ABSTRACT
The prohibitins (PHB) are SPFH domain-containing proteins found in the prokaryotes to eukaryotes. The plant PHBs are associated with a wide range of biological processes, including senescence, development, and responses to biotic and abiotic stresses. The PHB proteins are identified and characterized in the number of plant species, such as Arabidopsis, rice, maize, and soybean. However, no systematic identification of PHB proteins was performed in Solanum lycopersicum. In this study, we identified 16 PHB proteins in the tomato genome. The analysis of conserved motifs and gene structure validated the phylogenetic classification of tomato PHB proteins. It was observed that various members of tomato PHB proteins undergo purifying selection based on the Ka/Ks ratio and are targeted by four families of miRNAs. Moreover, SlPHB proteins displayed a very unique expression pattern in different plant parts including fruits at various development stages. It was found that SlPHBs processed various development-related and phytohormone responsive cis-regulatory elements in their promoter regions. Furthermore, the exogenous phytohormones treatments (Abscisic acid, indole-3-acetic acid, gibberellic acid, methyl jasmonate) salt and drought stresses induce the expression of SlPHB. Moreover, the subcellular localization assay revealed that SlPHB5 and SlPHB10 were located in the mitochondria. This study systematically summarized the general characterization of SlPHBs in the tomato genome and provides a foundation for the functional characterization of PHB genes in tomato and other plant species.
1. Introduction
The prohibitins (PHB) genes concede highly conserved stomatin/prohibitin/flotillin/HflK/C (SPFH) domain in their protein sequence also recognized as band_7 domain proteins.Citation1 PHBs proteins are ubiquitous proteins and are associated with a variety of biological processes including cell cycle, apoptosis, and respiration.Citation2–5 PHBs have been identified from eukaryotes, fungi, plants, and animals.Citation6,Citation7 In humans, the PHB proteins act as transcriptional regulators interacting with PSF3, retinoblastoma proteins (Rb), and E2F.Citation8,Citation9 PHB genes were observed to be linked with the breast cancer phenotype, where they localize in the nucleus of breast cancer cell lines as a transcriptional regulator via interaction with P53, RB and E2F to regulate the expression of downstream genes. PHBs were also identified in lipid raft, a key constituent of cell membrane.Citation10–13 Similarly, PHBs found in plasma membrane were considered to act as a target for small molecules in the inflammatory reponses as well as to regulate the membrane receptor and iron channels.Citation14,Citation15 In short, PHB genes play crucial roles in different biological processes and are associated with various disease phenotypes. However, less is known about the role of PHB proteins in the plant kingdom.
PHB proteins are classified into type I and type II and both are complimentary for stability and functioning of PHB protein.Citation17 In mammals, PHB1 and PHB2 have been well characterized and shown to form a 1–2 KDa protein complex on the inner mitochondrial membrane. In addition, the absence of any of these two proteins failed to produce this protein complex in Caenorhabditis elegens, resulted in decreased PHB proteins. PHB complex have been physically and functionally linked with the matrix-ATPase related to diverse cellular activities (m-AA) to regulate the degradation of respiratory chain proteins in mitochondria.Citation18 PHB and PHB2/REA were found to be involved in maintaining cellular survival via Ras–Raf–MEK–Erk pathway.Citation19 These findings suggest that both types of PHB are required for stable complex formation and proper functioning.Citation5,Citation20,Citation21Recently, various studies reported the role of PHB in plants. These proteins play a pivotal role not only in plant development and senescence but also in responses to abiotic and biotic stresses.Citation22,Citation23 PHB3 and PHB4 are the most broadly studied PHB genes from Arabidopsis thaliana, where they primarily expressed both in root and shoot proliferative tissues. Arabidopsis mutant, atphb3 exhibited severely retarded growth phenotypes, decreased stem, root proliferation, and declined cell division in root and stem apices.Citation24 Overexpression of Arabidopsis PHB (AtPHB3/AtPHB4) exhibited irregular leaf shape and extensive branching phenotype.Citation24 Notably, atphb3/4 double knockout mutants were not viable, suggesting that PHBs play important role in plant development.Citation24 Similar results were obtained in petunia and tobacco, where PHB-silenced genes showed decreased cell production and prolonged senescence.Citation25,Citation26 In tobacco, suppression of NbPHB2 delays growth and promotes leaf senescence and apoptosis.Citation26 Moreover, the cells in silenced flowers were larger as compared to control flowers, suggesting a significant decrease in the number of cell division that occurs during corolla development. PHB proteins directly or indirectly interact with mitochondrial DNA (mtDNA) to regulate the reactive oxygen species (ROS) formation and oxidative phosphorylation (OXPHOS), which potentially lead to senescence phenotype both in C. elegans and plants.Citation20,Citation25–27 Furthermore, PHB protein might also involve in maintaining crista morphology to employ proteins into the inner membrane.Citation21,Citation28 The abovementioned finding indicates that PHB play key functioning in cell proliferation. Several studies have shown that PHB proteins play key roles not only in plant development and senescence but also in response to salinity, defense and plant hormones. For instance, Arabidopsis eer3-1(atphb3) mutant showed an etiolated seedling phenotype upon constitutive exposure to ethylene with suppressed the expression of various ethylene inducible genes (Arabidopsis ethylene-responsive element binding protein [AtEBP], plant defensin [PDF 1.2]), indicating the dual role of AtPHB3 in Arabidopsis.Citation29 Additionally, AtPHB3 acted downstream of ethylene insensitive 2 (EIN2) and EIN3. A loss of function mutant atphb3-3 failed to affect diverse biological processes such as nitric oxide (NO) signaling, ABA (abscisic acid) induced stomatal closure, IAA (auxin) induced root formation.Citation30 This mutant resulted from the substitution of Gly at position 165 with Asp of AtPHB3 protein. However, another Arabidopsis PHB (At5g64870) induced under cold, salinity, and drought but suppressed in response to hormones such as gibberellin (GA), methyl jasmonate (MeJA), and ABA.Citation31 PHB proteins have been identified in various plant species including 17 in Arabidopsis, 19 in rice,Citation31 24 in Glycine max,Citation32 and Zea mays with 16.Citation17 The knowledge about PHB genes in tomato is insufficient. In this study, a total of 16 PHB genes were identified in the tomato genome. Phylogenetic analysis, gene structure, in silico subcellular location prediction, cis-regulatory elements, MEME motif scan, and protein chromosome location were also conducted. In addition, tissues/organ-specific expression profiling under normal conditions was evaluated. Moreover, differential expression patterns under salt, drought, and hormone-induced expression were analyzed. This study enables us to provide a foundation for the functional characterization of PHB genes in tomato.
