ABSTRACT
Potato virus Y (PVY) is a deadly environmental constraint that damages productivity of potato (Solanum tuberosum) around the globe. One of the major challenges is to develop resistance against PVY. Emerging clustered regularly short palindromic repeat (CRISPR)/Cas systems have the potential to develop resistance against PVY. In the current research, CRISPR-Cas13 has been exploited to target multiple strains of PVYN, PVYO, and PVYNTN. Multiple genes PI, HC-Pro, P3, Cl1, Cl2, and VPg genes of PVY were targeted by CRISPR/Cas13a. Multiplex gRNA cassettes were developed on the conserved regions of the PVY-genes. Three independent CRISPR/Cas13 transgenic potato lines were developed by applying an optimized concentration of trans-ribo zeatin and indole acetic acid at callus development, rooting, and shooting growth stages. The level of resistance in transgenic plants was confirmed through double-antibody sandwich enzyme-linked immunosorbent assay and real-time quantitative PCR. Our results have shown that efficiency of PVY inhibition was positively correlated with the Cas13a/sgRNA expression. Finding provides the specific functionality of Cas13 with specific gRNA cassette and engineering the potential resistance in potato crop against multiple strains of PVY.
1. Background
Potato is the most important consumable non-grain crop. It is the most consumable vegetable crop of 37 million ton production in the year of 2019 around the globe, and production rate of Pakistan was 4.8 million tons (http://www.fao.org/faostat/en/#data/QC). Sustainable potato production is affected by several biotic and abiotic environmental constraints. Among biotic factors, potato viruses like Potato virus Y (PVY), Potato leaf roll virus (PLRV), Potato virus A (PVA), Potato mop-top virus, Potato virus X, and Potato virus S (PVS) are reported. PVY is a significant devastating factor for the potato crop. PVY belongs to the family Potyviridae and has multiple strains, and it has affected the tuber quality and yield of crop losses up to 80%.Citation1 The PVY genome is single-stranded (ss) positive-sense RNA with approximately 9.7 kb in length. The translated protein 3061 amino acid from the viral genome is processed by viral proteases into a small reading frame (ORF) P1, HC-Pro, P3 (PIPO) 6K1, CI, 6K2, NIa (VPg plus Pro), NIb (viral replicase), and CP (capsid protein).Citation2
The mutations in the PVY genomes lead to a higher rate of recombination and diversity that developed an effective mechanism to escape from plant natural immunity. The most significant strains PVYO, PVYN, PVYC, including recombinant strains like PVYNTN and PVYN:O have devastated the potato crop.Citation3,Citation4 Previously, several transgenic approaches were applied to develop resistance against PVY, including over-expression of resistance eIF4E allele, mutating the susceptible eIF4E gene by CRISPR/Cas9, and utilization of RNAi strategy.Citation5,Citation6 Furthermore, RNAi approaches have off-target effects due to constitutive siRNA expression.Citation6,Citation7 Limitations of several transgenic approaches provided the reasons to develop new strategies for generating broad-spectrum resistance against dominating multiple PVY strains.
The clustered regularly short palindromic repeat (CRISPR)/Cas systems imparted molecular immunity to bacteria, Archaea and invading conjugative plasmid and phages. The immunity attain stepwise 1) acquisition of spacer (gRNA) from invader’s genome, 2) biogenesis and processing of CRISPR RNAs (crRNA), and 3) interference with the invader’s genome.Citation8,Citation9 The CRISPR/Cas9, Cas12, and Cas14 have ability to cleave the double strandedCitation10 DNA or single-stranded (ss) DNA, while Cas13 has unique ability to cleave the single-stranded (ss) RNA including viral genomes to provide defense.Citation11,Citation12
Based on function, the CRISPR/Cas13 belongs to single multi-domain effectors with 900–1300bp range with 4-subtypes (A-D). These subtypes have low sequence homology. Cas13 cleaves the Serna with nonspecific manners with the help of crRNA. Cas13 crRNA possesses a simple structure 20–30 nucleotide guide sequence with a protospacer flanking site of A, U, or C as a preference for the functional activity of Cas13a.Citation13–15 The functional activity of Cas13 is greatly affected by the secondary structure of the target sequences. Further, the knockdown of RNA transcript through Cas13a is comparable to the RNA-interference mechanism, but it has reduced off-target effects.Citation12 LshCas13a has been exploited to gain resistance in monocot grains plant against Southern rice black-streaked dwarf virus (SRBSDV) and Rice Stripe Mosaic Virus (RSMV).Citation16 Potentially, CRISPR/Cas13a system can develop stable resistance against significant RNA viral disease in major crops.Citation17 In fields, the plants are exposed to multiple viruses and their strains. More fruitful results would be obtained if conserved regions of multiple viruses or strains would be targeted to attain resistance. The key advantage of using CRISPR/Cas is the simplicity and ease of this system even for targeting multiple sites simultaneously. Multiplexed genome editing (MGE) approach involves more than one gRNA to target multiple regions in the genome. In plants, different strategies have been harnessed to deliver a combination of gRNAs. A construct carrying different gRNAs under the influence of their separate promoters has been used for MGE in various plants, e.g. Arabidopsis, maize, wheat, tomato, and rice.Citation18–21
In current research, CRISPR/Cas13a has been employed to target the multiple strains of PVY. The response of multiple gRNA cassettes with direct-repeat processing was exploited to develop resistance by targeting conserved coding regions of different PVY strains. We have demonstrated that the CRISPR/Cas13a system can be engineered to confer broad-spectrum resistance in transgenic potato plants against multiple PVY strains. The agrobacterium-mediated transformation protocol was optimized by altering the amount of acetosyringone and plant growth regulators (PGRs) at the time of transformation. The cytokinin (CK) and indole acetic acid (IAA) concentrations were optimized to generate quick stable transgenic lines. The expression of Cas13 has been confirmed by RT-PCR. Functional specificity of CRISPR/Cas13a system was confirmed by treating Cas13 lines with PLRV.
