ABSTRACT
The comprehensiveness of the allergen database used to bioinformatically compare a novel food protein with known allergens is critical to the ability to assess the allergenic risk of newly expressed proteins in genetically engineered crops. The strength of the relationship between a candidate GE protein’s amino acid sequence and that of known allergens is used to predict cross-reactive risk. The number of truly novel allergen sequences added annually to the COMPARE database reflects on the comprehensiveness of our knowledge of allergen amino acid sequence diversity. Here, we investigated the most recent five years of updates to the COMPARE allergen database for truly novel entries. Results indicate that few truly novel sequences are added each year, suggesting that the database and our knowledge of allergen sequence diversity is currently quite comprehensive, and that current in silico prediction of allergenic risk for novel food proteins is robust.
Introduction
Newly expressed proteins in genetically engineered (GE) crops are evaluated for allergenic risk. One powerful tool for evaluating this risk involves comparing the amino acid sequence of the GE protein with that of known allergens using bioinformatic tools.Citation1 This evaluation helps predict if the GE protein might be sufficiently similar to a known allergen to confer cross reactivity in previously sensitized individuals, or if the GE protein might possess similar sensitization properties compared with known allergens by virtue of its similar structure. The power of this approach rests heavily on the comprehensiveness of a database of known allergen sequences. One way to evaluate the comprehensiveness of an allergen database is to investigate the number of sequences added each year that are unrelated to those already in the database, referred to here as novel sequences.
The bioinformatic tools used to evaluate the potential cross reactivity of a GE protein with that of known allergens have been found to be very conservative for detecting true allergen cross reactivity.Citation2–4 From a regulatory perspective, sequences sharing >35% amino acid identity over a sliding window of ≥80 amino acids are considered a cross-reactive risk for a GE protein. However, the wider scientific literature suggests that statistical thresholds for amino acid similarity (E-values) across the entire sequence are equally conservative, yet have a much lower false-positive rate (fewer predictions of allergenicity for sequences with a history of no allergenicity).Citation5,Citation6 Together, these approaches can help determine the cross-reactivity risk for a GE protein.
The COMPARE allergen database is currently the most widely used by developers of GE crops for the bioinformatic investigation of newly expressed proteins in GE crops.Citation7 This database was initiated in 2017 based largely on the existing version of the Allergen Online (AOL) database.Citation8 Curated updates have been subsequently made to the COMPARE database annually since that time, allowing the series of five additional groups of sequences added in each update (2018 to 2022) to be investigated for novelty compared with entries in the previous versions of the database (https://comparedatabase.org/). Because the annual updates to the COMPARE database are based on the most recent publicly available scientific information, this analysis should help determine the comprehensiveness of the previous database and any trends observed over time in our knowledge of allergen amino acid sequence diversity. Since many of the sequences in the database are members of structurally related groups of allergens,Citation3 it is expected that most new sequences added to the database are not truly novel, but are instead members of allergen families already represented in the database, and thus, the majority of newly added sequences are expected to be largely redundant in their utility for detecting relationships with candidate GE proteins based on current bioinformatic approaches. As the annual addition rate of clinically relevant allergens with truly novel sequences approaches zero, the in-silico prediction of allergenic risk becomes very robust.
Here, we evaluate the bioinformatic novelty of the new sequence entries in each annual version of the COMPARE database and discuss how this informs on the comprehensiveness of the database. Further, we discuss the implications of these results toward elucidating the power of bioinformatic approaches for predicting the allergenic risk of GE proteins.
Methods and Materials
To assess the novelty of amino acid sequences newly added to the COMPARE allergen database in each of the five years from 2018 to 2022, an in silico bioinformatic analysis was conducted. For each year, the previous year’s version of the database was queried using each of the newly added sequences in the present year. For each newly added sequence, it was determined whether a sliding window FASTA (version 36, using default settings) alignment of 80 amino acids shared >35% identity with a member in the previous year’s version of the database. An adjustment was made for alignments of less than 80 amino acids in which ≥29 amino acid identity was considered passing the sliding window criteria (29/80 > 35%). Because this threshold is known to produce many false detections of cross-reactivity, the E-value for the alignment with the best matched sequence (lowest E-value) derived from full-length FASTA was also used to determine the significance of the best alignment. An E-value above E-7 was considered too poor to reliably establish lack of novelty for the new sequence.Citation1
In addition to full-length protein sequences in the COMPARE database, there are also partial (incomplete) amino acid sequences, which are included when the full-length sequence is unknown. This is a complicating factor for the aforementioned analysis of the novelty of newly added allergen sequences. First, partial sequences may not represent allergenic risk since they may not contain the structural features necessary to cause allergy (e.g., may contain no IgE epitopes). Second, sequences <29 amino acids long cannot mathematically produce an alignment of >35% identity over 80 amino acids (28/80 = 35%). Finally, short sequences with high identity alignments are less likely to produce highly significant (low E-value) alignments. For these reasons, new short-length sequences (<100 amino acids) not meeting the sliding window and E-value criteria were examined for their partial or full-length status and for their top sequence alignment (lowest E-value using whole available sequence) with the previous year’s database. Sequences >100 amino acids in length were generally assumed to be full-length sequences even though a subset of these sequences are almost certainly only partial sequences.
