Abstract
ABSTRACT. The CRISPR/Cas9 system is rapidly becoming the reagent of choice for targeted mutagenesis and gene editing in crop species. There are currently intense research efforts in the crop sciences to identify efficient CRISPR/Cas9 platforms to carry out targeted mutagenesis and gene editing projects. These efforts typically result in the incremental tweaking of various platform components including the identification of crop-specific promoters and terminators for optimal expression of the Cas9 enzyme and identification of promoters for expression of the CRISPR guide RNA. In this report, we demonstrate the development of an online web tool for fast identification of CRISPR/Cas9 target loci within soybean gene models, and generic DNA sequences. The web-tool described in this work can quickly identify a high number of potential CRISPR/Cas9 target sites, including restriction enzyme sites that can facilitate the detection of new mutations. In conjunction with the web tool, a soybean codon-optimized CRISPR/Cas9 platform was designed to direct double-stranded breaks to the targeted loci in hairy root transformed cells. The modified Cas9 enzyme was shown to successfully mutate target genes in somatic cells of 2 legume species, soybean and Medicago truncatula. These new tools may help facilitate targeted mutagenesis in legume and other plant species.
ABBREVIATIONS
| GUS | = | β-glucuronidase |
| ZFNs | = | zinc-finger nucleases |
| TALENs | = | transcription activator–like effector nucleases |
| CRISPR | = | clustered regularly interspaced short palindromic repeat |
| NHEJ | = | non-homologous end joining |
| crRNA | = | CRISPR RNA |
| tracrRNA | = | trans-activating crRNA |
| GS1 | = | Glutamine Synthase |
| NLS | = | nuclear localization signal |
| CAPS | = | Cleaved Amplified Polymorphic Sequences |
| CaMV | = | Cauliflower mosaic virus |
| UBQ10 | = | ubiquitin10 promoter |
| GmCas9 | = | soybean codon optimized Cas9 gene |
| NOS | = | nopaline synthase |
| PAM | = | protospacer adjacent motif |
BACKGROUND
Traditional mutagenesis using chemical or radiation treatments generates random DNA alterations genome-wide. Despite these inefficiencies, thousands of released cultivars and breeding lines with unique or desirable characteristics have been developed with these methods (Maluszynski et al. Citation1995). Recent developments in molecular biology are changing the mutagenesis platforms, allowing for more targeted and precise DNA alterations using zinc-finger nucleases (ZFNs), transcription activator–like effector nucleases (TALENs), and clustered, regularly interspaced, short palindromic repeat (CRISPR)–associated protein (Cas9) (Bibikova et al., Citation2002; Christian et al., Citation2010; Cong et al., Citation2013; Kim et al., Citation1996; Mali et al., Citation2013; Miller et al., Citation2011; Sander et al., Citation2011). Double-stranded breaks generated by these systems stimulate the host cell's DNA repair response, the non-homologous end joining (NHEJ) repair pathway, which repairs the break. However, this mechanism is often error-prone, resulting in the introduction of indels that range from single to hundreds of base pairs (Voytas, Citation2013). The capacity to create indels and disrupt the reading frame of a particular gene has been used to create mutations in model and crop species, typically for functional genomics applications. Furthermore, after confirming the desired germline mutation, the transgene used to generate these mutations can easily be removed in many plant species by standard genetic segregation to create null segregants (Curtin et al., Citation2012).
The CRISPR/Cas9 system is the most recent of these targeted mutagenesis platforms and was discovered as a component of the immune system from the bacterium Streptococcus pyogenes. In bacteria, the CRISPR system is employed to defend the host from invading viral (phage) and plasmid DNA species (Bhaya et al., Citation2011). One of the core enzymes of this bacterial defense system is the endonuclease Cas9 (SpCas9). Cas9 forms a complex with a duplex CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA) that can introduce sequence specific double-stranded DNA breaks (Cong et al., Citation2013; Jinek et al., Citation2012; Mali et al., Citation2013). By retrofitting the crRNA to target a specific DNA sequence, the user can introduce targeted mutations in a wide variety of eukaryotic cells, including human, yeast, mouse, fish and plants (Cho et al., Citation2013; Dicarlo et al., Citation2013; Hwang et al., Citation2013; Jiang et al., Citation2013; Li et al., Citation2013; Shan et al., Citation2013). The CRISPR/Cas9 system is rapidly becoming the method of choice for targeted mutagenesis and gene editing in crop species due to its ease of assembly and increased frequency of bi-allelic mutations (Baltes and Voytas, Citation2015; Canver et al., Citation2014).