2. Results
2.1. Construction Preparation
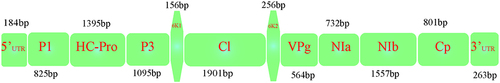
The PVY genome has high mutation, recombination rate, and holding genetic diversity among the strains ( and supplementary Table S1).Citation3 Alignment of 100 sequences of PVY was conducted using mega muscle alignment tool, based on which gRNAs were designed. A total of six gRNAs were designed based on conserved regions with 28bp sequence and specific PSF-preferences of the following genes as potyviral membrane protein (PI, P3), HC-Pro, cytoplasmic laminate inclusion (Cl, Cl2), and viral genome linked protein (VPg), respectively (, ). Further off-targeting of the gRNA was aligned with the manually against potato whole genome (https://www.plantbreeding.wur.nl/Solyntus/). No off-targets were found with all six targets with 28bp sequence. We constructed the binary vector that harboring the Cas13 (LshCas13a, WP_018451595.1) under the CAMV-35S-promoter and sgRNAs and multiplex-sgRNA vector with nuclear export signals expressed under U6-promoter. The construct confirmed through the PCR, restriction analysis, and Sanger sequencing ().
Table 1. List of gRNAs.
Figure 1. Representation of PVY sequence information. The indication of genes in the PVY genome with respective size.
2.2. Confirmation of Multiplex Targeting Construct
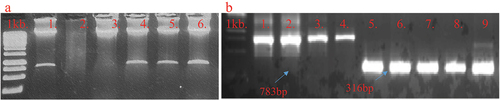
Multiplex guide RNA cassette was confirmed by restriction analyses. Restriction of plasmid with Hind III yielded a product of 792bp () indicative of the multiplex gRNA, sequence represented in supplementary data (Supplementary sequence 1, ). The restricted product through HindIII was ligated into Pk2GW7-LshCas13 vector through restriction-ligation protocols. In the clone, PVY-cassette was confirmed by using specific PCR primers with 783bp product size (: PVY-cassette confirmation), while Cas13 was confirmed by specific primers that yield a 316bp product size (primer’s sequence in # Cas13 confirmation ). Final confirmation was carried out by Sanger sequencing.
Table 2. Primers for transgene confirmation.
2.3. Transformation of Potato with Optimized PGRs
The internode cuttings from the potato variety Kruda were transformed via Agrobacterium-mediated transformation (GV3101 strain). Approximately 20 transformed internodes were remained viable on kanamycin selection plate of 25 internodes. The transformed calli were shifted to callus induction medium (CIM) up to 40 days, and healthy surviving calli were transferred to regeneration medium. Regenerated plantlets were moved from shooting medium to rooting medium with 0.2 mg/L and 0.35 mg/L CK and IAA, respectively. Parallel, 4 concentrations of 10 mg/mL, 15 mg/mL, 20 mg/mL, and 30 mg/mL of acetosyringone were added in the simple MS-liquid media at the time of transformation. The 10 mg/mL of acetosyringone has not affected the rate of transformation, while internode treated with 30 mg/mL showed bacterial growth during callus induction stage. The 20 mg/mL of acetosyringone enhanced the transformation efficiency to 10 mg/mL and 30 mg/mL. The optimal AS-treatment duration was 20 min for the internode-transformation ().
Table 3. Concentration of PGRs and media’s composition for optimized tissue culture.
The IAA and Cytokinin (CK trans-ribo zeatin) work antagonistically. We exploited the 100 μL/L, 250 μL/L, 350 μL/L, and 500 μL/L from 100 mg/mL stock of IAA, and 1 mL/L, 2 mL/L, 5 mL/L, and 6 mL/L from 1 mg/mL stock of CK in the plant full MS-media for callus induction (). We found the best callus development with the application of 350 μL/L and 5 mL/L to IAA and CK, respectively. The application of 0.2 mg/mL CK at shooting-stage provided strong secondary growth.
The developmental stages from internodes to plants are shown in . The transgenic lines were confirmed by PCR and Sanger sequencing (). The 15 independent transgenic lines were obtained and confirmed by PCR by using Cas13 detection primers (Cas13 F: 5ʹATGGGCAACCTCTTTGGCCATAAGCGTT3’ and Cas13 R: 5ʹCCACTTCTTCAGTCTCGAGGAAATCGTC3’ with 480bp product. To proceed further study, RNA-isolation from these lines and expression of Cas13 were identified. Among them, 3 transgenic lines 13.1, 13.2, and 13.3 were highly expressed Cas13, and these lines were multiplied. Confirmed transgenic lines were shifted to soil and kept in greenhouse under the controlled conditions for establishment of roots growth from media to soil. After 1 month of hardening, the resistance efficiency was verified by phenotypic assay.
