Abstract
A glycoside hydrolase family 32 exo-inulinase gene was cloned from Arthrobacter sp. HJ7 isolated from saline soil located in Heijing town. The gene encodes an 892-residue polypeptide with a calculated mass of 95.1 kDa and a high total frequency of amino acid residues G, A, and V (30.0%). Escherichia coli BL21 (DE3) cells were used as hosts to express the exo-inulinase gene. The recombinant exo-inulinase (rInuAHJ7) showed an apparently maximal activity at pH 5.0–5.5 and 40–45°C. The addition of 1.0 and 10.0 mM Zn2+ and Pb2+ had little or no effect on the enzyme activity. rInuAHJ7 exhibited good salt tolerance, retaining more than 98% inulinase activity at a concentration of 3.0%–20.0% (w/v) NaCl. Fructose was the main product of inulin, levan, and Jerusalem artichoke tubers hydrolyzed by the enzyme. The present study is the first to report the identification and characterization of an Arthrobacter sp exo-inulinase showing a high molecular mass of 95.1 kDa and NaCl tolerance. These results suggest that the exo-inulinase might be an alternative material for potential applications in processing seafood and other foods with high saline contents, such as marine algae, pickles, and sauces.
Keywords:
Introduction
Inulin, a reserve polysaccharide of plant origin, is a linear chain composed of multiple d-fructofuranose units linked primarily by β-(2,1)-glycosidic bonds and a terminal glucose residue linked by α-(1,2)-glycosidic bond.Citation1 Inulin is an important agricultural product planted in China, and it is found in the roots and tubers of many plants such as Jerusalem artichoke, Cichorium endivia, Dahlia pinnata, and Arctium lappa.Citation1 Inulin has the potential to be a renewable raw material for ultra-high-fructose syrup or fructo-oligosaccharide production by inulinase hydrolysis.Citation1,2 These hydrolysis products can be further converted into products with higher margins by fermentation, such as butanediol, lactic acid, sugar alcohols, bioethanol, single-cell oil, single-cell protein, and citric acid.Citation3
Exo-inulinases (EC 3.2.1.80; fructan β-fructosidases, exo-β-d-fructosidases) hydrolyze terminal, non-reducing d-fructofuranose residues linked by β-(2,1)-glycosidic bonds. As such, exo-inulinases are capable of efficiently hydrolyzing inulin to produce fructose yields as high as 90%–95%.Citation4 In addition, many exo-inulinases from bacteria also hydrolyze levan, sucrose, raffinose, stachyose, and starch.Citation1,2,5 Based on amino acid sequence similarities, exo-inulinases are classified into the glycoside hydrolase (GH) family 32.Citation6
Enzymes obtained from extremophilic sources have numerous potential applications. We previously sampled the saline soil from an abandoned salt mine located in Heijing town, aka the ‘‘town of salt’’, in Yunnan Province, China.Citation7 The saline soil provides novel genetic resources and salt-tolerant xylanases that are active and stable at high concentrations of NaCl and could be used in harsh industrial processes.Citation7,8 In addition, food processing, biosynthesis, or fermentation under high-salt conditions can reduce total costs by removing the sterilization process.Citation9 In this study, an exo-inulinase gene from Arthrobacter sp. HJ7 was cloned and expressed in Escherichia coli BL21 (DE3). The recombinant enzyme showed a molecular mass of approximately 91.5 kDa and salt tolerance.
Results
Strain identification
Comparison of the partial 16S rDNA sequence from strain HJ7 (710 bp; JQ863112) with that in GenBank showed nucleotide identity of 99% with Arthrobacter globiformis (Accession No. NR_112192), Arthrobacter humicola (NR_041546), Arthrobacter pascens (NR_026191), and Arthrobacter oxydans (NR_026236). Thus, strain HJ7 was classified under the genus Arthrobacter.