Recent reports in soybean (Glycine max) have demonstrated the use of CRISPR/Cas9 using hairy-root composite plants, soybean protoplasts, and whole soybean plants (Cai et al., Citation2015; Jacobs et al., Citation2015; Li et al., Citation2015; Sun et al., Citation2015). In this work, we used a combination of a streamlined CRISPR/Cas9 design website and a Glycine max (soybean) codon optimized Cas9 protein to create somatic mutations in both soybean and Medicago truncatula.
RESULTS AND DISCUSSION
Identification of Gene Targets for CRISPR/Cas9
We created a webserver (http://stuparcrispr.cfans.umn.edu/CRISPR/) that rapidly identifies CRISPR/Cas9 target sites (GN20GG) using either raw sequence information or a soybean gene model (the unique “Glyma” soybean gene identifier) as input. If raw sequence is entered, the website returns all forward and reverse strand target sites, the location of each target, the restriction sites associated with the target, and any primers that will be needed to design and assemble the CRISPR construct. The restriction site information may be useful for designing Cleaved Amplified Polymorphic Sequences (CAPS) assays, which greatly facilitates the screening of mutated alleles in transformed tissues.
If a Glyma gene model is entered into the webserver, the website again returns the forward and reverse strand target sites, the location of each target, the restriction sites associated with the target, and the primers that will be needed to design and assemble the CRISPR construct. However, in this case, the output will also include text highlighting for UTRs, introns and exons. Furthermore, only CRISPR targets located within exons will be identified.
Mutagenesis of 2 Soybean Genes in Hairy Root Tissues
The webserver was used to design CRISPR target sites for a soybean Glutamine Synthase (GS1) gene (gene identifier Glyma18g04660 in the version 1 genome assembly (Schmutz et al., Citation2010). and Glyma.18g041100 in the version 2 assembly) and a chalcone-flavanone isomerase gene (CHI20, gene identifiers Glyma20g38560 and Glyma.20g241500, respectively). To test these targets, a codon-optimized Cas9 (GmCas9) was designed based on the preferred codon usage of soybean. The GmCas9 enzyme has an amino acid sequence that matches that of the Arabidopsis thaliana codon-optimized Cas9 enzyme described by Li et al. Citation(2013) however there are 3 key distinguishing characteristics between the two 2 systems. 1) Due to the differing preferred codon usage of the 2 species, the Cas9 enzyme-encoding DNA sequences differ at 448 out of 4,103 bp (11%) positions. 2) The Arabidopsis thaliana codon-optimized Cas9 contains an IV2 intron whereas the GmCas9 does not include any intron. 3) The GmCas9 contains a single NLS at the C-terminus while the Arabidopsis thaliana version contains NLS at both the N-terminus and C-terminus.
The MDC32/GUS/GmCas9 destination vector was used to target GS1, which included the soybean codon-optimized Cas9 cassette and a β-glucuronidase (GUS) reporter gene (; ). The expression vector containing the target sequence was transformed in soybean hairy roots as a way to quickly screen for effectiveness of the expression vector. Transformed hairy roots were stained with 5-bromo-4-chloro-3-indolyl glucuronide and genomic DNA was extracted from roots that stained blue (). DNA from a stained root was extracted from 3 different regions of the root (top, middle and bottom). CAPS assays were run on each of the 3 samples, using a BsmA I restriction enzyme (). A 2-bp deletion was identified in each of the 3 samples, indicating that this mutation was found throughout the root (; Fig. S1).