Figure 5. Developmental stages of transformed internodes into plants: The plants internodes were transformed by the Agrobacterium-mediated transformation (GV3101). A: the callus development stage, B: Shoot initiation stage C & D: Shoot development stage E&F: Root initiation and development stage.
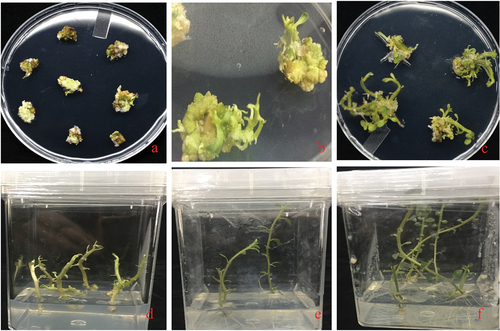
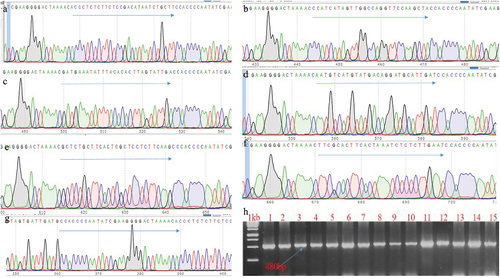
Figure 6. Transgene confirmation and gRNA-component in the plants. Transformed transgenic lines confirmed through PCR and Sanger sequencing to check the presence of gRNA; A is gRNA for the PI, B; HC-Pro, C: P3, D: Cl1, E: Cl2, F: Vpg, G: Direct repeat of Cas13 and H: representing the PCR confirmation of transgenic plants.
2.4. PVY-Bioassay and DAS-ELISA
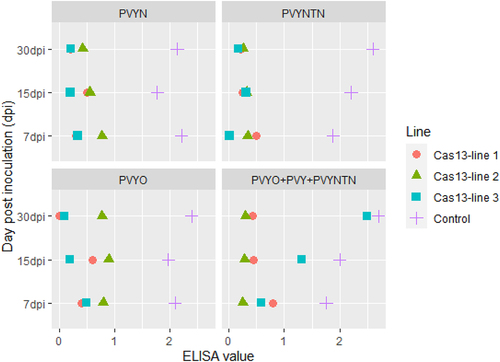
The confirmed strains PVYO, PVYN, and PVYNTN were applied for the bioassay. To find out the viral load, the bioassay confirmation through DAS-ELISA and real-time qPCR were performed after regular intervals, i.e. 7 days post inoculation (dpi), 15 dpi, and 30 dpi. The significant phenotypic symptoms of PVY like mosaic, mottled, and crinkled leaves and vein necrosis were observed in wild-type susceptible control lines (). The three strains of PVYo (HQ912865), PVYN (X97895), and PVYNTN (M95491) were exposed to the transgenic and control lines to check the resistance of the transgenic plants. The resistance assay was checked after regular interval of 7 dpi, 15 dpi, and 30 dpi to monitor the disease symptoms of PVY-strains. Mosaic and vein necrosis were observed in control lines, while no symptoms appeared in transgenic lines Cas13.1, Cas13.2, and Cas13.3, as shown in and supplementary Figure S2. The data from DAS-ELISA indicated the reduced level of PVY-accumulation in the transgenic lines as compared to the control lines (in graphic chart ).
2.5. Expression of Cas13 Inversely Proportional to PVY Viral Load
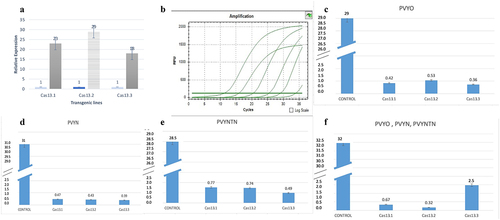
To find out the correlation of PVY viral load and Cas13a expression, the RNA-purified from PVY-treated control and transgenic lines were isolated. The cDNA-synthesized real-time qPCR was performed by St-AC and ST-AC97 endogenous primers for Actin97. For the Cas13 expression analysis, Cas13 primers were used (). The expression of LshCas13 was observed by the relative RT-qPCR for co-relation between the expression of LshCas13 and level of inhibition of PVY-accumulation. The DAS-ELISA and RT-qPCR strongly supported that the higher expression of Cas13 lines holding the higher resistance, as shown in . The results have shown a positive co-relation of Cas13 expression and inhibition of PVY systemic spreading as crRNA exploited for the multiplexed targeting the conserved regions of the three viral strains. The highest expression of Cas13 was observed in line 13.2. The symptoms observed in this line were near to wild-type untreated control plants. This was also confirmed by the DAS-ELISA where we found strong resistance and no PVY-accumulation was observed. Similar pattern was found in the lines 13.1 and 13.3, where symptoms were moderate that include yellow mosaic pattern. The titer of virus was slightly high as shown by DAS-ELISA and expression of Cas13 was low in Cas13.1 and 13.3. Interestingly, we identified mixed inoculation of PVYo, PVYN, and PVYNTN on all transgenic lines. Approximately, all lines showed minute to zero viral load, confirmed by RT-PCR by using targeted gene specific primers HC-Pro and VPg (). These findings suggest that expression of Cas13 has inverse relation with the symptoms and virus titer. ( supplementary Figure S3, Table S2 for additional information).