2. Material and Method
2.1. The Tomato PHB Gene Discovery
To predict PHB genes in the tomato genome, the Arabidopsis, rice, Zea mays, and Glycine max PHB peptide sequences were retrieved from the TAIR genome database (https://www.arabidopsis.org/),Citation33 rice genome annotation project (http://rice.plantbiology.msu.edu), phytozome database (https://phytozome.jgi.doe.gov/), respectively. These sequences were used as a query in the SOL genome network (https://solgenomics.net).Citation34 The candidates’ sequences were analyzed for SPFH Domain (PF01145) in the SMART (http://smart.embl-heidelberg.de)Citation35 and NCBI conserved domain database (CDD, https://ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi).Citation36 Moreover, PHB protein features including the isoelectric point (pI), the grand average of hydropathy (GRAVY), molecular weight (kDa) of each protein were calculated in sequence manipulation suite (SMS, bioinformatics.org/sms2).Citation37 The deduced PHB proteins were named in their order on the tomato chromosomes.
2.2. Phylogenetic Analysis and Ka/Ks Analysis of Duplications
Clustal Omega (ClustalO, https://www.ebi.ac.uk/Tools/msa/clustalo/)Citation38 program was used to generate SlPHB peptide sequences alignment. For the phylogenetic relationship, SlPHBs peptide sequences from rice, Arabidopsis, Zea mays, and soybean were retrieved from phytozome (https://phytozome.jgi.doe.gov). An unrooted neighbor-joiningCitation39 tree was constructed using MEGAX softwareCitation40 with the parameters set as follows: Poisson correlation of model; pairwise deletion of gaps/missing data; random seed of phylogeny test and bootstrap was set at 1000 replicates. The non-synonymous (Ka), synonymous (Ks) nucleotide substitution rates and the Ka/Ks ratio were predicted using k-estimator (http://en.bio-soft.net/format/KEstimator.html).Citation41 The divergence time (T) was calculated as follows: T = Ks/2y (y = 6.56 x 10−9).Citation42
2.3. Chromosome Location, Subcellular Location Prediction, and miRNA Target Prediction
The chromosome position of each SlPHB gene was obtained from the SOL genome and visualized in the MAPGene2Chromsome program (http://mg2c.iask.in/mg2c_v2.0/). In silico subcellular location, prediction analysis was performed in the WoLFPSORT program (https://wolfpsort.hgc.jp).Citation43 To predict miRNAs targeted putative PHBs, the cDNA sequences of each SlPHBs were submitted to psRNATargetCitation44 against all tomato miRNAs reported in miRbase.Citation45
2.4. Gene Structure Analysis, Conserved Motif Scan, and Cis-Regulatory Motif Prediction
The retrieved tomato SlPHBs coding sequences (CDS) and genomic sequences were submitted to the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn)Citation46 for intron and exon distribution in each gene. MEME suite (http://meme-suite.org)Citation47 was used to predict conserved motifs in SlPHB protein sequences with a parameter set as follows: (i) a maximum number of motifs – 10, (ii) number of repetitions – any, (iii) optimum motif width set to ≥10 and ≤50. A 1000bp 5`UTR nucleotide sequences from the start codon (ATG) of each SlPHB gene were retrieved from the SOL genome and scanned in the PlantCRAE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)Citation48 for cis-regulatory elements prediction.
2.5. Plant Material, Abiotic Stress, and Phytohormone Treatment
Tomato cv. Micro-Tom seedlings were grown in the College of Agriculture and life sciences, Kunming University, under controlled greenhouse conditions (25°C/20°C, day/night, 14 h/10 h light/dark photoperiod with relative humidity 80%). For tissue/organ-specific expression analysis of various plant parts such as root, leaves, stem, and flowers were collected from a six-week-old plant. For expression in fruit tissues, 1/2/3/cm, mature green fruit, breaker fruit, and ten days breaker fruits were harvested.Citation49 For salinity, drought, and phytohormone-induced stresses, six-week-old plants were treated with 200 mM NaCl, 0.01 mM ABA, GA3, IAA, MeJA, and PEG as described previously.Citation50 Roots and shoots (including stem and leaves) were harvested at 0 h, 3 h, 6 h, 12 h, and 24 h interval after treatment. All the samples were collected in triplicate and store immediately at −80°C.
2.6. Total RNA Extraction, cDNA Preparation, and qRT-PCR Analysis
Total RNA was extracted from selected samples using TRIZOL reagent according to the manufacturer’s instruction. RNA was quantified using nanodrop lite (Thermo USA) and RNA integrity was assessed by running 2% agar agarose gel electrophoresis. The cDNA was synthesized with a PrimerScript Real-Time (RT) reagent kit (Takara, Japan) according to the manufacturer’s protocol as described previously.Citation51–53 RT-qPCR was conducted in ABI 7500 Fast Real-Time system (AB, USA) using the iTaq™ Universal SYBR® Green Supermix (BIO-RAD, USA) according to the manufacturer’s protocol. The RT-qPCR was conducted in triplicate. Tomato SlUBQ (Solyc01g056940) gene was used as an internal control. The relative expression of tomato SlPHBs was calculated using the 2−ΔΔCt methodCitation54 and heat maps were generated with heat mapper program (http://www1.heatmapper.ca/expression/).
2.7. Subcellular Localization of SlPHB5 and SlPHB10
The full-length sequences of SlPHB5 and SlPHB10 excluding stop codon were fused into the vector p35S-GFP as explained previously.Citation51,Citation55 The Arabidopsis protoplast isolation and transformation were carried out as described by Sheen.Citation56 After 18–20 h of transformation, the protoplast was visualized by confocal laser scanning microscope and the images were processed using photoshop.