FIGURE 1. CRISPR/Cas9 cassette and constructs. (A) A map of the codon optimized Glycine max Cas9 (GmCas9) gene is shown along with surrounding components and restriction sites. (B) A map of the guide RNA with designated restriction site in red for insertion of target recognition sequence. (C) MDC32/Cas9 construct used for targeted mutagenesis in Medicago truncatula. (D) MDC32/GUS/Cas9 construct with GUS reporter used for targeted mutagenesis in soybean.
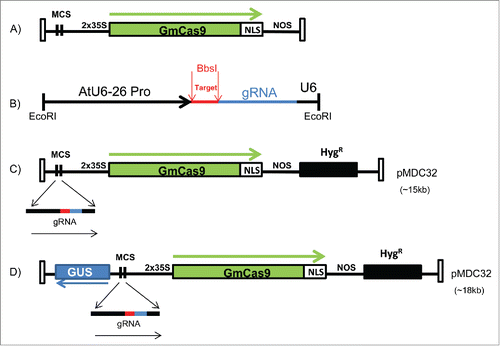
FIGURE 2. Detection of CRISPR/GmCas9-induced mutations for the GS1 gene in soybean hairy-root tissue. (A) Transgenic GUS expression in a soybean hairy root after GUS staining is shown. (B) CAPS-PCR analysis was used to detect mutations in soybean hairy roots, based on PCR amplicon digestion with the BsmA I restriction enzyme. wt and wt/D indicate undigested and digested wild-type controls. Assays were performed on 3 different root segments taken from the top (lane 1), middle (lane 2) and bottom (lane 3) portion of the transgenic hairy-root. (C) Sequencing of the mutation induced by the CRISPR/GmCas9 system revealed a 2-bp deletion. The underlined sequences indicate the restriction enzyme and PAM sites, respectively.
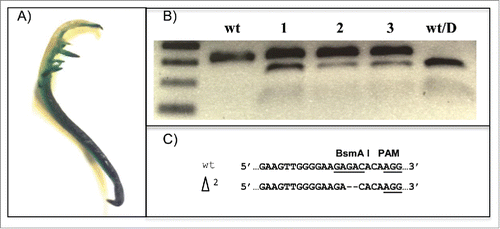
TABLE 1. The expression vectors and guide RNA sequences used to target a GUS transgene in genes in Medicago truncatula and endogenous soybean genes
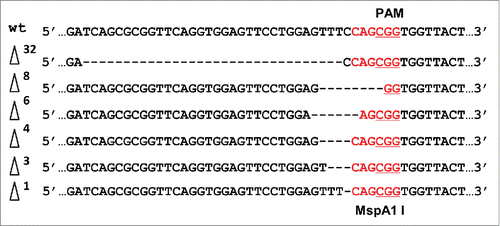
Similar hairy root transformations were performed for the CRISPR/Cas9 construct targeting the CHI20 gene (). DNA from a GUS-positive transformed root was tested by CAPS analysis using an MspA1 I restriction enzyme recognition site (CAGCGG) in the target sequence, which includes the PAM site (CGG). Sequencing of cloned PCR products of the target region revealed 2 mutations that disrupted the MspA1 I and 4 mutations that did not disrupt this site (). Altogether, 6 different deletion types were detected, including those of 1, 3, 4, 6, 8 and 32 bp.
Mutagenesis of a Medicago Truncatula GUS Transgene
To test the soybean codon-optimized Cas9 in a different legume, we transformed hairy roots from a M. truncaluta line that is transgenic for the GUS gene. The MDC32/GmCas9 construct carried a CRISPR guide RNA sequence targeting the GUS transgene (; ). This experiment was designed to test whether this CRISPR/Cas9 system could cause mutations in the GUS transgene of M. truncatula cells.