Figure 9. RT-PCR analysis; A; Relative expression of Cas13a in the transgenic lines were represented as compared to control line, error bar is representing the technical repeats. Line Cas13.2 strongly expressing as compared to Cas13.1, Cas13.3. B: Copy number of PVY were determined by qRT-PCR: B; Control dilutions parameters, while C, D, E and F expressing virus titer in the transgenic lines against PVYO, PVYN, PVYNTN, and co-inoculations of PVYO, PVYN, PVYNTN respectively.
2.6. Identification of Collateral Activity of Cas13
In addition, the CRISPR/Cas13a transgenic lines were treated with PLRV to check the collateral activity of Cas13a. Plants which were inoculated with PLRV showed typical symptoms at 15 dpi that include rolling of the top leaves followed by yellowing (). These symptoms were indistinguishable from that of wild-type control plants. On later stages, plants also showed stunted growth. These plants were subjected to DAS-ELISA for the confirmation, and they showed the presence of PLRV as confirmed through DAS-ELISA. Our results have shown that Cas13a plants failed to provide resistance against heterologous virus, i.e. PLRV. Thus, the resistance developed by Cas13a is specific for PVY.
3. Discussion
PVY has the potential to devastate the yield of potato crops a large scale. Several conventional approaches have been applied to develop resistance which were met with limited success. Many bacterial and archaeal species protect themselves against invading phages and nucleic acid by activating various forms of Cas proteins to attain immunity. To gain resistance and immunity against DNA and RNA viruses, CRISPR/Cas systems have been widely applied. The enzymes Cas9, Cas12, and Cas14 were applied to target the dsDNA and ssDNA viruses. In case of Cas13a, it cleaves single-stranded (ss) RNA followed by activating and maturation of crRNA and making targeted effector complex.Citation22 Inconclusively, Cas13 has the potential ability to develop resistance against ssRNA viruses. In the current study, the CRISPR/Cas13 has been exploited to cleave the ssRNA of multiple strains of PVY as demonstrated in the theoretical model (Supplementary Figure S4).
The Cas13 from Leptotrichia shahii (LshCas13a) was used to develop resistance against tobacco turnip mosaic RNA virus (TuMV) in model plants Nicotiana benthamiana and Arabidopsis thaliana. They employed green fluorescent protein fused with TuMV to explore the functional potency of LshCas13a against TuMV. Cas13a reduced the expression of HC-Pro more effectively than Coat protein (CP) of TuMV.Citation23 Previously, it has been shown that dual and distinct functions of Cas13 for processing the pre-crRNA and degradation of ssRNA moreover, multiple genes can be targeted to develop resistance against pathogens.Citation24 In current study, multiple genes, i.e. P3, Cl, HC-Pro, and VPg were targeted by Cas13 to develop broad-spectrum resistance. We targeted multiple strains of PVY simultaneously by CRISPR arrays harboring 28-nt targeting the PI, HC-Pro, Cl, and VPg on the conserved regions of the PVY. The gRNAs were assembled with regular interval of 28-nt direct repeats (DRs). These targeted genes are essential for the integrity of viral genome and proper functioning as PI & P3 potyviral membrane protein for the virus replication and systemic spreading and pathogenicity.Citation1,Citation25 The CI have been involved in the virus infection and movement.Citation2,Citation10 The VPg is essential for interaction with the host-machinery and responsible for the viral genome replication and systemic spreading.Citation1,Citation26 In this study, the multiple gRNA-targeting Cas13a harboring vector (14.5kb) has assembled to develop resistance against recombinant strains of PVY.
Recently Desiree and King Edward potato lines were developed against bacterial blight resistance by targeting DND1 and DMR6-1 genes at multiple sites.Citation27 Transformation efficiencies were improved by optimizing several factors affecting regeneration, including the quality of the starting plant material, acetosyringone, and different concentrations of the PGRs. The plant-derived phenolic compounds work as chemo-attractants for the Agrobacterium. The significant phenolics are 3,5 dimethoxyacetophenone (acetosyringone) and hydroxyacetosyringone and act as inducers during Agrobacterium-mediated transformation. The VirA/VirG component of Ti-plasmid senses the acetosyringone as host signals and activates the vir-gene expression.Citation28 The transfer and copy number replication of the Ti-plasmid from Agrobacterium to plants is regulated by the phenolic signals.Citation29 A specific protocol was designed to alleviate and improve the growth vigor and resulted in a 4- to 10-fold increase in transformation efficiency.Citation30,Citation31 The phytohormones potentially develop the signaling interaction to regulate several metabolic processes. During the transformation, the IAA and ethylene levels elevated in the potato internodes. The IAA is a substantial requirement for growth and development, while a higher concentration over 50 μM enhanced the suppression of vir-gene that affects the rate of transformation.Citation32 The crosstalk between several hormones is a crucial regulatory network in defense reactions against environmental constraints. Previously, it has been shown that a higher level of CK provides immunity and exogenous application of CK increased the level of SA-dependent gene expression during pathogen infection.Citation33 CK involve in callus greening and shoot induction in the explant.Citation34 Synthetic IAA has been used for root-development, proper leaf growth, and identification of several auxin responsive pathways.Citation35,Citation36 IAA and CK play crucial role for regeneration and development.Citation37 Basically, the plant’s signals activate the virulence genes of Agrobacterium to transfer and integrate the T-DNA to plant nucleus. That expression of foreign T-DNA leads to the expression of IAA and CK. In the current study, we exploited the various amount of PGRs for the optimization, and we noticed the rate of callus development and growth was enhanced by the application of 0.5 mg/L and 0.2 mg/L of trans-ribo zeatin, respectively. We identified that the concentration from 0.1 mg/mL to 0.35 mg/L of IAA promotes the shooting and rooting growth. Upon the induction of acetosyringone, the large size binary vectors significantly improved transformation in potato as previous reported into rice cells.Citation38 We observed an improved transformation efficiency with 20 mg/mL of acetosyringone during the transformation of internodes.