3. Results
3.1. Identification of SlPHB Genes
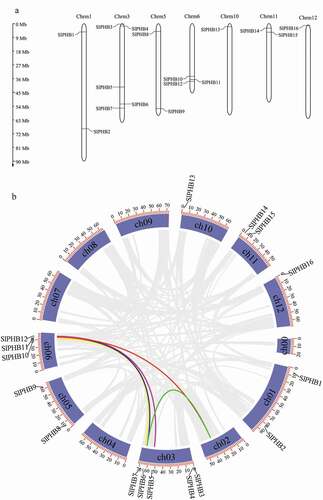
The Arabidopsis, rice, Zea mays, and glycine max PHB protein sequences were used as a query in the SOL genome to identify all putative PHB protein sequences in the tomato genome. A total of 16 non-redundant genes were identified. The Pfam, SMART, and NCBI CDD searches were used to verify the SPFH domain in all SlPHBs protein sequences. The tomato PHB genes were named as SlPHB1 to SlPHB16 in order of their position in chromosomes. The peptide length to the molecular weight of SlPHBs ranged from 261 aa (SlPHB8) to 518 aa (SlPHB7), and 30.08 kDa (SlPHB1) to 57.75 kDa (SlPHB7). The GRAVY values of all the SlPHB proteins were negatively exhibiting indicating that these proteins are hydrophilic except SlPHB15 (Solyc11g013260) which show a positive GRAVY score. The deduced SlPHB genes were distributed in seven chromosomes ()). A pair of genes SlPHB1 and SlPHB2, SlPHB8 and SlPHB9, SlPHB14, and SlPHB15 were located on chromosomes 1, 5, and 11 each, respectively. SlPHB3, SlPHB4, SlPHB5, SlPHB6, and SlPHB7 were located on chromosome 3. Three genes (SlPHB10, SlPHB11, SlPHB12) were located on chromosome 6 while a single gene was located on chromosome 10 (SlPHB13) and chromosome 12 (SlPHB16) each. In silico subcellular location, prediction indicated that SlPHBs were localized in the cytoplasm, mitochondria, and chloroplast (). Tomato PHB genes displayed segmental duplication and five segmental gene duplication (eight genes) were found in tomato as shown in ).
Table 1. The characteristic features of tomato SlPHB proteins in tomato genome
Figure 1. Chromosomal location and synteny of PHB genes in the tomato genome. (a) The chromosome location of tomato SLPHB genes. The scale of chromosomes is in megabases (MB). (b) Circos plot presenting gene segmental duplication events of PHB genes. Segmental duplication pairs are indicated with different color lines.
3.2. Phylogeny, Strong Purifying Selection, and Conserved Motif Analysis of SlPHB Proteins
To unveil the phylogenetic relationship of tomato SlPHB proteins with PHBs from other plant species such as Arabidopsis, rice, maize, and soybean, an unrooted neighbor-joining phylogenetic tree was generated. It was observed that all PHB proteins were divided into four major clades (II, III, IV, and V). The subclade of each group contains 7–15 members from different species. SlPHBs were found in all clades such as five SlPHBs in group IV (2 in IV B and 3 in IV A). Similarly, three in subclade V B and single in V A subclade of major clade V. Moreover, clade III has four, and clade II contained two tomato SlPHBs. Similar trends of PHB distribution were observed for other species (). Furthermore, three sister pairs of SlPHB genes were detected in the phylogenetic tree such as SlPHB14/SlPHB15 in clade IV A, SlPHB2/SlPHB3 in subclade V B of major clade V, and SlPHB11/SlPHB6 in clade III. It was observed that SlPHBs localized in chloroplast were clustered together as shown in ).
Figure 2. The phylogeny of the PHB proteins. An unrooted neighbor-joining phylogenetic tree of PHB proteins from Arabidopsis, rice, maize, soybean, and tomato was generated in the MEGA program with a bootstrap value set as 1000 replicates. The tree was clustered into various clades and subclades. The black dots represent tomato SlPHB proteins.
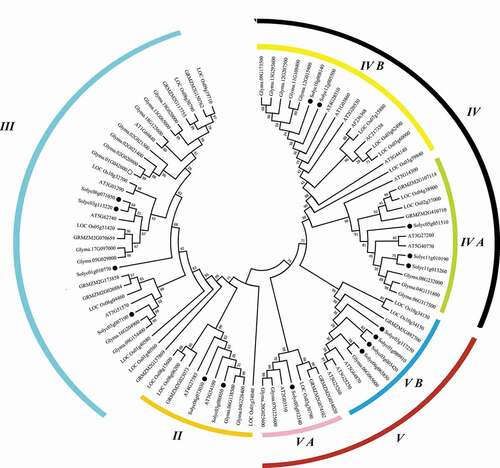
Figure 3. Phylogeny, gene exon/intron distribution, and conserved motif analysis of 16 tomato SlPHB genes. (a) An unrooted neighbor-joining phylogenetic tree of PHB proteins with bootstrap set at 1000 replicates and clustered into different clades and subclades. (b) Tomato SlPHB gene intron and exon distribution. The scale at the bottom is corresponding to gene size in kb. (c) The putative conserved motifs in 16 tomato PHB proteins identified using the MEME suite. A total of ten motifs (1 to 10) were identified and each color of the box is corresponding to a motif. The scale at the bottom represents the protein size in kb.
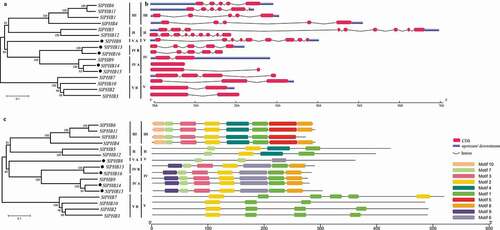
A comparison of the gene structure of each tomato PHB revealed a diverse structure. The number of intron and exon ranged from one to nine exons and zero to eight introns. The exon/intron pattern was similar in different clades and subclades. For example, five exon and four introns were found in clade III, nine exons and eight introns in clade II, and clade V. Similarly, five exons in subclade IV B and two in IV A clade. Besides, the length and positions of exons were also highly similar in clades and subclade ). We identified ten conserved motifs in SlPHBs using the MEME server. It was observed that the motifs pattern was also similar within clades ()). For instance, motif 1 and motif 2 found in clade V B; motif 1, motif 2, motif 3, motif 7, motif 8, and motif 9 in clade IV. SlPHBs in clade III contained all motifs except motif 9. To explore the fate of divergence of these genes in the tomato genome, the Ka/Ks values were estimated for three duplicate SlPHB gene pairs. The Ks was used in estimating the divergence time of each SlPHB gene pairs (). Our results showed that the Ka/Ks ratio of duplicated genes pairs was more than 0.04. This suggesting that the purifying selection pressure was a major factor that occurs during the evolution, function divergence was limited after duplication and was estimated to occur between 36.8 and 101.55 million years ago (Mya).