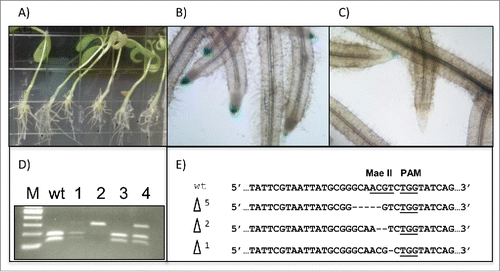
A hairy root protocol that has had prior success with RNA interference in Ivashta et al. Citation(2005) and Boisson-Dernier et al. Citation(2001) was utilized for this purpose (. To compare visual indications of disruption to the GUS gene, hairy roots transformed with a control expression vector with no guide RNA target were compared to hairy roots transformed with the MDC32/GmCas9 construct (). The control roots consistently showed GUS staining while some roots from the MDC32/GmCas9 transformation showed a loss of GUS staining. Genomic DNA from hairy roots of transgenic composite plants were extracted and assayed for mutations. An enrichment PCR CAPS assay (see Methods section for details) consisted of using a Mae II restriction site, designed 3–5 bp upstream of the protospacer adjacent motif (PAM) region. Electrophoresis of the CAPS bands indicated that some roots contained PCR products that were recalcitrant to Mae II digestion, indicating new mutations at this locus (). Cloning and sequencing of these bands confirmed the frame-shift mutations (). Three different sized deletions were obtained from hairy root transformation, indicating successful mutagenesis at the target GUS locus using the MDC32/GmCas9 expression vector.
FIGURE 4. Induction of mutations in Medicago truncatula hairy roots. (A) Representative Medicago truncatula composite plants are shown. (B) Histochemical staining for β-glucuronidase activity in hairy roots from composite plants containing Cas9 with no guide RNA shows consistent GUS expression. (C) Histochemical staining for β-glucuronidase activity in hairy roots from composite plants containing the MDC32/Cas9 construct with GUS guide RNA target shows loss of GUS expression in some roots. (D) CAPS-PCR analysis was used to detect mutations in M. truncatula hairy roots, based on amplicon digestions with the Mae II restriction enzyme. wt indicates the digested wild-type control and lanes 1, 2, 3 and 4 represent 4 digestions of PCR product from the hairy roots of different composite plants. (E) Alignments of nucleotide sequences between mutant and wild-type alleles for CRISPR M. truncatula GUS targets shows a range of mutations. The underlined sequences indicate the restriction enzyme and PAM sites, respectively.
CONCLUSIONS
In this report, we demonstrated targeted mutagenesis using a new web-tool for identifying target sites and a soybean codon-optimized CRISPR/Cas9 system to induce mutations in soybean and M. trucatula. This is promising evidence that these tools can also be used to design new constructs and generate heritable mutations in whole soybean and M. trucatula plants. One of the benefits of using the hairy-root ex vitro assays in M. trucatula and soybean is the rapid turn-around in generating transgenic material for testing various configurations of platform components. For example, there is currently ongoing work to identify improved promoters for the expression of the Cas9 in legumes, such as the Cauliflower mosaic virus (CaMV) 35S, the Glycine max ubiquitin (Gmubi) and the Arabidopsis thaliana ubiquitin10 promoter (UBQ10). In addition, improved expression of the single guide RNA has also been a major research focus, such as the use of U6 promoters from various species including Arabidopsis, soybean and M. trucatula. While mutations were identified in this study, we have no evidence that the soybean codon-optimized Cas9 is necessarily better than other Cas9s for plant targeted mutagenesis. Such assessments would require more structured comparisons, as it is likely that the optimal mutagenesis system will vary among different plant species. As such, the present work describes new tools that can continue to be optimized moving forward.