Results
Initial Categorization of Novelty for Newly Added Allergen Sequences
In the sequential five years from 2018 to 2022, 67, 50, 188, 104, and 118 new sequences, respectively, were added to the COMPARE allergen database. Of these sequences, 48, 33, 55, 35, and 45 sequences were initially categorized as similar to those already in the database because they share >35% identity over a sliding window alignment of ≥80 amino acids and also have a highly significant full-sequence alignment (E-value <E-7). The remaining sequences, 19, 17, 133, 69, and 73, in each of the five consecutive years, were evaluated for their status as partial or full-length sequences, and each was evaluated for novelty based on the E-value for the best (lowest E-value) alignment with a sequence in the previous version of the database. The number of partial sequences in each consecutive year was found to be 2, 4, 121, 64, and 72, respectively.
Value of Partial Allergen Sequences
The presence of newly added partial sequences in the allergen database complicates the task of determining the comprehensiveness of the database because partial sequences may contain no structural features that are critical to the full-length protein’s allergenicity (e.g., IgE epitopes), making bioinformatic matches to a GE protein potentially irrelevant to allergenic risk. However, the bioinformatic relationship of the partial sequence to allergen sequences in the previous version of the database still provides some information on the relevance of the partial novel sequences to the comprehensiveness of the database. Most regulatory agencies consider short identical contiguous matches of ≥8 amino acids between a GE protein and an allergen to be a cross-reactive risk, even though such matches alone have been shown to be of negligible value in predicting allergen cross reactivity.Citation9,Citation10 For these reasons, partial sequences not meeting the aforementioned >35%-identity sliding-window and E-value criteria were further investigated.
Specific Findings for Novel Partial Sequences
In 2018, two partial sequences of the 19 identified as potentially novel were present in the group added to the database. Accession C0HKC0 had a highly significant alignment (E-value = 8E-8) across all 20 amino acids with a preexisting sequence (P86888.1) despite its short length, indicating that the partial sequence was not truly a novel addition. Similarly, accession C0HJX6.1 had a highly significant (6.7E-7) 21-amino-acid alignment with accession P85984.1 indicating that this partial sequence was again not truly a novel addition.
In 2019, four partial sequences were identified as potentially novel (COMPARE004, COMPARE005, COMPARE002, & COMPARE003). While none of these sequences were found to be highly similar to existing sequences in the database, their short length (18, 20, 39, & 39 amino acids) made the likelihood that they contain sufficient unique information relating to the allergenicity of the full-length protein low. As such, these sequences were not categorized as truly novel allergen sequences.
In 2020, a large number of partial sequences (121) were added to the database, all of which were ≤50 amino acids in length. Each sequence between 20 and 50 amino acids in length was individually checked as to its status as a partial sequence (all found to be partial sequences), while those <20 amino acids in length were assumed to be partial sequences. Although a few of these sequences shared significant homology with existing sequences in the database due to high identity and/or similarity over their short length, none were categorized as truly novel sequences due to the likelihood that the peptides represented by these short partial sequences do not have key allergenic features of the full-length proteins.
Again in 2021, a large number of partial sequences (64) were added to the database, all but one of which was <35 amino acids in length. One partial sequence (BAM22586.1a) was 82 amino acids in length and had an E-value of 5.2e-05 over an alignment of 69 amino acids with an existing sequence (AAC61261.1) in the database. Again, these partial sequences do not appear to represent truly novel allergen sequence additions.
Many partial sequences (72) were also added to the COMPARE database in 2022. All but three of these sequences were <35 amino acids in length. The three longer partial sequences, COMPARE00323, COMPARE00324, and COMPARE00325, are 60, 90, and 193 amino acids long with the longest sequence having an E-value of 1.6e-08 for an alignment of 164 amino acids with an existing sequence (CAI43283.4) in the previous version of the database. Again, these partial sequences were not considered truly novel additions to the database.