Table 2. The Ka/Ks of tomato SlPHB paralogs
3.3. Bioinformatics Analysis of SlPHB Promoter Sequences
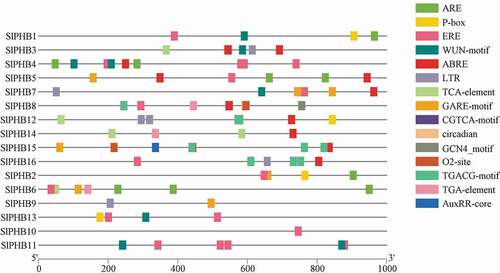
The cis-acting elements of potential tomato SlPHB genes were predicted by searching a 1000 bp region from the transcriptional activation site (ATG) of each gene against the PlantCARE database. As shown in , several putative cis-regulatory sequences were identified in SlPHB genes. For an instance, four different kinds of development-related cis-regulatory elements such as circadian control (circadian), meristem development (CAT-box), endosperm development (GCN4_motif), and zein metabolism regulation (O2-site) were predicted in the promoter region of some of the SlPHBs, suggesting that these genes may play roles in organ/tissue-specific development and growth. Moreover, a various stress-responsive element such as the MYB binding site involved in drought-inducibility (MBS), WRKY binding site involved in abiotic stress and defense response (W-box), anaerobic induction element (ARE), defense- and stress-responsive element (TC-rich repeats), low-temperature-responsive element (LTR), wound-responsive element (WUN-motif), and element for maximal elicitor-mediated activation (AT-rich sequence) were also detected. The promoters of tomato SlPHB genes possessed cis-regulatory sequences related to ethylene (ERE), suggesting that these genes may involve in ethylene responses (). In addition, various hormone-related responsive elements related to gibberellin (GARE-motif), methyl jasmonate (MeJA, CGTCA-motif), abscisic acid (ABRE), and salicylic acid (TCA-element) were also detected, implying that these genes may respond to phytohormone as well (). The promoters of tomato SlPHB genes possessed cis-regulatory sequences related to ethylene (ERE), suggesting that these genes may involve in ethylene responses.
Figure 4. The putative cis-regulatory sequences were identified in 16 tomato SlPHB genes by submitting their corresponding promoter sequences to the PlantCARE database. Different cis-regulatory elements circadian control (circadian), meristem development (CAT-box), endosperm development (GCN4_motif), zein metabolism regulation (O2-site), MYB binding site involved in drought-inducibility (MBS), WRKY binding site involved in abiotic stress and defense response (W-box), anaerobic induction element (ARE), defense- and stress-responsive element (TC-rich repeats), low-temperature-responsive element (LTR), wound-responsive element (WUN-motif), element for maximal elicitor-mediated activation (AT-rich sequence) ethylene (ERE), gibberellin (GARE-motif), methyl jasmonate (MeJA, CGTCA-motif), abscisic acid (ABRE), and salicylic acid (TCA-element) and son on was detected.
3.4. miRNAs Targeting the PHB Family Members of the Tomato
To find out miRNAs targeting the SlPHBs of tomato, the sequences were subjected to the miRNA database. The psRNATarget predicted that four SlPHBs gene family members were targeted by conserved miRNAs belongs to different miRNAs gene families each. SlPHB7 was targeted by the sly-miRNA869 family and sly-miRNA4239 cause the cleavage of SlPHB3. A single member from sly-miR396 and sly-miR397 family member target to cleavage of SlPHB15 and SlPHB13 gene, respectively (Table S1).
3.5. Expression Analysis of SlPHB Genes in Different Plant Parts
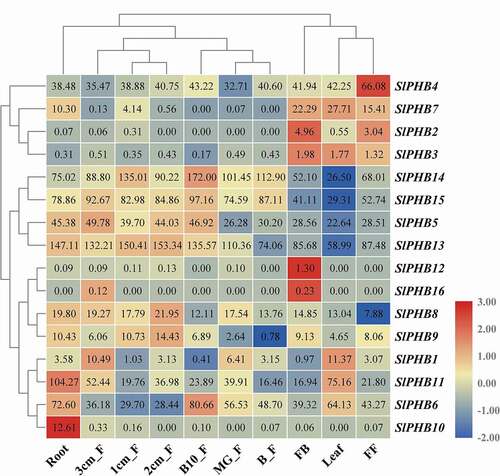
To understand the role of putative SlPHBs in tomato plant growth and development, the expression profile analysis of SlPHBs in various plant parts was evaluated. The SlPHBs exhibited a diverse expression pattern among various plant parts. It was found that two SlPHBs were expressed in leaves, and root tissues. One SlPHB gene had high expression levels in fully opened flower and three expressed in flower at bud condition. It was observed that the number of genes was expressed in fruit at different development stages with more and less expression levels. For example, SlPHB1 in 3 cm fruit, SlPHB6 in ten days fruit breaker, SlPHB8, and SlPHB9 in 2 cm fruit. However, SlPHB5, SlPHB14, and SlPHB15 exhibited increasing expression during fruit development and ripening (2 cm fruit till ten days breaker fruit) (). The results showed that tomato SlPHB genes play an important role in the growth and development of specific plant parts or tissues.
Figure 5. The endogenous expression profile of 16 tomato SlPHB genes in various plant parts including root, leaves, FB (flower bud), FF (fully opened flower), 1/2/3 cm fruit, mature green fruit (MG_F), breaker fruit (B_F), and 10 days breaker fruit (B10_F). A log2 transformed heatmap was generated using heatmapper program. Blue, white, and red color is corresponding to low, moderate, and high expressions. The genes were clustered by applying the Euclidean method.