METHODS
Construction of Soybean Codon-Optimized CRISPR/Cas9 Expression Cassettes
The amino acid sequence of the Cas9 protein was based on published work in Mail et al. Citation(2013) Codon optimization for this protein was performed using a custom Perl script designed to utilize the preferred codon usage of soybean (Murray et al., Citation1989). The Cas9 cassette design consisted of a 2x35S CaMV promoter (Odell et al., Citation1985), the soybean codon optimized Cas9 gene (GmCas9), a SV40 NLS, and a nopaline synthase terminator (NOS) (Fig. 1A). The Cas9 gene with flanking Sal I and Xba I restriction sites was then synthesized through Life Technologies gene art facility along with the guide-RNA expression cassette (gRNA), which was flanked by the Arabidopsis U6 promoter and terminator. The gRNA expression cassette was inserted into the pBluescript KS+ backbone using the EcoR I site to create a shuttle vector ().
Identification of gRNA Target Sites
The CRISPR design website was programmed in a combination of Python 2 and PHP (http://stuparcrispr.cfans.umn.edu/CRISPR/). The website also depends on the Phytozome database for soybean genome information and specific highlighting. The tool was tested on the browsers Google Chrome, Firefox and Safari.
Assembly of CRISPR/Cas9 Destination Vectors
The GmCas9 gene was ligated into a pMDC32 vector backbone digested with the Asn I and Pac I restriction enzymes. The pMDC32 backbone was used for both cassettes based on previous reports of successful plant gene transformation (Curtis and Grossniklaus, Citation2003). The GmCas9 coding region was subcloned as an Asc I/Pac I fragment to replace the ccdB gene in the pMDC32 vector. Both restriction sites were already present (flanking the GmCas9 ORF) in the plasmid from Life Technologies. The guide RNA and the 2x35s, GmCas9, NOS, MDC32 vector were digested with EcoRI and ligated together to create the GmCas9/MDC32 expression vector (). The MDC32/GUS/GmCas9 expression vector was created by amplifying a GUS gene with flanking Pme I sites and inserting it into the MDC32/GUS/GmCas9 expression vector (). The guide RNA for both vectors consisted of the commonly used Arabidopsis U6 promoter and terminator as wells as a palindromic Bbs I type II restriction site for insertion of the CRISPR target site sequence. The guide RNA for the soybean transformation was designed to target Glutamine synthase (GS1) and chalcone-flavanone isomerase (CHI20) genes, while the guide RNA for the M. truncatula transformation was designed to target a GUS transgene.
Hairy Root Transformation of Soybean and Medicago Truncatula
The MDC32/GUS/GmCas9 construct was transformed into Agrobacterium rhizogenes strain ARqual (Quandt et al., Citation1993) and introduced into soybean cultivar ‘Bert’ (derived from the individual ‘Bert-MN-01’) coytledons using a modified in vitro hairy root transformation assay (Supplementary Text S1) (Govindarajulu et al., Citation2008). The MDC32/GmCas9 construct was transformed into the Agrobacterium rhizogenes strain ARqual for hairy-root transformation of a M. truncatula accession containing a GUS gene. The method was based on a previously reported ex vitro composite plant transformation assay (Boisson-Dernier et al., Citation2001).
Identification of Mutations Using a PCR-Restriction Enzyme Assay
The CAPS-PCR protocol used to detect the presence of targeted mutations in soybean and M. truncatula was adapted from previous published work (Konieczny and Ausubel, Citation1993). For soybean, a transformed hairy root was identified by GUS staining. This root transformed with the construct targeting GS1 was divided into 3 segments: top (1), middle (2), and bottom (3). Genomic DNA was obtained for the 3 segments using the Qiagen DNeasy Mini Kit and a 302 bp fragment that flanks the GS1 target was directly amplified by PCR (Fig. S2). The PCR was performed using Qiagen HotStar Taq Plus master mix in a thermal cycler at 95°C for 5 minutes, 95°C for 45 seconds, 54°C for 45 seconds, 72°C for 1 minute, repeat steps 2–4 32 times, 72°C for 5 minutes, 4°C forever. PCR buffer and fragments were then removed using the Qiagen PCR cleanup kit. The PCR product was then digested with 100 units of BsmA I restriction enzyme (designed to cut 3–5bp upstream of the PAM) at 37°C for 4 hours, 65°C for 15 minutes, and 4°C forever. Digested DNA was then run on a 1% agrose gel in 1x TBE for 30 minutes at 100 V, then imaged on a UV gel imager. Gel bands that appeared to be undigested were then extracted using the Qiagen gel extraction kit, cloned with the pGEM-T easy vector for direct T/A cloning of PCR products and sequenced using Sanger sequencing.