The functionality of Cas13a to combat the RNA in the cytoplasm was identified by targeting three different genes in rice by LwaCas13a. Transformed protoplasts exhibited more than 50% knockdown after 48 hours of transformation.Citation12 The level of resistance was directly correlated with the expression efficiency of Cas13a. The Lsh-Cas13a was exploited by generating stable transgenic lines to attain resistance against PVYo, PVYNTN, and PVYN. We tested Cas13 lines against three strains of PVY and PLRV to find out the level of resistance, interference, and specificity of Cas13a with guide RNA. Our results are in line with previous studies where CRISPR/Cas13a transgenic Solanum tuberosum cv. Desiree was developed having resistance against PVY exhibiting no effects against PVA and PVS.Citation39 We stably developed the transgenic plants with multiple gRNAs targeting the conserved regions of the PVY-strains. The multiple gRNA cassette with alternative DRs. These repeats processed as self-cleaving ribozyme,Citation40 Csy4 RNase,Citation27 or t-RNA processing enzyme,Citation41 and Cas13 can process its own pre-crRNA.Citation22,Citation42 The latest strategy shows very promising efficiency for MGE in plants and successfully confirmed and provides resistance against multiple recombinant strains of PVY.
In previous studies, the RNA viruses target by Cas9 from Francisella novicida (FnCas9) to develop resistance in tobacco and Arabidopsis.Citation43 Currently, we generated the resistance in tetraploid potato against three strains of PVY. To further test the inheritability of the resistance against PVYO, PVYN, and PVYNTN into the successive generations, T2 of the transgenic Cas13 were harvested and would be challenged to various strains of PVY. Overexpressing of crRNA–LshCas13a specifically targeting the viral genome was an effective way to generate stable RNA virus resistance with multiple DRs, which was helpful in over-expression of Cas13a. The qualitative expression of Cas13 was observed in all three resistance lines to check the expression of Cas13 relative to resistance. The RT-PCR indicated the Cas13.3 highly expressed as compared to 13.2 and 13.1 lines. The 13.3 transgenic line exhibited higher resistance against individual PVYO, PVYN, and PVYNTN assays, while less resistance level was observed when the mixture of strains was applied, which indicates discovering more about the emergence of novel strain or weak expression of gRNA cassette under U6 promoter. The 13.2 line exhibited lowest viral load of PVY in individual treatments and mixed treatment as compared to control, transgenic 13.1, and 13.3 lines. The transgenic line 13.2 possess the higher resistance as compared to transgenic lines 13.1 and 13.3 by qRT-PCR screening assays. Previously reported that the Cas13 have collateral activity after recognition and cleavage of target site, leading to nonspecific degradation of RNA regardless of complementarity to the spacer.Citation44 However, these lines failed to show resistance against PLRV. These finding confirm that the resistance produced by Cas13a is highly specific and it does not work against heterologous viruses. PVY-resistant lines have not exhibited collateral activity.
Our finding showed that the RNA-targeting CRISPR/Cas’s system has provided the potential benefits over the DNA genome editing as expanding the functional capacity improving anti-viral immunity and avoiding the pleiotropic effects of genome editing. This research potentially provides new avenues to develop resistance against pathogens especially with RNA genomes.
4. Conclusion and Future Prospects
Potato is an important staple food that has a substantial role to feed the burgeoning population of the world. Molecular breeding and genetics have provided tremendous solutions against viral diseases, and the development of CRISPR/Cas systems has speed up the generation and development of resistance. The LshCas13 has provided resistance against three strains of PVY simultaneously. This system is widely used for the transcriptome engineering and would be helpful for identification of solutions against ssRNA viruses. Interestingly, the co-evolution of the viruses and microbes propelled the diversification of CRISPR-Cas defense systems to combat novel emerging recombinant strains. The functionality of Cas13 widely exploited in the RNA-base conversions in mammalian cellsCitation45 that would open the new avenues in the plant editing field. The ability of Cas13 to process pre-crRNA into mature crRNA, which targets the ss RNA of phage genome during viral interference, would be utilized for tracking of editing and signaling pathways in plants.Citation46 That eventually helpful in detection of many diseases and exploited in improving crop yield.
5. Materials and Methods
5.1. Plants, Virus, and Viral Strains
Potato (S. tuberosum cultivar Kruda) plants were grown in a growth chamber with conditions mentioned earlier.Citation47 The purity of PVY was confirmed by DAS-ELISA. The Local-PVYo, PVYN, and PVYNTN strains were confirmed by DAS-ELISA (Catalog # V093); most crop damaging strains in Pakistan were used for this study.