3.6. Expression Profile of Tomato SlPHB in Response to Salinity and Drought Stress
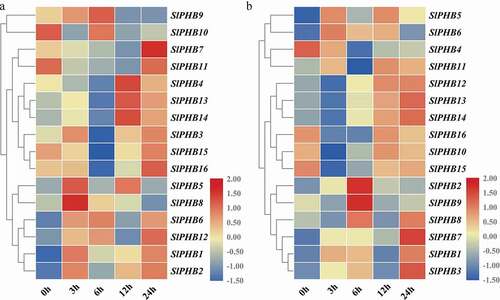
To further investigate the role of SlPHB in tomato against abiotic stresses, the expression profile of SlPHB in response to salt and drought was analyzed at various time points. It was observed that under salt stress, the transcript abundance of SlPHB9 was sharply increased at 3 h and peak at 6 h time point and subsequently declined at 12 h and 24 h time points. SlPHB7 and SlPHB11 had maximum transcript levels at 24 h while, SlPHB4, SlPHB13, and SlPHB14 exhibited transcript abundance at 12 h time point. SlPHB5 and SlPHB8 induced only at 3 h after treatment but SlPHB10 induced at 6 h time point ()). Under drought conditions, the majority of genes were expressed at the late time point (12 h and 24 h). SlPHB2 and SlPHB9 induced only at 6 h after treatment ()). In comparison, SlPHB5, SlPHB13, SlPHB14, SlPHB15, SlPHB9, and SlPHB7 showed similar trends of expression under both drought and salinity stresses but SlPHB4, SlPHB2, and SlPHB8 exhibited opposite trends under both stresses (). These results suggest that tomato SlPHB genes may play a key role in regulating abiotic stress responses.
Figure 6. Abiotic stress-induced expression profile of SlPHB genes. (a) salt (b) drought (PEG) induced expression profile at 0 h, 3 h, 6 h, 12 h, and 24 h time points. A log2 transformed heatmap was generated using heatmapper program. Blue, white, and red color is corresponding to low, moderate, and high expressions. The genes were clustered by applying the Euclidean method.
3.7. Phytohormone Induced Expression Profile Analysis of SlPHBs in Tomato
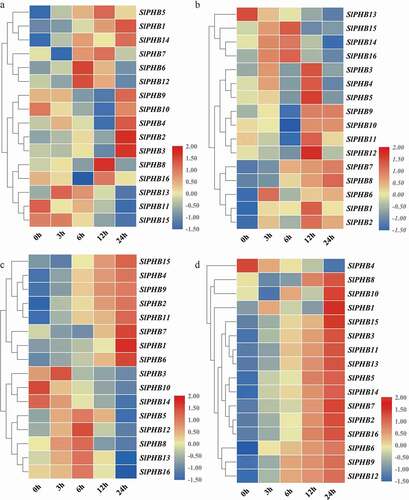
To check the effectiveness of exogenous phytohormone application, the expression profile of tomato SlPHB under various hormones such as abscisic acid, gibberellin, auxin, and methyl jasmonate was examined. For ABA treatment, SlPHB13 and SlPHB15 were induced at 3 h time points while SlPHB6 and SlPHB12 were upregulated at 6 h after application with decreased expression in later time points. SlPHB11 expression was downregulated upon treatment with ABA but SlPHB8 and SlPHB16 were induced only at 12 h after treatment. Moreover, SlPHB9, SlPHB10, SlPHB4, SlPHB12, and SlPHB3 was upregulated at 24-time points ()). SlPHB5, SlPHB14, and SlPHB16 transcript levels were sharply induced at 3 h interval and reach a maximum at 6 h time point but decreased in subsequent time intervals to GA3. SlPHB3, SlPHB4, SlPHB5, SlPHB11, SlPHB12, SlPHB1, and SlPHB2 were induced with maximum transcript levels at 12 h after exposure to GA3 ()). The transcript abundance of SlPHB7 and SlPHB8 was increased temporally but SlPHB13 expression was downregulated upon treatment with GA3. For auxin, SlPHB10 and SlPHB14 genes were downregulated after application but SlPHB3 showed maximum transcript accumulation at 3 h point interval. SlPHB5, SlPHB12, SlPHB8, SlPHB13, and SlPHB16 was upregulated with time and reached maximum expression at 6 h after treatment while, SlPHB15, SlPHB4, SlPHB9, SlPHB2, and SlPHB11 expression levels were upregulated across 6 h to 24 h time points and showed maximum expression at 24 h interval ()). The SlPHBs exhibited a unique expression profile upon exposure to MeJA. It was observed that all the genes were upregulated temporally across all time intervals and have high transcript accumulation at 24 h time point except for SlPHB4 ()). The data suggest that tomato SlPHB genes may play various important roles in cross-talk with different kinds of hormones signaling.
Figure 7. Phytohormone induced expression profile of SlPHB genes. (a) abscisic acid (ABA), (b) gibberellin (GA3), (c) auxin (IAA), (d) methyl jasmonate (MeJA) induced expression profile at 0 h, 3 h, 6 h, 12 h, and 24 h time points. A log2 transformed heatmap was generated using heatmapper program. Blue, white, and red color is corresponding to low, moderate, and high expressions. The genes were clustered by applying the Euclidean method.
3.8. Subcellular Localization Assay
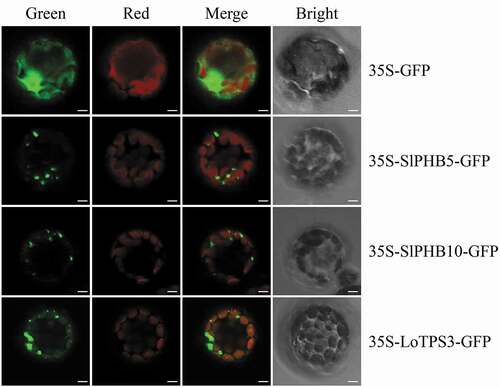
The amino acid sequence of SlPHB5 and SlPHB10 was submitted to the WoLFPSORT (https://wolfpsort.hgc.jp/) to predict subcellular localization. The predicted results showed that both SlPHB proteins were expressed in the mitochondria. To experimentally verify, full-length sequences of candidate SlPHB5 and SlPHB10 were fused to a GFP reporter gene and transferred to Arabidopsis protoplast (). Subcellular localization experiment results revealed that both proteins were localized in the mitochondria as predicted. LoTPS3 protein from Lilium Siberia was used as a positive control.Citation51 Scale bar 5 µm.
Figure 8. Subcellular localization images of SlPHB5 and SlPHB10 in Arabidopsis protoplasts. The full-length sequences of SlPHB5 and SlPHB10 were fused in the pro35S vector to generate p35S-SlPHBs/GFP constructs. The images were observed via confocal laser scanning microscopy. The LoTPS3 form Lilium ‘Siberia’ was used as red mitochondrial control for SlPHB5 and SlPHB10. The green, red, merged and BF represents the GFP fluorescence, chlorophyll autofluorescence, combined chlorophyll autofluorescence, and GFP fluorescence and bright field respectively. Scale bars 5 µm.