Similar methods were used to identify mutations in the hairy root transformed with the construct targeting CHI20. However, in this case, a single DNA extraction was prepared from the whole root. PCR amplification of a 316 bp regions surrounding the targeted interval was performed on this DNA sample (Fig. S2) and PCR products were cloned into the pGEM-T easy vector. Colony PCR was performed on a subset of clones and the PCR products were digested for 3 hours with MspA1 I. The PCR inserts from all the undigested clones and some digested clones were sequenced by Sanger methods to detect gene mutations.
The CAPS-PCR analysis to detect mutations in M. trucatula was performed using a similar method as described above, with the following modifications. DNA was extracted by sampling all of the hairy roots for a given transgenic composite plant; multiple composite plants were independently sampled (e.g. lanes 1–4 in represent samplings from different composite plants). The genomic DNA samples were pre-digested with 100 units of Mae II restriction enzyme (designed to cut 3–5bp upstream of the PAM) at 37°C for 4 hours, 65°C for 15 minutes, and 4°C forever. This step enriches the subsequent PCR for mutated sequences, as unmutated sequences will presumably be digested and unable to amplify during the PCR step. A 320 bp fragment that flanks the GUS target was amplified by PCR (Fig. S2). The PCR products were purified and digested with 100 units of Mae II as described above. PCR buffer and fragments were then removed using the Qiagen PCR cleanup kit. The CAPS-PCR products were visualized as bands on agarose gels. Undigested bands were extracted, cloned and sequenced to identify mutations as described in the previous paragraph.
Resource Information and Availability
The web-tool used for CRISPR target identification is available at http://stuparcrispr.cfans.umn.edu/CRISPR/. The nucleotide sequence information for both the GmCas9 and guide RNA cassettes shown in are provided in GenBank accession numbers KT827738 and KT827739. The essential components of the constructs, replacing the Hygromycin and GUS selectable markers with bialaphos resistance, can be ordered through Addgene accession numbers 59184 and 59188. Inquiries about the exact vectors used in this study, which utilize Hygromycin and GUS selectable markers, can be made to the corresponding author.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
SUPPLEMENTAL MATERIAL
Supplemental data for this article can be accessed on the publisher's website.
Supplemental_Material.zip
Download Zip (631.4 KB)Acknowledgments
The authors would like to thank Justin Anderson for providing the soybean codon-optimization script and Colby Starker for helpful guidance. The authors are also grateful to the University of Minnesota Genomics Center for sequencing support and the Minnesota Supercomputing Institute for computational support.
Funding
This work was generously supported by the Minnesota Soybean Research & Promotion Council, the United Soybean Board, and the National Natural Science Foundation of China (project 31201226).