Gene cloning and sequence analysis
A fragment of inuAHJ7 (360 bp) was amplified by PCR using the degenerate primers eInu32F and eInu32R. DNA fragments amplified by TAIL-PCR were assembled with the inuAHJ7 fragment. The results showed that the open reading frame of inuAHJ7 (JQ863113, 2,679 bp) starts with the initiation codon ATG, ends with the termination codon TAG, and encodes an 892-residue polypeptide with a calculated mass of 95.1 kDa.
The signal peptide was predicted from M1 to A41 followed by a catalytic domain of GH 32 from H55 to G835 of InuAHJ7. The consensus pattern H-x(2)-[PV]-x(4)-[LIVMA]-N-D-P-N-[GA] (http://prosite.expasy.org/PS00609) of GH 32 was also detected in InuAHJ7 (HYTPQQNWMNDPNG in ). The two amino acids D65 and E248 (), corresponding to D41 and E241 of the exo-inulinase from Aspergillus awamori var. 2250 (1Y4W),Citation10 are the putative nucleophile and catalytic acid/base, respectively.
Figure 1. Partial amino acid sequence alignment of InuAHJ7 with GH 32 exo-inulinases. Sequence names are shown with accession numbers, except InuAHJ7, as follows: exo-inulinases from A. awamori var. 2250 (1Y4W),Citation10 Paenibacillus polymyxa ZJ-9 (AHN08014),Citation21 Geobacillus stearothermophilus KP1289 (BAC45010),Citation15 and Penicillium sp. TN-88 (BAC16218).Citation22 Identical residues are shaded in black and conserved residues are shaded in gray. The black bars indicate blocks used for designing degenerate primers. The asterisks show the putative catalytic residues.
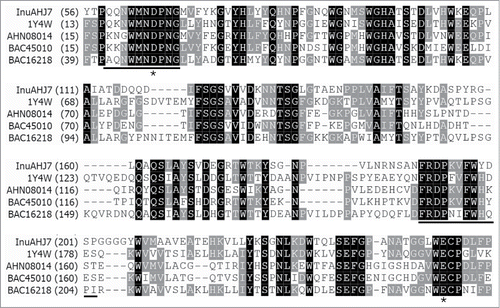
BLASTP results revealed that InuAHJ7 was most similar (57%–92% identity) to a number of putative GH 32 glycosidases with high molecular masses obtained by genome sequencing of Arthrobacter spp., such as the putative glycosidases from Arthrobacter globiformis NBRC 12137 (Accession Number WP_003799354; 825 residues), Arthrobacter sp 162MFSha1.1 (WP_018769218; 902 residues), and Arthrobacter sp. M2012083 (WP_020372846; 867 residues). Aligned with the characterized GH 32 enzymes, such as the GH 32 levanase from Actinomyces naeslundii T14V (AAA67876),Citation11 InuAHJ7 showed less than 45% identity.
Expression, purification, and identification of rInuAHJ7
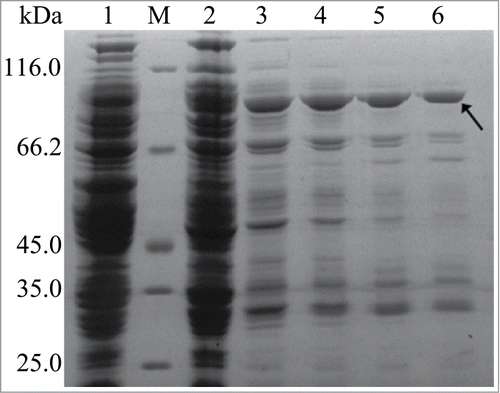
rInuAHJ7 was effectively expressed in E. coli BL21 (DE3). SDS-PAGE indicated that the recombinant enzyme could not be purified to electrophoretic homogeneity (). However, elution with 500 mM imidazole led to the reduction of most non-specific proteins and a distinct band with a molecular mass of approximately 91.5 kDa (), which is close to the calculated value of rInuAHJ7 (93.1 kDa). The MALDI-TOF MS spectrum of the specific band matched the molecular mass of the internal peptides of rInuAHJ7 (Fig. S1), confirming that the protein in the band was indeed rInuAHJ7. Therefore, the impure rInuAHJ7 eluted by 500 mM imidazole was used for further studies.