5.2. Multiplex Designing and Construction of LshCas13a/sgRNA Cassettes
For the development of CRISPR/Cas13a with its multiplex gRNA cassette, specific gRNAs were designed. For this, 100 of PVY (9.7Kb) sequences were aligned by using the Mega-6 muscle alignment software. The multiple 28bp gRNA were chosen on the conserved regions with respect to PFS preferences. The gRNAs targeting the PVY genome were arranged in the Cas13ʹs gRNA Cassette as PI, HC-Pro, P3, Cl1, Cl2, and VPg (). The gRNAs were manually blast with whole potato genome to identify the off-targets; there were no off-targets found on the potato genome. The multiple gRNAs with alternative 28bp DRs encoded under Arabidopsis U6 promoter; cassette was commercially synthesized. In pK2GW7-pCas13a vector, the Cas13a was expressed under the 35S promoter. pK2GW7-pCas13a vector restricted by HindIII and treated with calf intestinal alkaline phosphatase (CIP) to avoid self-ligation and gRNA cassette was restricted from the PTZ57Rkana vector by HindIII, specific cassette eluted from gel-purification, and ligated with linear Cas13 vector.
5.3. Transformation of Solanum Tuberosum through Agrobacterium
Constructs harboring the LshCas13a clone with multiple gRNA Cassette were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Briefly, overnight-grown cultures were harvested by centrifugation and suspended in liquid MS-media. The suspension was washed with simple liquid half MS to remove the traces of antibiotics, and final concentrations of O.D600, 0.6 was used for potato internodal transformation. Transgenic plants were identified by their resistance to kanamycin.
5.4. Optimizing the Transformation Protocols through PGRs at Developmental Stages
The transformation protocol was optimized as 0.6 O. D was measured and culture was pellet down and washed by MS-media to remove the traces of rifampicin and spectinomycin antibiotics. Final O.D for the transformation should be 0.5–0.8 measured and added to the acetosyringone for enhancing the rate of transformation. The concentration of trans-zeatin (naturally CK) and IAA were optimized to get rapid cell division during callus development and early roots and shoot development ().
5.5. Transgene Confirmation
Genomic DNA from transformed Solanum tuberosum lines were extracted through CTAB Method.Citation48 PCR amplifications were performed to confirm the presence of Cas13 in transgenic control line Cas13.0 without gRNA cassette and transgenic lines with presence of gRNA cassette with Cas13 by specific primers (). The presence of construct in the transgenic plants was confirmed by Sanger sequencing in three independent transgenic lines 13.1, 13.2, and 13.3.Citation49
5.6. Mechanical Inoculation of Viruses and DAS-ELISA Assay
Transgenic and wild-type control plants were challenged with the individual and mixed strains of PVYO, PVYN, and PVYNTN. The confirmed transgenic lines were multiplied before shifting to soil. At the 8–10 leaf stages, plants were exposed to viral strain by mechanical rubbing with carborundum powder.Citation50 The development of symptoms in transgenic and control lines was analyzed, and samples were collected at the 7-day, 15-day, and 30-day intervals from the inoculated and uninoculated leaves for confirmation of systemic spreading. DAS-ELISA with PVY specific antibodies (Agdia, Elkhart, IN) were performed to analyze the virus accumulation in the inoculated and non-inoculated leaves.Citation51
5.7. RNA-extraction and qRT-PCR Analyses
The expression of LshCas13a and copy number of as determined by of PVYO, PVYN, and PVYNTN have been analyzed by RT-qPCR assays by using specific primers. Total RNA was isolated by TRIzol reagentCitation52 and treated with DNase I (amplification grade, catalog number 18068015). The synthesis of cDNA was carried out by using reverse transcriptase kit (RevertAid First Strand cDNA synthesis Kit: Catalog number: K1622), and oligo (dT) primers. The fold expression of Cas13 was analyzed by qualitative RT-PCR as compared to wild-type control by using Cas13 detection primers in . PVY titer confirmed by amplification of HC-Pro with 167bp and VPg 176bp and CP 147bp product size with 56°C annealing temperature. Primer sequences for qRT-PCR are listed in . The technical repeats were performed for each biological replicates. The actin 97 gene was exploited as reference gene.
Author’s Contribution
S. M. conceive the idea. A.N. designed and conducted the experiment. M.Z.K. and S.H. helped in experiments. T.Z., N.A., and I.A. analyzed the RT-PCR results. A.N. prepared the manuscript. I.A. and S. M. reviewed the manuscript. All the authors read and approved the manuscript.
Ethical Approval and Consent to Participate
All authors read and approved the MS.
Ethical Statement for Conducting Study
The study was approved by Institutional Biosafety Committee (IBC) of the National Institute for Biotechnology and Genetic Engineering. The approval was given under the number IBC No. NIBG-49-112. All genetic manipulation in plants is approved by Institutional Biosafety Committee, duly registered with National Biosafety Committee (NBC). The case NIBG-49-112 was recommended by Technical Advisory Committee and approved by NBC.
