Abstract
N6-carboxymethyl-NAD (N6-CM-NAD) can be used to immobilize NAD onto a substrate containing terminal primary amines. We previously immobilized N6-CM-NAD onto sepharose beads and showed that Thermotoga maritima glycerol dehydrogenase could use the immobilized cofactor with cofactor recycling. We now show that Saccharomyces cerevisiae alcohol dehydrogenase, rabbit muscle L-lactate dehydrogenase (type XI), bovine liver L-glutamic dehydrogenase (type III), Leuconostoc mesenteroides glucose-6-phosphate dehydro-genase, and Thermotoga maritima mannitol dehydrogenase are active with soluble N6-CM-NAD. The products of all enzymes but 6-phospho-D-glucono-1,5-lactone were formed when sepharose-immobilized N6-CM-NAD was recycled by T. maritima glycerol dehydrogenase, indicating that N6-immobilized NAD is suitable for use by a variety of different dehydrogenases. Observations of the enzyme active sites suggest that steric hindrance plays a greater role in limiting or allowing activity with the modified cofactor than do polarity and charge of the residues surrounding the N6-amine group on NAD.
Abbreviations
| NAD | = | nicotinamide adenine dinucleotide |
| N6-CM-NAD | = | N6-carboxymethyl-NAD |
| TmGlyDH | = | Thermotoga maritima glycerol dehydrogenase |
| TmMtDH | = | Thermotoga maritima mannitol dehydrogenase |
| yADH | = | Saccharomyces cerevisiae alcohol dehydrogenase |
| rLDH | = | rabbit muscle type XI L-lactate dehydrogenase |
| bGDH | = | bovine liver type III L-glutamic dehydrogenase |
| LmG6PDH | = | Leuconostoc mesenteroides glucose-6-phosphate dehydrogenase |
| DHA | = | dihydroxyacetone |
Introduction
NAD(P)-dependent dehydrogenases participate in a wide variety of metabolic pathways. They catalyze over 12% of all metabolic reactions in Escherichia coli.Citation1 The requirement that the cofactor be provided in stoichiometric amounts is the greatest limitation to using dehydrogenases as catalysts. Widespread industrial use will require regeneration of the cofactor. Immobilization of the cofactor allows it to be retained in a reaction vessel or recovered more easily, allowing the same cofactor to be used for many cycles. As a proof of concept, we previously synthesized the NAD analog N6-carboxymethyl-NAD (N6-CM-NAD), in which the N6 primary amine in the adenine moiety is now –NH-CH2-COO-, and we immobilized it on sepharose beads using a diamino linker. The immobilized cofactor was tested with Thermotoga maritima glycerol dehydrogenase (TmGlyDH) in 2 simultaneous reactions: glycerol oxidation to dihydroxyacetone (DHA), and hydroxyacetone reduction to (R)-1,2-propanediol. TmGlyDH could use and recycle the immobilized cofactor for more than 1,800 turnovers to allow measurable product accumulation.Citation2
Ottolina and colleagues (1990) used a similar N6 linkage to modify NAD.Citation3 They studied the effects of strong cationic and anionic substituents as well as that of a large polymer (PEG, 20 kDa) on the properties of the cofactor. The eleven enzymes tested generally showed increased Km and decreased Vmax with the modified NADs. Enzyme activity with PEG-NAD varied from as high as 66% of the activity with native NAD to undetectably low. A strong anionic substituent led to a stronger reduction in activity (8–43% of native NAD) than a strong cationic substituent (4–93% of native NAD). The large differences in activity of the various enzymes with the NAD analogs were likely caused by the different shapes and chemistries of these enzymes' NAD binding sites.
NAD(P)-dependent dehydrogenases share a common NAD-binding domain and Rossman fold.Citation4 However, the nature and positioning of residues around the NAD binding site can vary significantly from one enzyme to the next, causing variations in enzymes' affinity for the cofactor, determining which of NADP and NAD is the preferred cofactor, and changing which areas of the cofactor are solvent-accessible. Here we test 5 NAD-dependent dehydrogenases for activity with Sepharose-bead immobilized N6-CM-NAD to address 2 questions. Is our immobilization method suitable for use with a broad set of enzymes? And can structural information on the NAD-binding site and cofactor solvent accessibility be used to predict activity with immobilized N6-CM-NAD? We selected 4 commercially-available NAD-dependent dehydrogenases whose crystal structure in complex with NAD is known, as well as T. maritima mannitol dehydrogenase (TmMtDH), which we had studied previously.Citation5
Results and Discussion
The Sigma-Aldrich website was searched for commercially available dehydrogenases and NAD-dependent oxidoreductases. The list of enzymes generated was then cross-referenced against the Protein Data Bank (PDB). Of the 10 enzymes that had a crystal structure in complex with NAD, 4 () were selected and purchased from Sigma-Aldrich for our study. TmMtDH was added to the study because it was already available as a purified, recombinant enzyme in our laboratory.Citation5 A 3D model of TmMtDH in complex with NAD was built ().