Potential Novel Full-length Sequences
There were 17, 13, 12, 5, and 1 new potentially full-length sequences (>100 amino acids) in each of the five consecutive years evaluated from 2018 to 2022, respectively, that did not meet both the sliding window and E-value criteria. In 2018, one accession (BAG93480.1) of the 17 potentially novel full-length sequences, while not meeting the sliding-window criteria, had highly significant alignments (E-values <E-7) to an existing sequence in the database (AAD38942.1) which indicates that this sequence was not truly a novel addition. Similarly, p13080 did not meet the sliding window criteria but also had a highly significant alignment (E-value <E-7). Of the remaining 15 potentially novel sequences, four pairs of highly related sequences (1510259A & adv71357.1: BAW03243.1 & BAW03242.1: Q7X7E6.1 & Q7X8H9.1: AK242260.1 & BAH01262.1) and one highly related group of three sequences (AK068307.1, BAG95020.1, & XP_015646887.1) were added in 2018 indicating that this group of 11 sequences represents only five truly distinct novel sequence types. Combined with the four distinct novel sequences added to the 2018 database (BAG88472.1, BAV90601.1, L7UZ85.1, & NP_001036878.1), a total of nine truly novel sequences were added to the 2018 database.
Of the 13 potentially novel full-length sequences identified in 2019, two pairs of highly related sequences (ATI08931.1 & ATI08932.1: BBE74942.1 & COMPARE010) and one highly related group of seven sequences (ARQ16437.1, ARQ16438.1, ARQ16439.1, ARQ16440.1, ARQ16441.1, ARQ16442.1, & ARQ16443.1) were added in 2019 indicating that this group of 11 sequences represent only three truly distinct novel sequence types. Combined with the two distinct novel sequences added to the 2019 database (COMPARE013 & Q4W1G2), a total of five truly novel sequences were added to the 2019 database. It is noteworthy that an additional unique sequence (COMPARE008) was added to the database in 2019 but was excluded from our analysis as it was removed as a potential allergen in the 2022 update cycle (http://db.comparedatabase.org/docs/COMPARE-2022-Documentation.pdf).
Of the 13 potentially novel full-length sequences identified in 2020, one group of three highly related sequences (AAX84656.1, AIO08861.1, & QAT18644.1) was added, indicating that this group of three sequences only represents one truly distinct novel sequence type. Combined with the nine distinct novel sequences added to the 2020 database (ACT37323.1, ADM53099.1, BAF43535.1, CAA67128.1, CAY85463.1, NP_001138311.1, QAT18643.1, XP_392204.2, & ACS49840.1), a total of 10 truly novel sequences were added to the 2020 database.
In 2021, five unrelated full-length sequences were identified (QBP14757.1, QFI57017.1, QIJ32297.1, QDO73345.1, & AAD32205.1), each representing a distinct novel sequence type. In 2022, one full-length sequence addition (P62927) was identified as truly novel.
Discussion
Role of Comprehensive Allergen Database in Allergenicity Assessment
The allergenicity risk of novel food proteins is assessed using a weight-of-evidence approach since there is no single characteristic or indicator of protein allergenic risk, and unlike an assessment for toxicity, no reliable animal models are available that predict allergenicity.Citation11 While protein characteristics like thermal and digestive stability have been suggested as differentiating features of allergens, these characteristics have not been shown to correlate with the allergenic status of proteins.Citation12–15 A history of exposure to a protein or an organism that contains that protein, with no reports of allergy, is an important consideration in establishing a low risk of allergenicity. However, this history may be lacking for some newly expressed proteins in GE crops. As such, an assessment of the degree of similarity between a novel food protein and known allergens may represent the single best tool for predicting allergenic risk. Conservative bioinformatic tools and thresholds have been developed to predict the cross-reactive risk among allergens.Citation2,Citation3 Therefore, the robustness of these tools for predicting the allergenic risk of novel food proteins is largely dependent on the comprehensiveness of the allergen database that is used for the analysis.