4. Discussion
PHB, a highly conserved multigene family has been identified in many organisms from humans to various plant species playing essential roles in various aspects of growth and development. In plants, the PHB gene family has been reported from Arabidopsis (17), rice (19),Citation31 Glycine max (24),Citation32 and Zea mays with 16.Citation17 However, no genome-wide identification of the PHB gene family has been reported in the tomato genome. In this study, a total of 16 PHB genes were identified in the tomato genome (). The tomato genome size (960Mb) is 7.68 folds of the Arabidopsis genome (125 Mb), 2.46 folds of rice (389 Mb) but 2.3 folds less of maize (2300 Mb) and 1.14 folds less than soybean (1100 Mb) genome. However, the number putative PHBs in the tomato genome even lower than Arabidopsis and riceCitation31 but equal to reported in maize.Citation17
Gene duplication either segmental or tandem plays an important role in the expansion of the genome. The expansion of the PHB gene family in Arabidopsis, rice, and soybean was caused by segmental duplication while tandem duplication was another cause of an increasing number of PHB genes in Arabidopsis but was absent in tomato. This implying that gene duplication of the PHB gene family in tomato was different from Arabidopsis. We have analyzed Ka/Ks values of three pairs of SlPHB gene duplication and found that tomato PHB genes undergo purifying selection ().
The PHB genes from fungi and mammals including humans were clustered in five phylogenetic clades. However, like Arabidopsis, rice,Citation31 Glycine max,Citation32 and Zea mays,Citation17 tomato SlPHBs were also clustered in four clades. The genes sharing clades and subclades displayed a similar gene structure and conserved motifs patterns. PHB genes are involved in various aspects of plant growth and development. In this study, cis-regulatory sequences were predicted. It was observed that tomato SlPHB genes contained various development, abiotic stress, and phytohormone responsive elements in their promoter regions (). It has been well documented that PHB genes involved in leaf yellowing, hormone signal transduction pathways, and abiotic stress responses. For example, Arabidopsis AtPHB3/4 causes proliferation of root and shoot tissues.Citation24 Similarly, petunia PHBs, tobacco NbPHB1/2 promote leaf senescence.Citation25,Citation26 In this study, the expression profile of SLPHBs in various parts of tomato plant was also investigated. Tomato SlPHB genes showed diverse expression patterns in different parts such as SlPHB4 and SlPHB10 was expressed in flower and root tissues, respectively. Two genes (SlPHB8, SlPHB9) were highly expressed in 2 cm fruit while SlPHB5, SlPHB14, and SlPHB15 showed increasing expression pattern with the fruit development stages (). These results suggest the crucial role of SlPHB genes in development of these organs in tomato plant.
In this study, cis-regulatory elements involved in diverse signaling pathways were identified. Most PHBs contain cis-regulatory elements involved in ABA, GA, JA, and ethylene. In addition, cis-elements involved in abiotic stresses, such as MBS (MYB binding site involved in drought-inducibility), LTR (low-temperature responsiveness element), HSE (heat stress responsiveness element), were also observed in the promoter regions of SlPHB genes (). In Glycine max, most of PHBs contained numerous hormone-responsive, development and stress-related cis-regulatory elements in the GmPHB promoters.Citation32 It was observed that the expression of SlPHB genes was altered under these stresses. For salt treatment, SlPHB5, SlPHB8, SlPHB9, and SlPHB10 were upregulated at early time points (3 h and 6 h) while, SlPHB7, SlPHB11, SlPHB4, SlPHB13, SlPHB14, and SlPHB12 were induced at 12 h and 24 h after treatment ()). Similar response was observed in Arabidopsis, where PHBs were involved in abiotic stimulus and phytohormones functioning.Citation16,Citation29 SlPHB2 and SlPHB9 genes were induced under drought at 6 h time point but SlPHB4 was downregulated ()). SlPHB1, SlPHB14, SlPHB9, SlPHB10, SlPHB4, and SlPHB3 were upregulated after 24 h exposure to ABA but SlPHB11 and SlPHB15 were downregulated upon exposure ()). Moreover, SlPHB13, SlPHB14, SlPHB15, and SlPHB16 were suppressed in late intervals of GA3 exposure but the rest of the genes were upregulated ()). SlPHB10 and SlPHB14 were downregulated after auxin application but, SlPHB3 was induced after 3 h of treatment. SlPHB7, SlPHB1, and SlPHB6 exhibited maximum expression at a 24 h time point ()). For MeJA treatment, all the genes were induced sharply along with all the time points and peaked at 24 h after treatment except for SlPHB4, which was suppressed upon exposure to MeJA ()). Likewise, Atphb3 mutant was highly responsive to ethylene in etiolated seedlings. One Arabidopsis prohibitin (At5g64870) was down-regulated under some hormones (GA, MeJA and ABA), while highly upregulated under salt, drought and cold treatment.Citation31 In Capsicum annum, hypersensitive-induced reaction (HIR) proteins (PHB encoding proteins), such as CaHIR1, maize ZmHIR1-3, barley HvHIR1/3 and AtHIR1-3 were induced under abiotic stresses.Citation57–59 Our findings are in line with previous studies that PHB genes showed differential expression pattern under different development stages as well as under different stimulus.Citation5,Citation25,Citation26,Citation30,Citation31 The above-mentioned findings highlighted the potential diverse role of PHB genes.
5. Conclusion
In short, this study provides knowledge about the PHB gene family in the tomato genome. All the identified SlPHBs were clustered in four clades according to the phylogenetic tree. The gene structure and conserved motifs distribution patterns in each clade validated the phylogenetic classification of tomato SlPHBs. Cis-regulatory sequences prediction in combination with complex regulation of tomato PHB genes family expression against salinity, drought, and various phytohormones such as ABA, IAA, GA, and MeJA provide a foundation for further functional characterization of these genes in tomato and other plant species.
Disclosure Of Potential Conflicts Of Interest
The author(s) declare neither financial nor non-financial conflict of interest.
Author Contributions
All authors contributed to the study conception and design.