Figure 2. SDS-PAGE analysis of rInuAHJ7. Lanes: 1, cell extract from an induced transformant harboring the empty plasmid pEASY-E1; M, low-molecular weight marker; 2, crude rInuAHJ7; 3, 4, 5, and 6, rInuAHJ7 purified by Ni2+-NTA chelating affinity chromatography with imidazole gradient of 100, 200, 300, and 500 mM, respectively. The arrow indicates the band cut for MALDI-TOF MS analysis.
Enzyme characterization
The activities of the impure rInuAHJ7 (determined at pH 5.3 and 45°C) toward substrates of 0.5% (w/v) inulin, levan, sucrose, raffinose, and stachyose were 44.40 ± 0.64, 35.63 ± 5.07, 23.53 ± 0.52, 18.61 ± 0.68, and 9.15 ± 0.29 U/mL, respectively. Consequently, the activity ratio of inulinase/sucrose was approximately 1.9, and the result revealed that rInuAHJ7 is an inulinase.Citation1
When assayed at 37°C, rInuAHJ7 showed an apparently optimal inulinase activity over the range of pH 5.0–5.5, and exhibited over 60% of its maximal inulinase activity at pH 4.5–7.0 (). The enzyme was stable in the pH range 6.0–10.0; it retained more than 70% of its initial activity after 60 min pre-incubation ().
Figure 3. Characterization of rInuAHJ7. (A) Effect of pH on rInuAHJ7. (B) pH stability of rInuAHJ7. (C) Thermoactivity of rInuAHJ7. (D) Thermostability assay. The error bars represent the mean ± SD (n = 3).
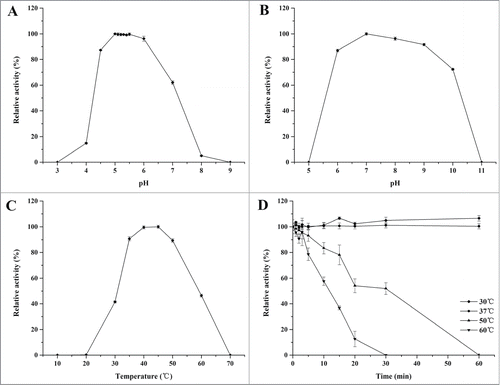
The activity of rInuAHJ7 was apparently optimal at 40°C to 45°C (). The enzyme was stable at 30°C and 37°C for more than 60 min. Half-lives of the enzyme were approximately 30 and 10 min at 50°C and 60°C, respectively ().
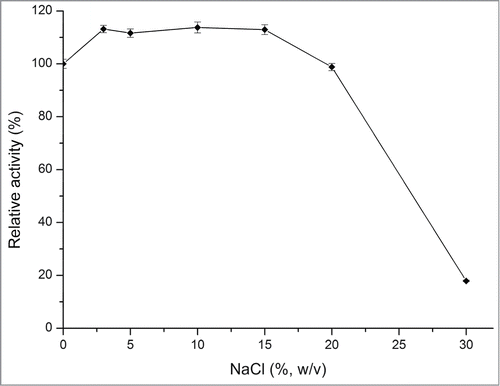
The inulinase activity of rInuAHJ7 was strongly or partially inhibited by 1.0 and 10.0 mM Hg2+, SDS, and EDTA, as well as 30% (w/v) NaCl (retaining less than 63.8% activity). Meanwhile, the addition of other reagents, including Zn2+ and Pb2+ had little or no effect on enzyme activity (retaining 75.0%–112.2% activity) (; ). Notably, rInuAHJ7 exhibited good salt tolerance, retaining more than 98% inulinase activity at a concentration of 3.0%–20.0% (w/v) NaCl ().
Table 1 Primers used in this study
Table 2 Effect of metal ions and organic reagents on the activity of rInuAHJ7.