Rationale for Investigative Approach
The COMPARE allergen database was established in 2017 and its contents have been updated annually through 2022 thus far. The number of truly novel sequences added during these years is a reflection of the current understanding of allergen sequence diversity and can be used as a measure of the comprehensiveness of the database since preexisting sequences in the database are likely to make highly related additional sequences redundant for detecting the relationship between a novel food protein and known allergens. Although it is acknowledged that the degree of similarity and novelty of a protein sequence is a matter of degree, and designating a sequence as novel is necessarily subjective, we attempted to automate an initial categorization of non-novelty based on a sequence sharing both >35% identity over a ≥80 amino acid sliding window and a sequence alignment with an E-value <E-7 derived from a full-length FASTA. The remaining sequences were designated potentially novel and further examined individually. We believe that this process was reasonable for identifying additions to the database that truly represent novel sequences potentially useful for uniquely identifying the allergenic risk of novel food protein using current bioinformatic approaches. However, the practice of including sequences based solely on in vitro IgE binding and including partial sequences, while conservative for risk assessment, necessarily lead to an overestimate of the number of newly identified sequences that are clinically relevant to allergy.Citation7
Data Trends
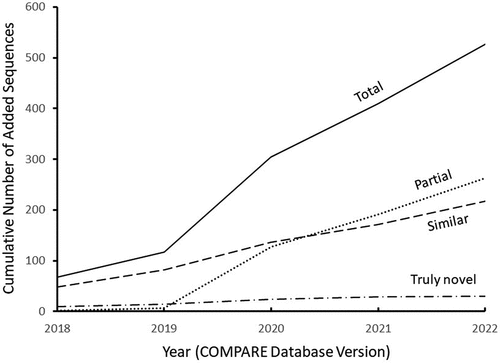
The number of total sequences added to the COMPARE allergen database stepped up to a higher level during the latter three years (2020, 2021, & 2022) which appears to be mostly driven by the addition of partial sequences (). The number of added sequences that were considered highly similar to existing sequences in the database was relatively consistent across years and represented a significant portion of the total as well. The number of truly novel sequence types was a minor component of the total number of sequences added each year and again was fairly consistent over time, with the possible exception of the most recent sequence additions (2022) where only one truly novel sequence was added. It is noteworthy that the single truly novel sequence identified in the 2022 COMPARE update (P62927 from pea, Pisum sativum) appears to have very weak evidence of allergenicity based on the paper cited in support of its inclusion in the database.Citation16 In this paper, the authors describe this protein as being present in pea total protein extract but having little evidence of IgE binding using serum from pea-allergic children. This observation seems to support the conclusion that the annual addition rate of truly novel allergen sequences to the COMPARE database is approaching zero and that the in-silico prediction of allergenic risk is robust based on our current knowledge of allergen sequence diversity.
Conclusions
During the consecutive years 2018 through 2022, we found that 9, 5, 10, 5, and 1 truly novel sequences were added (), respectively, out of 2,463 total allergen sequences in the 2022 version of the COMPARE allergen database. This relatively low number of additional novel sequences in combination with limited evidence for their clinical relevance supports the comprehensiveness of our contemporary knowledge of allergen amino acid sequence diversity (as reflected in the current version of the COMPARE database) and the robustness of the current bioinformatic prediction of allergenicity for novel food proteins such as those newly expressed in GE crops. This is likely a major contributor to the observation that no newly expressed protein in any GE crop has been shown to cause allergy in anyone.Citation17
Table 1. Unique additions to COMPARE database with most unique member representing closely related groups of added sequences (largest E-value for alignment with closest preexisting target)
Acknowledgments
We thank Liisa Koski and Lucilia Mouriès of the Health and Environmental Sciences Institute’s COMPARE Database program for providing data files used for this analysis. We further thank Andre Silvanovich of Bayer Crop Science and Lucilia Mouriès of the Health and Environmental Sciences Institute’s COMPARE Database program for insightful review of the draft manuscript.
Disclosure Statement
The authors are employed by Corteva Agriscience which develops and markets transgenic seed.
Additional information
Funding
References
- Ladics GS, Cressman RF, Herouet-Guicheney C, Herman RA, Privalle L, Song P, Ward JM, McClain S. Bioinformatics and the allergy assessment of agricultural biotechnology products: industry practices and recommendations. Regul Toxicol Pharmacol. 2011;60(1):46–53. doi:10.1016/j.yrtph.2011.02.004.
- EFSA Panel on Genetically Modified Organisms, E. Mullins, J-L. Bresson, T. Dalmay, I. C. Dewhurst, M. M. Epstein, L. George Firbank, P. Guerche, J. Hejatko, H. Naegeli, F. Nogué, et al. Scientific opinion on development needs for the allergenicity and protein safety assessment of food and feed products derived from biotechnology. EFSA J. 2022;20(1):e07044. doi:10.2903/j.efsa.2022.7044.