Table 1. Enzymes used in this study.
Figure 1. Zoom view of enzyme-bound NAD (8-iodo-NAD for yADH). A. TmMtDH, B. TmGlyDH, C. yADH, D. LmG6PDH, E. rLDH, F. bGDH. PDB numbers are listed in . Several 3D models of TmMtDH were generated using Modeler softwareCitation8 and the online homology modeling server I-TASSER.Citation9-11 The different modeling approaches used single and multiple templates that each showed over 25% identity and below 10% gaps in alignments with TmMtDH. Models were analyzed using the scoring methods DOPE, DFIRE, and OPUS.Citation12-14 The best model was produced by I-TASSER using the structures of the silverleaf whitefly sorbitol dehydrogenase (PDB # 1E3J), human sorbitol dehydrogenase (PDB # 1PL8), Sulfolobus solfataricus glucose dehydrogenase (PDB # 2CDC), Thermus thermophilus threonine 3-dehydrogenase (PDB # 2DQ4), and mouse class II alcohol dehydrogenase (PDB # 1E3I) as templates. The structure was minimized using the CHARMM force fieldCitation15 and NAD was imported into TmMtDH's active site using the coordinates of NAD in human sorbitol dehydrogenase. Enzyme surfaces were visualized using The PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC.
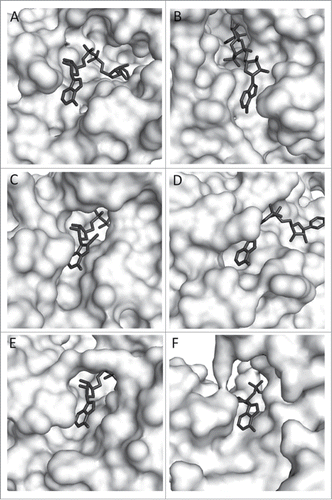
The adenine moiety binding portion of the NAD binding pocket of the 5 enzymes and of TmGlyDH was visualized (). The residues and pocket structure around the adenine varied greatly among the enzymes. The configuration of NAD is similar in TmMtDH, yADH, rLDH, bGDH, and LmG6PDH, with the adenine's N6-amine pointing toward the solvent. In TmGlyDH, NAD's adenine moiety is flipped, with the N6-amine pointing toward the enzyme's surface. Since NAD's N6-amine is less solvent exposed in TmGlyDH than in the other enzymes, we expected TmGlyDH to have the lowest activity with N6-CM-NAD. The specific activity of the mesophilic enzymes with NAD and N6-CM-NAD and the Vmax values with the cofactor and analog of TmGlyDH and TmMtDH were compared (). TmMtDH's Vmax with N6-CM-NAD was almost 70% of that with NAD and the Km increased only 1.8-fold. In contrast, TmGlyDH's Vmax with N6-CM-NAD was only 2% of that with NAD and the Km increased 15-fold. With the exception of LmG6PDH, the N6-amine solvent-accessible area () correlated well (R2 = 0.95 for linear regression, not shown) with the percent activity with N6-CM-NAD () (R2 = 0.61 when LmG6PDH is included). In contrast, the type of residues (i.e., charged, polar, or non-polar) surrounding the N6-amine () showed no relationship to activity with N6-CM-NAD ().
Table 2. Specific activity of the selected enzymes with 100 μM NAD and N6-CM-NAD and enzyme ranking by polarity/charge and openness of the area surrounding NAD's N6-amine. Specific activities were tested at 25°C by following the increase in A340nm in quartz cuvettes containing 100 mM substrate (L-lactate, glucose-6-phosphate, L-glutamate, or ethanol), 100 μM NAD or N6-CM-NAD in 50 mM sodium phosphate (pH 7.5). Reactions were started by adding 1.25 μg (rLDH), 1.08 μg (yADH) 0.201 μg (LmG6PDH), or 4.22 μg (bGDH) enzyme. Using the crystal structures or 3D models of the enzymes visualized in PyMOL, amino acid side-chains and backbone groups within 5 Å of the 6N amine were assessed for polarity and charge. The N6-amine solvent-accessible area was calculated using PyMOL.