Avalibility of Data and Materials
The materials used in this study would be available from the Dr. Imran Amin (PS); Molecular Virology and Gene Silencing Lab, Agricultural Biotechnology Division (ABD), NIBGE, Faisalabad, Pakistan.
Abbreviations
Supplemental Material
Download MS Word (11 MB)Acknowledgments
This research was funded by a research grant from Higher Education Commission (HEC-project# 7308) and National Centre for Industrial Biotechnology (NCIB) for establishment of emerging CRISPR/Cas’s techniques in tetraploid Solanum tuberosum. We are thankful to our Collaborator Prof. Magdy M. Mahfouz (King Abdullah University of Science and Technology) for providing Cas13a gene for conducting this study.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
Supplementary Material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21645698.2022.2080481
Additional information
Funding
References
- Quenouille J, Vassilakos N, Moury B. P otato virus Y: a major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Mol Plant Pathol. 2013;14(5):439–52. doi:10.1111/mpp.12024.
- Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 2010;6(6):e1000962. doi:10.1371/journal.ppat.1000962.
- Davie K, Holmes R, Pickup J, Lacomme C. Dynamics of PVY strains in field grown potato: impact of strain competition and ability to overcome host resistance mechanisms. Virus Res. 2017;241:95–104. doi:10.1016/j.virusres.2017.06.012.
- Karasev AV, Gray SM. Continuous and emerging challenges of Potato virus Y in potato. Annu Rev Phytopathol. 2013;51(1):571–86. doi:10.1146/annurev-phyto-082712-102332.
- Takakura Y, Udagawa H, Shinjo A, Koga K. Mutation of a Nicotiana tabacum L. eukaryotic translation-initiation factor gene reduces susceptibility to a resistance-breaking strain of potato virus Y. Mol Plant Pathol. 2018;19(9):2124–33. doi:10.1111/mpp.12686.
- Pooggin MM. RNAi-mediated resistance to viruses: a critical assessment of methodologies. Curr Opin Virol. 2017;26:28–35. doi:10.1016/j.coviro.2017.07.010.
- Kenesi E, Carbonell A, Lózsa R, Vértessy B, Lakatos L. A viral suppressor of RNA silencing inhibits ARGONAUTE 1 function by precluding target RNA binding to pre-assembled RISC. Nucleic Acids Res. 2017;45(13):7736–50. doi:10.1093/nar/gkx379.
- Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans Royal Soc B Biol Sci. 2016;371(1707):20150496. doi:10.1098/rstb.2015.0496.
- Van Der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat Rev Microbiol. 2014;12(7):479–92. doi:10.1038/nrmicro3279.
- Edwardson J. Inclusion bodies. Potyvirus Taxon. 1992;5:25–30. ISBN : 978-3-211-82353-8.
- Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF, Doudna JA, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–42. doi:10.1126/science.aav4294.
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. RNA targeting with CRISPR–Cas13. Nature. 2017;550(7675):280–84. doi:10.1038/nature24049.
- Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463(7280):568–71. doi:10.1038/nature08703.
- Cox DB, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–27. doi:10.1126/science.aaq0180.
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70(2):327–339. e5. doi:10.1016/j.molcel.2018.02.028.
- Zhang T, Zhao Y, Ye J, Cao X, Xu C, Chen B, An H, Jiao Y, Zhang F, Yang X, et al. Establishing CRISPR /Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol J. 2019;17(7):1185–87. doi:10.1111/pbi.13095.
- Khan MZ, Amin I, Hameed A, Mansoor S. CRISPR–Cas13a: prospects for plant virus resistance. Trends Biotechnol. 2018;36(12):1207–10. doi:10.1016/j.tibtech.2018.05.005.
- Li J-F, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31(8):688. doi:10.1038/nbt.2654.
- Xing H-L, Dong L, Wang Z-P, Zhang H-Y, Han C-Y, Liu B, Wang X-C, Chen Q-J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14(1):327. doi:10.1186/s12870-014-0327-y.
- Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42(17):10903–14. doi:10.1093/nar/gku806.
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–84. doi:10.1016/j.molp.2015.04.007.
- O’Connell MR. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J Mol Biol. 2019;431(1):66–87. doi:10.1016/j.jmb.2018.06.029.
- Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding S, Mahfouz M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018;19(1):1. doi:10.1186/s13059-017-1381-1.
- East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, Doudna JA. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–73. doi:10.1038/nature19802.
- Cui X, Wei T, Chowda-Reddy RV, Sun G, Wang A. The Tobacco etch virus P3 protein forms mobile inclusions via the early secretory pathway and traffics along actin microfilaments. Virology. 2010;397(1):56–63. doi:10.1016/j.virol.2009.11.015.
- Andersen K, Johansen IE. A single conserved amino acid in the coat protein gene of pea seed-borne mosaic potyvirus modulates the ability of the virus to move systemically inChenopodium quinoa. Virology. 1998;241(2):304–11. doi:10.1006/viro.1997.8967.
- Kieu NP, Lenman M, Wang ES, Petersen BL, Andreasson E. Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci Rep. 2021;11(1):1–12. doi:10.1038/s41598-021-83972-w.
- Subramoni S, Nathoo N, Klimov E, Yuan Z-C. Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front Plant Sci. 2014;5:322. doi:10.3389/fpls.2014.00322.
- Cho H, Winans SC. VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc Nat Acad Sci. 2005;102(41):14843–48. doi:10.1073/pnas.0503458102.
