tangsathapornpong A, Kosalaraksa P, et al. Immunogenicity and safety profiles of a new MAV/06 strain varicella vaccine in healthy children: a multinational, multicenter, randomized, double-blinded, active-controlled phase III study. Vaccine. 2021;39(12):1758–64. doi:10.1016/j.vaccine.2021.02.013. PMID: 33627245.
- Kim MH, Jeon JS, Kim IK, Park JS, Park H, Shin OS, Lee CH. Characterization and phylogenetic analysis of Varicella-zoster virus strains isolated from Korean patients. J Microbiol. 2017;55(8):665–72. doi:10.1007/s12275-017-7171-3. PMID: 28752294.
- Zell R, Taudien S, Pfaff F, Wutzler P, Platzer M, Sauerbrei A. Sequencing of 21 varicella-zoster virus genomes reveals two novel genotypes and evidence of recombination. J Virol. 2012;86(3):1608–22. doi:10.1128/JVI.06233-11. PMID: 22130537.
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–26. doi:10.1093/oxfordjournals.molbev.a040023. PMID: 8336541.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. doi:10.1093/molbev/msy096. PMID: 29722887.
- Shin D, Shin Y, Kim E, Nam H, Nan H, Lee J. Immunological characteristics of MAV/06 strain of varicella-zoster virus vaccine in an animal model. BMC Immunol. 2022;23(1):27. doi:10.1186/s12865-022-00503-6. PMID: 35658899.
- Lee YH, Choe YJ, Lee J, Kim E, Lee JY, Hong K, Yoon Y, Kim Y-K. Global varicella vaccination programs. Clin Exp Pediatr. 2022;65(12):555–62. doi:10.3345/cep.2021.01564. PMID: 36457198.
- Marin M, Leung J, Anderson TC, Lopez AS. Monitoring varicella vaccine impact on varicella incidence in the United States: surveillance challenges and changing epidemiology, 1995–2019. J Infect Dis. 2022;226(Supplement_4):S392–9. doi:10.1093/infdis/jiac22. PMID: 36265855.
- Varela FH, Pinto LA, Scotta MC. Global impact of varicella vaccination programs. Human Vacc Immunother. 2019;15(3):645–57. doi:10.1080/21645515.2018.1546525. PMID: 30427766.
- Service HIRA. Healthcare bigdata hub [Internet]. Wonju (Korea): Health Insurance Review & Assessment Service; 2022 [accessed 2021 Sep 30]. https://opendata.hira.or.kr/home.do.
- Hattori F, Miura H, Sugata K, Yoshikawa A, Ihira M, Yahata Y, Kamiya H, Tanaka-Taya K, Yoshikawa T. Evaluating the effectiveness of the universal immunization program against varicella in Japanese children. Vaccine. 2017;35(37):4936–41. doi:10.1016/j.vaccine.2017.07.090. PMID: 28784281.
- Huang WC, Huang LM, Chang IS, Tsai FY, Chang LY. Varicella breakthrough infection and vaccine effectiveness in Taiwan. Vaccine. 2011;29(15):2756–60. doi:10.1016/j.vaccine.2011.01.092. PMID: 21315697.