Hydrolysis products
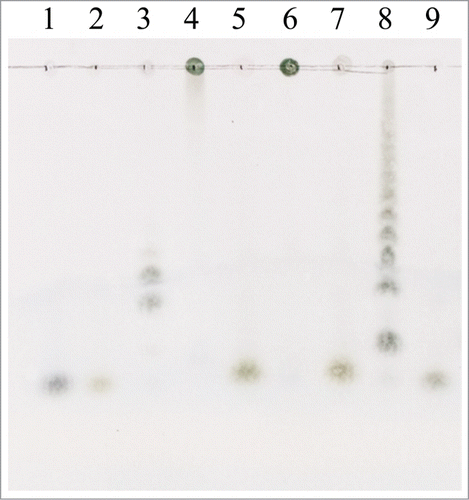
The hydrolysis products of 0.5% (w/v) inulin, 0.5% (w/v) levan, and 10.0% (w/v) Jerusalem artichoke tuber were analyzed by TLC (). Fructose was the primary product released from fructans and fructo-oligosaccharides in these substrates. The results revealed the exo-acting mode of rInuAHJ7.
Figure 5. TLC chromatograms of the hydrolysis products of 0.5% (w/v) inulin, 0.5% (w/v) levan, and 10.0% (w/v) Jerusalem artichoke tuber powder. Lanes: 1, glucose; 2, fructose; 3, fructo-oligosaccharide mixture (kestose, nystose, and fructofuranosylnystose); 4, 6, and 8, inulin, levan, and artichoke powder with the inactivated (90°C for 5 min) rInuAHJ7, respectively; 5, 7, and 9, inulin levan, and artichoke powder hydrolyzed by rInuAHJ7 for 3 h, respectively.
Discussion
Several studies have been published on salt-tolerant enzymes,Citation5,7,8 including the salt-tolerant exo-inulinase InuAMN8 from Arthrobacter sp MN8, which shows 127.9%–88.4% activity at concentrations between 3.5% and 15.0% (w/v) NaCl and 49.8% activity at 20.0% (w/v) NaCl.Citation5 However, among the published studies to date, salt-tolerant inulinases have rarely been reported.Citation5 The inulinase from Aspergillus fumigatus MTCC 3009 retains 77% activity in the presence of approximately 0.006% (w/v) NaCl,Citation12 and the inulinase from Kluyveromyces marxianus var bulgaricus retains 50% activity in the presence of approximately 0.3% (w/v) NaCl.Citation13 In this study, rInuAHJ7 retained even higher relative activity (98.8%) at high NaCl concentration of 20.0% (w/v) than the salt-tolerant exo-inulinase InuAMN8. The results indicate that rInuAHJ7 is a novel salt-tolerant exo-inulinase.
Compared with their homologs, salt-tolerant enzymes have a higher proportion of amino acid residues G, A, and V.Citation14 The total frequencies of amino acid residues G, A, and V of 15 characterized inulinases are over the range of 15.4%–26.6% (Accession Numbers BAC45010,Citation15 AAF44125,Citation16 CAA04518,Citation17 AIO03624,Citation18 AAD27873,Citation19 AAL82575,Citation20 AHN08014,Citation21 BAC16218,Citation22 ACC61059,Citation23 CAC44220,Citation10 AEO12135,Citation24 CAB14645,Citation25 BAD88632,Citation26 BAL70276,Citation27 and AAK00768Citation28). Compared with these inulinases, the salt-tolerant exo-inulinases InuAMN8 (JQ863111, 29.5%)Citation5 and InuAHJ7 (30.0%) present higher proportions of amino acid residues G, A, and V. The results suggest that amino acid residues G, A, and V may contribute to the salt tolerances of inulinases.
Many exo-inulinases are inhibited by 1 or 5 mM Zn+ and 5 mM Pb2+ Citation16,17,19,20,24,26,29 except the exo-inulinases from Bacillus subtilis subsp. 168,Citation25 Penicillium sp TN-88,Citation22 and InuAMN8.Citation5 Like the salt-tolerant exo-inulinase InuAMN8, which also shows resistance to both lead and zinc,Citation5 rInuAHJ7 retained 99.6% and 90.8% activity in the presence of 10.0 mM Zn2+ and Pb2+, respectively.