- Herman RA, Song P. Validation of bioinformatic approaches for predicting allergen cross reactivity. Food Chem Toxicol. 2019;132:110656. doi:10.1016/j.fct.2019.110656.
- Kessenich C, Silvanovich A. Challenges of automation and scale: bioinformatics and the evaluation of proteins to support genetically modified product safety assessments. J Invertebr Pathol. 2021;186:107587. doi:10.1016/j.jip.2021.107587.
- Cressman RF, Ladics G. Further evaluation of the utility of “sliding window” FASTA in predicting cross-reactivity with allergenic proteins. Regul Toxicol Pharmacol. 2009;54(3):S20–S25. doi:10.1016/j.yrtph.2008.11.006.
- Herman RA, Song P, Mirsky HP, Roper JM. Evidence-based regulations for bioinformatic prediction of allergen cross-reactivity are needed. Regul Toxicol Pharmacol. 2021;120:104841. doi:10.1016/j.yrtph.2020.104841.
- van Ree R, Sapiter Ballerda D, Berin MC, Beuf L, Chang A, Gadermaier G, Guevera PA, Hoffmann-Sommergruber K, Islamovic E, Koski L. The COMPARE database: a public resource for allergen identification, adapted for continuous improvement. Front Allergy. 2021;2:700533. doi:10.3389/falgy.2021.700533.
- Goodman RE, Ebisawa M, Ferreira F, Sampson HA, van Ree R, Vieths S, Baumert JL, Bohle B, Lalithambika S, Wise J. AllergenOnline: a peer‐reviewed, curated allergen database to assess novel food proteins for potential cross-reactivity. Mol Nutr Food Res. 2016;60(5):1183–98. doi:10.1002/mnfr.201500769.
- Herman R, Song P, ThirumalaiswamySekhar A. Value of eight-amino-acid matches in predicting the allergenicity status of proteins: an empirical bioinformatic investigation. Clin Mol Allergy. 2009;7(1):1–7. doi:10.1186/1476-7961-7-9.
- Silvanovich A, Nemeth MA, Song P, Herman R, Tagliani L, Bannon GA. The value of short amino acid sequence matches for prediction of protein allergenicity. Toxicol Sci. 2006;90(1):252–58. doi:10.1093/toxsci/kfj068.
- Ladics GS. Current codex guidelines for assessment of potential protein allergenicity. Food Chem Toxicol. 2008;46(10):S20–S23. doi:10.1016/j.fct.2008.07.021.
- Bøgh KL, Madsen CB. Food allergens: is there a correlation between stability to digestion and allergenicity? Crit Rev Food Sci Nutr. 2016;56(9):1545–67. doi:10.1080/10408398.2013.779569.
- Herman RA, Roper JM, Zhang JX. Evidence runs contrary to digestive stability predicting protein allergenicity. Transgenic Res. 2020;29(1):105–07. doi:10.1007/s11248-019-00182-x.
- Privalle L, Bannon G, Herman R, Ladics G, McClain S, Stagg N, Ward J, Herouet-Guicheney C. Heat stability, its measurement, and its lack of utility in the assessment of the potential allergenicity of novel proteins. Regul Toxicol Pharmacol. 2011;61(3):292–95. doi:10.1016/j.yrtph.2011.08.009.
- Verhoeckx K, Bøgh KL, Dupont D, Egger L, Gadermaier G, Larré C, Mackie A, Menard O, Adel-Patient K, Picariello G. The relevance of a digestibility evaluation in the allergenicity risk assessment of novel proteins. Opinion of a joint initiative of COST action ImpARAS and COST action INFOGEST. Food Chem Toxicol. 2019;129:405–23. doi:10.1016/j.fct.2019.04.052.
- Popp J, Trendelenburg V, Niggemann B, Randow S, Völker E, Vogel L, Reuter A, Spiric J, Schiller D, Beyer K. Pea (Pisum sativum) allergy in children: pis s 1 is an immunodominant major pea allergen and presents IgE binding sites with potential diagnostic value. Clin Exp Allergy. 2020;50(5):625–35. doi:10.1111/cea.13590.
- Dunn SE, Vicini JL, Glenn KC, Fleischer DM, Greenhawt MJ. The allergenicity of genetically modified foods from genetically engineered crops: a narrative and systematic review. Ann Allergy Asthma Immunol. 2017;119(3):214–222. e213. doi:10.1016/j.anai.2017.07.010.