To test enzyme activity with Sepharose-bound N6-CM-NAD, we initially tested bGDH as the cofactor-recycling enzyme in recycling reactions between rLDH and bGDH and between LmG6PDH and bGDH. In these recycling reactions, rLDH and LmG6DH were set to reduce the cofactor, and bGDH to oxidize it. Neither of the 2 reactions yielded any product. We could not tell whether none of the enzymes used the immobilized N6-CM-NAD, or if the recycling enzyme (bGDH) was causing the problem. Note that bGDH showed very low specific activity with NAD in the conditions tested (). For these reasons, we repeated the recycling reactions with TmGlyDH as the cofactor-recycling enzyme. Even though TmGlyDH is poorly active at 25°C, we knew it to be active with Sepharose-bound N6-CM-NAD.Citation2 Recycling reactions were set between TmGlyDH and each of yADH, rLDH, bGDH, and TmMtDH, where yADH, rLDH, bGDH, and TmMtDH oxidized the cofactor and TmGlyDH reduced it while oxidizing glycerol to DHA. Reaction progress was monitored by measuring DHA accumulation with high-performance liquid chromatography. Because LmG6PDH's only commercially available substrate is glucose-6-phosphate, the recycling reaction between LmG6PDH and TmGlyDH was set with TmGlyDH producing NAD+ during DHA reduction to glycerol, and reaction progress was monitored by measuring glycerol accumulation by high-performance liquid chromatography. No product accumulation was observed in the LmG6PDH-TmGlyDH reaction (not shown). Even though LmG6PDH has the highest specific activity with soluble N6-CM-NAD of the 6 enzymes tested (), LmG6PDH could be inactive with immobilized N6-CM-NAD. Because TmGlyDH is inhibited by its products (Beauchamp, unpublished results), glycerol added as part of the LmG6PDH suspension (initial glycerol concentration in the reaction estimated at 138 mM) could also have inhibited glycerol production by TmGlyDH, preventing cofactor recycling. Product accumulation was observed for the recycling reactions involving yADH, rLDH, bGDH, and TmMtDH (). Although the solvent-accessible area of the N6-amine correlated well with activity for soluble N6-CM-NAD (), no such relationship was apparent after immobilization on Sepharose, with only small differences in DHA accumulation observed between the different reactions. Regardless, our results indicate that many different enzymes can use Sepharose-immobilized N6-CM-NAD as their cofactor.
Figure 2. A. DHA accumulation during recycling reactions with yADH (○), and bGDH (•). B. DHA accumulation during recycling reactions with rLDH (□) and TmMtDH (▪). Glycerol oxidation to DHA by TmGlyDH was used to regenerate NADH in the recycling reactions. Recycling reactions were set at 25°C for all mesophilic enzymes tested and at 50°C for TmMtDH. Reactions contained 200 mM glycerol (substrate for TmGlyDH) and 100 mM acetaldehyde, pyruvate, α-ketoglutarate and NH4Cl, or fructose , as well as 45 mg Sepharose-N6-CM-NAD in 50 mM sodium phosphate (pH 7.5) (yADH, bGDH, and TmMtDH) or 50 mM Tris-HCl (pH 7.5) (rLDH). Seventeen units (one unit = amount of enzyme required to produce 1 μmol of NAD(H) per min) of TmGlyDH were used to regenerate NADH for the mesophilic enzymes (8.5 units each), and 8.5 units TmGlyDH were used to regenerate NADH for TmMtDH (4.25 units). One mM ADP was added to the bGDH reaction to activate the enzyme. Samples were collected at increasing time points over a 24 hour period, and products were quantified using a Breeze high-performance liquid chromatograph (Waters, Milford, MA) equipped with an Aminex-87C carbohydrate analysis column (Bio-Rad, Hercules, CA).
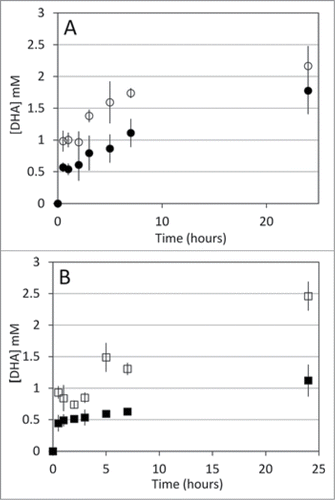
Note that we used TmGlyDH, the enzyme with the least activity with soluble N6-CM-NAD (), as the cofactor-recycling enzyme. TmMtDH, bGDH, and yADH showed the highest relative activities with soluble N6-CM-NAD (). bGDH used in combination with TmMtDH as the cofactor-recycling enzyme produced 2.6 times more product than in combination with TmGlyDH (data not shown), suggesting that poor activity of TmGlyDH with immobilized N6-CM-NAD could have limited product accumulation. In contrast, the yADH/TmMtDH combination produced only 20% as much product as the yADH/TmGlyDH combination. The reason behind this poor yield is unknown, since the 2 enzymes were not inhibited by each other's substrate (data not shown).