To our knowledge, most characterized exo-inulinases have a molecular mass lower than 60 kDa. For example, the molecular masses of 9 characterized inulinases are over the range of 49.8–59.1 kDa (Accession Numbers CAA04518,Citation17 BAL70276,Citation27 BAC45010,Citation15 AAK00768,Citation28 AHN08014,Citation21 ACC61059,Citation23 AAF44125,Citation16 AIO03624,Citation18 and CAC44220Citation10). Two reports are available on exo-inulinases from Penicillium janthinellum B01Citation24 and Lactobacillus casei IAM 1045Citation26 with molecular masses of 74.8 and 138.8 kDa, respectively. Characterization of the 2 exo-inulinases reveals that they are inhibited by 1 mM Zn2+,Citation24,26 and the exo-inulinase from P. janthinellum B01 is also inhibited by 1 mM Na+.Citation24 Compared with the above exo-inulinases, InuAHJ7 is different: the exo-inulinase has a molecular mass of 95.1 kDa and shows tolerance to 3.0%–20.0% (w/v) Na+ and 1.0 and 10.0 mM Zn2+ and Pb2+.
Conclusion
The present study is the first to report the identification and characterization of an Arthrobacter sp. exo-inulinase that has a high molecular mass of 95.1 kDa and shows tolerance to 3.0%–20.0% (w/v) Na+ and 1.0 and 10.0 mM Zn2+ and Pb2+. Considering its NaCl-tolerance, the exo-inulinase has potential uses in processing seafood and other foods with high saline contents, such as marine algae, pickles, and sauces.
Materials and Methods
Bacterial, vectors, and reagents
The strain HJ7 was isolated from the saline soil located in Heijing town. The method of strain isolation has been described in our previous study.Citation7 The taxon of HJ7 was identified by 16S rDNA sequence PCR-amplified using primers 27F and 1492R ().
The pMD 18-T vector from TaKaRa (6011) and the E. coli Trans1-T1 from TransGen (CD501) were used for gene cloning. The pEASY-E1 Expression Kit (CE101) and E. coli BL21 (DE3) (CD601) from TransGen were used for gene expression. Genomic DNA isolation (DP302), DNA purification (DP209), and plasmid isolation kits (DP103) were purchased from Tiangen. Nickel-NTA agarose from Qiagen (30210) was used for His-tagged protein purification. DNA polymerases Taq (R500A) and LA Taq (RR52A) and dNTPs (4030) were purchased from TaKaRa. Inulin was purchased from J&K (543181), levan from Advanced Technology & Industrial (1151239), stachyose from TCI (S0397), raffinose from Sigma (R0514), fructo-oligosaccharides set including kestose, nystose, and fructofuranosylnystose from Wako Pure Chemical (062-02381), and TLC Silica gel 60 plates from Merck KGaA (HX394049). All other reagents are analytical grade and commercially available.
Gene cloning
The genomic DNA of strain HJ7 was extracted using the Tiangen genomic DNA isolation kit following the manufacturer's instructions. The degenerate primer set eInu32F and eInu32R (),Citation5 corresponding to the conserved blocks—H-N-W-M-N-D-P-N-G and R-D-P-K-V-F-W-H-E-Q-S of GH 32 exo-inulinases (), respectively, was used to clone a partial exo-inulinase gene. The full-length exo-inulinase gene (inuAHJ7) was amplified by improved TAIL-PCR (thermal asymmetric interlaced-PCR) method described in a previous study.Citation30 The nested insertion-specific (SP) primers (inuAHJ7uSP1, inuAHJ7uSP2, inuAHJ7dSP1, inuAHJ7dSP2, inuAHJ7dSP3, and inuAHJ7dSP4) and annealing temperatures used for high-stringency step in TAIL-PCR are shown in . The PCR product was gel purified, ligated to pMD 18-T vector, transformed into E. coli Trans1-T1, and sequenced by Beijing Genomics Institute (Guangzhou, China).