REFERENCES
- Baltes NJ, Voytas DF. Enabling plant synthetic biology through genome engineering. Trends Biotechnol 2015; 33:120-31; PMID:25496918; http://dx.doi.org/10.1016/j.tibtech.2014.11.008
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas Systems in Bacteria and Archaea: Versatile Small RNAs for Adaptive Defense and Regulation. Annu Rev Genet 2011; 45:273-97; PMID:22060043; http://dx.doi.org/10.1146/annurev-genet-110410-132430
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 2002; 161:1169-75; PMID:12136019
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 2001; 14:695-700; PMID:11386364; http://dx.doi.org/10.1094/MPMI.2001.14.6.695
- Cai Y, Chen L, Liu X, Sun S, Wu C, Jiang B, Han T, Hou W. CRISPR/Cas9-Mediated Genome Editing in Soybean Hairy Roots. PLoS One 2015; 10:e0136064; PMID:26284791; http://dx.doi.org/10.1371/journal.pone.0136064
- Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, Orkin SH. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/cas9 nuclease system in mammalian cells. J Biol Chem 2014; 289:21312-24; PMID:24907273; http://dx.doi.org/10.1074/jbc.M114.564625
- Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013; 31:230-2; PMID:23360966; http://dx.doi.org/10.1038/nbt.2507
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 2010; 186:757-61; PMID:20660643; http://dx.doi.org/10.1534/genetics.110.120717
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013; 339:819-23; PMID:23287718; http://dx.doi.org/10.1126/science.1231143
- Curtin SJ, Voytas DF, Stupar RM. Genome Engineering of Crops with Designer Nucleases. Plant Genome 2012; 5:42; http://dx.doi.org/10.3835/plantgenome2012.06.0008
- Curtis MD, Grossniklaus U. A Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta. Plant Physiol 2003; 133:462-9; PMID:14555774; http://dx.doi.org/10.1104/pp.103.027979
- Dicarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 2013; 41:4336-43; PMID:23460208; http://dx.doi.org/10.1093/nar/gkt135
- Govindarajulu M, Elmore JM, Fester T, Taylor CG. Evaluation of constitutive viral promoters in transgenic soybean roots and nodules. Mol Plant Microbe Interact 2008; 21:1027-35; PMID:18616399; http://dx.doi.org/10.1094/MPMI-21-8-1027
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31:227-9; PMID:23360964; http://dx.doi.org/10.1038/nbt.2501
- Ivashuta S, Liu J, Lohar D. RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell 2005; 17:1-11; PMID:16199614; http://dx.doi.org/10.1105/tpc.105.035394
- Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 2015; 15:1-10; PMID:25636680; http://dx.doi.org/10.1186/s12896-015-0131-2
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 2013; 31:233-9; PMID:23360965; http://dx.doi.org/10.1038/nbt.2508
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. 2012; 337:816-21; PMID:22745249
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA 1996; 93:1156-60; PMID:8577732
- Konieczny A, Ausubel FM. A procedure for map-ping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 1993; 4:403-10; PMID:8106085; http://dx.doi.org/10.1046/j.1365-313X.1993.04020403.x
- Li J-F, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 2013; 31:688-91; PMID:23929339; http://dx.doi.org/10.1038/nbt.2654
- Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM. Cas9-guide RNA Directed Genome Editing in Soybean. Plant Physiol 2015; 169(2):960-70; PMID:26294043; http://dx.doi.org/10.1104/pp.15.00783
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science 2013; 339:823-6; PMID:23287722; http://dx.doi.org/10.1126/science.1232033
- Maluszynski M, Ahloowalia BS, Sigurbjörnsson B. Application of in vivo and in vitro mutation techniques for crop improvement. Euphytica 1995; 85:303-15; http://dx.doi.org/10.1007/BF00023960
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 2011; 29:143-8; PMID:21179091; http://dx.doi.org/10.1038/nbt.1755
- Murray EE, Lotzer J, Eberle M. Codon usage in plant genes. Nucleic Acids Res 1989; 17:477-498; PMID:2644621 http://dx.doi.org/10.1093/nar/17.2.477
- Odell JT, Nagy F, Chua N-H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985; 313:810-2; PMID:3974711; http://dx.doi.org/; http://dx.doi.org/10.1038/313810a0
- Quandt HJ, Puehler A, Broer I. Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 1993; 6:699-703; http://dx.doi.org/10.1094/MPMI-6-699
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 2011; 8:67-9; PMID:21151135; http://dx.doi.org/10.1038/nmeth.1542
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature 2010; 463:178-83; PMID:20075913; http://dx.doi.org/10.1038/nature08670
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu J-L, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 2013; 31:686-8; PMID:23929338; http://dx.doi.org/10.1038/nbt.2650
- Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep 2015; 5:10342; PMID:26022141; http://dx.doi.org/10.1038/srep10342
- Voytas DF. Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 2013; 64:327-50; PMID:23451779; http://dx.doi.org/10.1146/annurev-arplant-042811-105552