Conclusion
Our work demonstrates that N6-CM-NAD immobilized onto a large particle, such as Sepharose, can be used by a variety of enzymes. NAD's N6-amine solvent-accessible area in an enzyme·NAD complex is a good indicator of whether that enzyme will be active with N6-CM-NAD. The possibility of one enzyme's substrate inhibiting the enzyme used in combination highlights the need for an alternative cofactor regeneration method. Electrochemical regeneration could be a good alternative but requires efficient methods for cofactor and enzyme immobilization on electrodes. N6-CM-NAD could be used to create a bioelectronics linkage similar to that previously developed by Hassler and Worden.Citation6 They developed a renewable bioelectronics interface with the enzyme and cofactor immobilized on an electrode surface. The immobilization method used relied on a boronate linkage to NADP's cis-diol. The main limitation of the boronate linkage was that this linkage is relatively weak with a dissociation constant (Kd) of 5.9×10–3, and boronate shows high affinity for other cis-diols found on the substrates of many dehydrogenases.Citation7 The enzyme substrates would displace the cofactor from the electrode surface. The N6-immobilized NAD tested here would allow a new linkage to be produced that is stable in the presence of the enzyme substrates and products.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Phillip G. Gross for contributing to the kinetic studies of TmGlyDH. We thank Dr. Srinivasa Gopal for helping with CHARMM minimization and DOPE, DFIRE, and OPUS analysis of the TmMtDH model, and Dr. Kaillathe Padmanabhan for his help with PyMOL and solvent-accessible area determination.
Funding
This work was supported by grant # CBET 0756703 from the National Science Foundation, grant # 2008–35504–04611 from the United States Department of Agriculture Cooperative State Research, Education, and Extension Service National Research Initiative, grant # 2010–04061 from the United States Department of Agriculture National Institute for Food and Agriculture's Sustainable Bioenergy Research Program, and by Michigan State University startup funds.
References
- Reed JL, Vo TD, Schilling CH, Palsson BO. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 2003; 4:R54; PMID:12952533; http://dx.doi.org/10.1186/gb-2003-4-9-r54
- Beauchamp J, Gross PG, Vieille C. Characterization of Thermotoga maritima glycerol dehydrogenase for the enzymatic production of dihydroxyacetone. Appl Microbiol Biotechnol 2014; 98:7039-50; PMID:24664447; http://dx.doi.org/10.1007/s00253-014-5658-y
- Ottolina G, Carrea G, Riva S, Buckmann AF. Coenzymatic properties of low molecular-weight and macromolecular N6-derivatives of NAD+ and NADP+ with dehydrogenases of interest for organic synthesis. Enzyme Microb Technol 1990; 12:596-602; PMID:1366782; http://dx.doi.org/10.1016/0141-0229(90)90133-B
- Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J Mol Biol 1973; 76:241-56; PMID:4737475; http://dx.doi.org/10.1016/0022-2836(73)90388-4
- Song SH, Ahluwalia N, Leduc Y, Delbaere LT, Vieille C. Thermotoga maritima TM0298 is a highly thermostable mannitol dehydrogenase. Appl Microbiol Biotechnol 2008; 81:485-95; PMID:18719905; http://dx.doi.org/10.1007/s00253-008-1633-9
- Hassler BL, Kohli N, Zeikus JG, Lee I, Worden RM. Renewable dehydrogenase-based interfaces for bioelectronic applications. Langmuir 2007; 23:7127-33; PMID:17503864; http://dx.doi.org/10.1021/la7004437
- Bailon P, Ehrlich GK, Fung W-J, Berthold W, eds. Methods in Molecular Biology: Affinity Chromatography methods and protocols. Totowa NJ: Taylor & Francis, 2000.
- Sali A. Comparative protein modeling by satisfaction of spatial restraints. Mol Med Today 1995; 1:270-7; PMID:9415161; http://dx.doi.org/10.1016/S1357-4310(95)91170-7
- Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins 2007; 69 Suppl 8:108-17; PMID:17894355; http://dx.doi.org/10.1002/prot.21702
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008; 9:40; PMID:18215316; http://dx.doi.org/10.1186/1471-2105-9-40
- Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol 2007; 5:17; PMID:17488521; http://dx.doi.org/10.1186/1741-7007-5-17
- Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci 2006; 15:2507-24; PMID:17075131; http://dx.doi.org/10.1110/ps.062416606
- Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci 2002; 11:2714-26; PMID:12381853; http://dx.doi.org/10.1110/ps.0217002
- Lu M, Dousis AD, Ma J. OPUS-PSP: an orientation-dependent statistical all-atom potential derived from side-chain packing. J Mol Biol 2008; 376:288-301; PMID:18177896; http://dx.doi.org/10.1016/j.jmb.2007.11.033
- Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J Comput Chem 2009; 30:1545-614; PMID:19444816; http://dx.doi.org/10.1002/jcc.21287