- Wang ES, Kieu NP, Lenman M, Andreasson E. Tissue culture and refreshment techniques for improvement of transformation in local tetraploid and diploid potato with late blight resistance as an example. Plants. 2020;9(6):695. doi:10.3390/plants9060695.
- Bruce MA, Rupp JLS. Agrobacterium-mediated transformation of Solanum tuberosum L., potato. In: Transgenic plants. Springer; 2019. p. 203–23. doi:10.1007/978-1-4939-8778-8_15.
- Liu P, Nester EW. Indoleacetic acid, a product of transferred DNA, inhibits vir gene expression and growth of Agrobacterium tumefaciens C58. Proc Nat Acad Sci. 2006;103(12):4658–62. doi:10.1073/pnas.0600366103.
- Albrecht T, Argueso CT. Should I fight or should I grow now? The role of cytokinins in plant growth and immunity and in the growth–defence trade-off. Ann Bot. 2017;119(5):725–35. doi:10.1093/aob/mcw211.
- Hill K, Schaller GE. Enhancing plant regeneration in tissue culture: a molecular approach through manipulation of cytokinin sensitivity. Plant Signal Behav. 2013;8(10):212–24. doi:10.4161/psb.25709.
- Gaspar T, Kevers C, Penel C, Greppin H, Reid DM, Thorpe TA. Plant hormones and plant growth regulators in plant tissue culture. In Vitro Cell Dev Biol-Plant. 1996;32(4):272–89. doi:10.1007/BF02822700.
- Uzair M, Long H, Zafar SA, Patil SB, Chun Y, Li L, Fang J, Zhao J, Peng L, Yuan S, et al. Narrow Leaf21, encoding ribosomal protein RPS3A, controls leaf development in rice. Plant Physiol. 2021;186(1):497–518. doi:10.1093/plphys/kiab075.
- Sugimoto K, Meyerowitz EM. Regeneration in Arabidopsis tissue culture. Methods Mol Biol. 2013;959:265-75. doi:10.1007/978-1-62703-221-6_18. PMID: 23299682
- Xi J, Patel M, Dong S, Que Q, Qu R. Acetosyringone treatment duration affects large T-DNA molecule transfer to rice callus. BMC Biotechnol. 2018;18(1):1–8. doi:10.1186/s12896-018-0459-5.
- Zhan X, Zhang F, Zhong Z, Chen R, Wang Y, Chang L, Bock R, Nie B, Zhang J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol J. 2019;17(9):1814–22. doi:10.1111/pbi.13102.
- Gao Z, Herrera-Carrillo E, Berkhout B. Improvement of the CRISPR-Cpf1 system with ribozyme-processed crRNA. RNA Biol. 2018;15(12):1458–67. doi:10.1080/15476286.2018.1551703.
- Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Nat Acad Sci. 2015;112(11):3570–75. doi:10.1073/pnas.1420294112.
- Wolter F, Puchta H. The CRISPR/Cas revolution reaches the RNA world: cas13, a new Swiss Army knife for plant biologists. Plant J. 2018;94(5):767–75. doi:10.1111/tpj.13899.
- Zhang T, Zheng Q, Yi X, An H, Zhao Y, Ma S, Zhou G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol J. 2018;16(8):1415–23. doi:10.1111/pbi.12881.
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi:10.1126/science.aaf5573.
- Xu C, Zhou, Y., Xiao, Q, et al. Programmable RNA editing with compact CRISPR–Cas13 systems from uncultivated microbes. Nat Methods. 2021;18:499–506.
- Pandita D, Puli COR, Palakolanu SR. CRISPR/Cas13: A Novel and Emerging Tool for RNA Editing in Plants. In: Tang, G., Teotia, S., Tang, X., Singh, D. (eds) RNA-Based Technologies for Functional Genomics in Plants. Concepts and Strategies in Plant Sciences. Springer, Cham. https://doi.org/10.1007/978-3-030-64994-4_14
- Zhang J, Khan SA, Hasse C, Ruf S, Heckel DG, Bock R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science. 2015;347(6225):991–94. doi:10.1126/science.1261680.
- Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 1997;15(1):8–15. doi:10.1007/BF02772108.
- Chen L, Cai Y, Zhou G, Shi X, Su J, Chen G, Lin K. Rapid Sanger sequencing of the 16S rRNA gene for identification of some common pathogens. PloS one. 2014;9(2):e88886. doi:10.1371/journal.pone.0088886.
- Nie X, Singh RP, Singh M. Molecular and pathological characterization of N: o isolates of the Potato virus Y from Manitoba, Canada. Can J Plant Pathol. 2004;26(4):573–83. doi:10.1080/07060660409507178.
- Wang J, Meng F, Chen R, Liu J, Nie X, Nie B. RT-PCR differentiation, molecular and pathological characterization of Andean and ordinary strains of Potato virus S in potatoes in China. Plant Dis. 2016;100(8):1580–85. doi:10.1094/PDIS-11-15-1257-RE.
- Bilgin DD, DeLucia EH, Clough SJ. A robust plant RNA isolation method suitable for Affymetrix GeneChip analysis and quantitative real-time RT-PCR. Nat Protoc. 2009;4(3):333–40. doi:10.1038/nprot.2008.249.