Conceptualization: Yanguo Ke, Feiyang Huang and Lei Y; Methodology: Zhijiang Wang, Yan Ding and Muhammad Waseem; Formal analysis and investigation: Muhammad Waseem, Lei Yu, Xianjie Cai, Xianwen Ye, Umair Ashraf and Yanguo Ke; Writing - original draft preparation: Yanguo Ke, Farhat Abbas, Muhammad Waseem and Feiyang Huang; Writing - review and editing: Yanguo Ke, Xialong Chen, Umair Ashraf and Farhat Abbas; Funding acquisition: Xianjie Cai, Feiyang Huang and Yanguo ke; Resources: Yanguo Ke, Xiaolong Chen and Feiyang Huang.
Supplemental Material
Download MS Word (17.1 KB)Supplemental Material
Download MS Word (14.7 KB)Supplementary Material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Morrow IC, Parton RG. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic (Copenhagen, Denmark). 2005;6:725–40. doi:https://doi.org/10.1111/j.1600-0854.2005.00318.x.
- Lee JH, Nguyen KH, Mishra S, Nyomba BL. Prohibitin is expressed in pancreatic beta-cells and protects against oxidative and proapoptotic effects of ethanol. Febs J. 2010;277:488–500. doi:https://doi.org/10.1111/j.1742-4658.2009.07505.x.
- Sievers C, Billig G, Gottschalk K, Rudel T. Prohibitins are required for cancer cell proliferation and adhesion. PloS One. 2010;5:e12735. doi:https://doi.org/10.1371/journal.pone.0012735.
- Takata H, Matsunaga S, Morimoto A, Ma N, Kurihara D, Ono-Maniwa R, Nakagawa M, Azuma T, Uchiyama S, Fukui K, et al. PHB2 protects sister-chromatid cohesion in mitosis. Curr Biol. 2007;17:1356–61. doi:https://doi.org/10.1016/j.cub.2007.07.009.
- Van Aken O, Whelan J, Van Breusegem F. Prohibitins: mitochondrial partners in development and stress response. Trends Plant Sci. 2010;15:275–82. doi:https://doi.org/10.1016/j.tplants.2010.02.002.
- Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 2007;17:394–402. doi:https://doi.org/10.1016/j.tcb.2007.06.005.
- Tavernarakis N, Driscoll M, Kyrpides NC. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem Sci. 1999;24:425–27. doi:https://doi.org/10.1016/S0968-0004(99)01467-X.
- Lee S-J, Choi D, Rhim H, Choo H-J, Ko Y-G, Kim CG, Kang S. PHB2 interacts with RNF2 and represses CP2c-stimulated transcription. Mol Cell Biochem. 2008;319:69–77. doi:https://doi.org/10.1007/s11010-008-9878-2.
- Wang S, Zhang B, Faller DV. Prohibitin requires Brg-1 and Brm for the repression of E2F and cell growth. Embo J. 2002;21:3019–28. doi:https://doi.org/10.1093/emboj/cdf302.
- Rivera-Milla E, Stuermer C, Málaga-Trillo E. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cellular and Molecular Life Sciences CMLS. 2006;63:343–57. doi:https://doi.org/10.1007/s00018-005-5434-3.
- Wang S, Nath N, Fusaro G, Chellappan S. Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol Cell Biol. 1999;19:7447–60. doi:https://doi.org/10.1128/MCB.19.11.7447.
- Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853–61. doi:https://doi.org/10.1074/jbc.M305171200.
- Joshi B, Rastogi S, Morris M, Carastro L, DeCook C, Seto E, Chellappan S. Differential regulation of human YY1 and caspase 7 promoters by prohibitin through E2F1 and p53 binding sites. Biochem J. 2007;401:155–66. doi:https://doi.org/10.1042/BJ20060364.
- Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat Med. 2004;10:625–32. doi:https://doi.org/10.1038/nm1048.
- Sharma A, Qadri A. Vi polysaccharide of Salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc Natl Acad Sci. 2004;101:17492–97. doi:https://doi.org/10.1073/pnas.0407536101.
- De Diego JG, David Rodriguez F, Rodriguez Lorenzo JL, Cervantes E. The prohibitin genes in Arabidopsis thaliana: expression in seeds, hormonal regulation and possible role in cell cycle control during seed germination. J Plant Physiol. 2007;164:371–73. doi:https://doi.org/10.1016/j.jplph.2006.05.002.
- Wen X, Niu T, Kong X. In silico analysis of PHB gene family in maize. Plant Growth Regul. 2014;73:181–91. doi:https://doi.org/10.1007/s10725-013-9879-3.
- Nijtmans LG, de Jong L, Sanz MA, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane‐bound chaperone for the stabilization of mitochondrial proteins. Embo J. 2000;19:2444–51. doi:https://doi.org/10.1093/emboj/19.11.2444.
- Chowdhury I, Thompson WE, Thomas K. Prohibitins role in cellular survival through Ras‐Raf‐MEK‐ERK pathway. J Cell Physiol. 2014;229:998–1004. doi:https://doi.org/10.1002/jcp.24531.
- Artal-Sanz M, Tsang WY, Willems EM, Grivell LA, Lemire BD, van der Spek H, Nijtmans LGJ. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in caenorhabditis elegans. J Biol Chem. 2003;278:32091–99. doi:https://doi.org/10.1074/jbc.M304877200.
- Merkwirth C, Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim Biophys Acta. 2009;1793:27–32. doi:https://doi.org/10.1016/j.bbamcr.2008.05.013.
- Snedden WA, Fromm H. Characterization of the plant homologue of prohibitin, a gene associated with antiproliferative activity in mammalian cells. Plant Mol Biol. 1997;33:753–56. doi:https://doi.org/10.1023/A:1005737026289.
- Wen T-J, Hochholdinger F, Sauer M, Bruce W, Schnable PS. The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol. 2005;138:1637. doi:https://doi.org/10.1104/pp.105.062174.
- Van Aken O, Pecenkova T, van de Cotte B, De Rycke R, Eeckhout D, Fromm H, De Jaeger G, Witters E, Beemster GTS, Inzé D, et al. Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 2007;52:850–64. doi:https://doi.org/10.1111/j.1365-313X.2007.03276.x.
- Chen JC, Jiang CZ, Reid MS. Silencing a prohibitin alters plant development and senescence. Plant J. 2005;44:16–24. doi:https://doi.org/10.1111/j.1365-313X.2005.02505.x.
- Ahn CS, Lee JH, Reum Hwang A, Kim WT, Pai HS. Prohibitin is involved in mitochondrial biogenesis in plants. Plant J. 2006;46:658–67. doi:https://doi.org/10.1111/j.1365-313X.2006.02726.x.
- Artal-Sanz M, Tavernarakis N. Prohibitin couples diapause signalling to mitochondrial metabolism during ageing in C. Elegans Nature. 2009;461:793–97. doi:https://doi.org/10.1038/nature08466.
- Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT, von Kleist-retzow J-C, Waisman A, Westermann B, Langer T, et al. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22:476–88. doi:https://doi.org/10.1101/gad.460708.
- Christians MJ, Larsen PB. Mutational loss of the prohibitin AtPHB3 results in an extreme constitutive ethylene response phenotype coupled with partial loss of ethylene-inducible gene expression in Arabidopsis seedlings. J Exp Bot. 2007;58:2237–48. doi:https://doi.org/10.1093/jxb/erm086.
- Wang Y, Ries A, Wu K, Yang A, Crawford NM. The Arabidopsis prohibitin gene PHB3 functions in nitric oxidemediated responses and in hydrogen peroxide-induced nitric oxide accumulation. Plant Cell. 2010;22:249. doi:https://doi.org/10.1105/tpc.109.072066.
- Di C, Xu W, Su Z, Yuan JS. Comparative genome analysis of PHB gene family reveals deep evolutionary origins and diverse gene function. BMC Bioinform. 2010;11(Suppl 6):S22. doi:https://doi.org/10.1186/1471-2105-11-S6-S22.
- Song M, Peng X, Du C, Lei L, Zhang T, Xiang Y. Genome-wide analysis of the PHB gene family in Glycine max (L.). Merr Genes Genomics. 2017;39:1095–106. doi:https://doi.org/10.1007/s13258-017-0580-1.
- Reiser L, Subramaniam S, Li D, Huala E. Using the arabidopsis information resource (TAIR) to find information about arabidopsis genes. Curr Protoco Bioinf. 2017;60:1.11.1–1.45. doi:https://doi.org/10.1002/cpbi.36.
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, et al. The SOL genomics network: a comparative resource for Solanaceae biology and beyond. Plant Physiol. 2005;138:1310–17. doi:https://doi.org/10.1104/pp.105.060707.
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci. 1998;95:5857. doi:https://doi.org/10.1073/pnas.95.11.5857.
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–d3. doi:https://doi.org/10.1093/nar/gkw1129.
- Stothard P. The sequence manipulation suite: javaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000;28:1102, 4. doi:https://doi.org/10.2144/00286ir01.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;539. doi:https://doi.org/10.1038/msb.2011.75
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi:https://doi.org/10.1093/oxfordjournals.molbev.a040454.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–49. doi:https://doi.org/10.1093/molbev/msy096.
- Comeron JM. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–64. doi:https://doi.org/10.1093/bioinformatics/15.9.763.
- He Y, Liu X, Ye L, Pan C, Chen L, Zou T, Lu G. Genome-wide identification and expression analysis of two-component system genes in tomato. Int J Mol Sci. 2016;17:1204. doi:https://doi.org/10.3390/ijms17081204.
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–7. doi:https://doi.org/10.1093/nar/gkm259.
- Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–9. doi:https://doi.org/10.1093/nar/gkr319.
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi:https://doi.org/10.1093/nar/gkt1181.
- Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics (Oxford, England). 2015;31:1296–97. doi:https://doi.org/10.1093/bioinformatics/btu817.
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Intl Conf Intell Syst Mol Biol. 1994;2:28–36.
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–27. doi:https://doi.org/10.1093/nar/30.1.325.
- Waseem M, Ahmad F. The phosphoenolpyruvate carboxylase gene family identification and expression analysis under abiotic and phytohormone stresses in Solanum lycopersicum L. Gene. 2019;690:11–20. doi:https://doi.org/10.1016/j.gene.2018.12.033.
- Waseem M, Ahmad F, Habib S, Gao Y, Li Z. Genome-wide identification of FK506-binding domain protein gene family, its characterization, and expression analysis in tomato (Solanum lycopersicum L.). Gene. 2018;678:143–54. doi:https://doi.org/10.1016/j.gene.2018.08.021.
- Abbas F, Ke Y, Yu R, Fan Y. Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium ‘Siberia’. Planta. 2019;249:71–93. doi:https://doi.org/10.1007/s00425-018-3006-7.
- Abbas F, Ke Y, Zhou Y, Ashraf U, Li X, Yu Y, Yue Y, Ahmad KW, Yu R, Fan Y, et al. Molecular cloning, characterization and expression analysis of LoTPS2 and LoTPS4 involved in floral scent formation in oriental hybrid Lilium variety ‘Siberia’. Phytochemistry. 2020;173:112294. doi:https://doi.org/10.1016/j.phytochem.2020.112294.
- Ke Y, Abbas F, Zhou Y, Yu R, Yue Y, Li X, Yu Y, Fan Y. Genome-wide analysis and characterization of the Aux/IAA Family genes related to floral scent formation in hedychium coronarium. Int J Mol Sci. 2019;20:3235. doi:https://doi.org/10.3390/ijms20133235.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–08. doi:https://doi.org/10.1006/meth.2001.1262.
- Abbas F, Ke Y, Zhou Y, Waseem M, Yu Y, Ashraf U, Li X, Wang C, Yue Y, Yu R, et al. Cloning, functional characterization and expression analysis of LoTPS5 from Lilium ‘Siberia’. Gene. 2020;756:144921. doi:https://doi.org/10.1016/j.gene.2020.144921.
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–72. doi:https://doi.org/10.1038/nprot.2007.199.
- Nadimpalli R, Yalpani N, Johal GS, Simmons CR. Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J Biol Chem. 2000;275:29579–86. doi:https://doi.org/10.1074/jbc.M002339200.
- Yu X-M, Yu X-D, Qu Z-P, Huang X-J, Guo J, Han Q-M, Zhao J, Huang -L-L, Kang Z-S. Cloning of a putative hypersensitive induced reaction gene from wheat infected by stripe rust fungus. Gene. 2008;407:193–98. doi:https://doi.org/10.1016/j.gene.2007.10.010.
- Qi Y, Tsuda K, Nguyen LV, Wang X, Lin J, Murphy AS, Glazebrook J, Thordal-Christensen H, Katagiri F. Physical association of arabidopsis hypersensitive induced reaction proteins (HIRs) with the immune receptor RPS2. J Biol Chem. 2011;286:31297–307. doi:https://doi.org/10.1074/jbc.M110.211615.