Sequence analysis
The amino acid sequence (InuAHJ7) deduced from inuAHJ7 was loaded to SignalP (http://www.cbs.dtu.dk/services/SignalP/) for signal peptide analysis. The identity values of DNA and protein sequences were calculated with BLASTN and BLASTP programs, respectively (http://blast.ncbi.nlm.nih.gov/Blast/). The classification of the glycoside hydrolase family of protein was determined with the dbcan online tool (http://csbl.bmb.uga.edu/dbCAN/annotate.php). Other sequential analyses were performed using Vector NTI 10.3 software (InforMax, Gaithersburg, MD, USA).
Expression of inuAHJ7 in E. coli
The CDS of inuAHJ7 was amplified by PCR using LA Taq DNA polymerase and the primers rinuAHJ7EF and rinuAHJ7ER (). The resulting PCR product was directly cloned into the vector pEASY-E1 by T-A ligation. The recombinant plasmid (pEASY-E1-inuAHJ7) was transformed into E. coli BL21 (DE3) competent cells. Then, the positive transformants were identified by PCR analysis and confirmed by DNA sequencing. The transformant harboring pEASY-E1-inuAHJ7 was picked up from a single colony and grown overnight at 37°C in LB medium containing 0.1 mg/mL ampicillin. The culture was then inoculated at 1:100 dilution into culture-complex auto-inducing media (CAI) Citation31 containing ampicillin and was grown at 25°C for 24 h.
Purification and identification of recombinant inulinase
To purify recombinant InuAHJ7 (rInuAHJ7) tagged with His6, the cells were harvested by centrifugation at 5,000×g at room temperature for 5 min and resuspended in McIlvaine buffer (pH 7.0). The cells were disrupted by sonication (7 s, 150 W) on ice several times and centrifuged at 13,000×g for 10 min at 4°C. The supernatant was applied to a Ni2+-NTA agarose gel column for purification with a linear imidazole gradient of 20–500 mM in buffer A [20 mM Tris–HCl, 0.5 M NaCl, 10% (v/v) glycerol, pH 7.2].
The purified protein was detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10% running gel), and the in-gel band was cut and verified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) performed by Tianjin Biochip (Tianjin, China).
Enzyme assay
Exo-inulinase activity was determined by measuring the release of reducing sugars from the substrate. The standard reaction consisted of 900 μL of 0.5% (w/v) substrate in Mcllvaine buffer (pH 5.3) and 100 μL of appropriately diluted enzyme. After incubation at 37°C for 10 min, the reaction was stopped with 1.5 mL of 3,5-dinitrosalicylic acid (DNS) reagent and the mixture was boiled at 100°C for 5 min to produce a measurable reddish brown (540 nm) product. One unit (U) of exo-inulinase activity was defined as the amount of enzyme that catalyzed the formation of 1.0 μmol of reducing sugars equivalent to d-fructose per minute. The enzyme activity was assayed by following this standard procedure unless otherwise noted.
Biochemical characterization
To identify the substrate specificity of rInuAHJ7, 0.5% (w/v) inulin, levan, sucrose, raffinose, or stachyose (determined at pH 5.3 and 45°C) was added to each reaction solution. Any further biochemical characterizations were determined using inulin as the substrate.
The optimal pH for the inulinase activity was determined at 37°C in different buffers, with pH ranging from 3.0 to 9.0. The pH stability of the enzyme were determined by incubating the enzyme solution in various buffers at 37°C for 1 h, and the residual enzyme activity was measured under the standard assay conditions. The buffers used were McIlvaine buffer for pH 3.0–8.0 and 0.1 M glycine–NaOH for pH 9.0–11.0.
The thermoactivity of rInuAHJ7 was determined over the range of 10–70°C in McIlvaine buffer (pH 5.3). Thermostability of the enzyme was determined under standard assay conditions following pre-incubation of the enzyme for 60 min at 30°C, 37°C, 50°C, or 60°C, with the untreated enzyme defining 100% activity.
The effects of different metal ions and organic reagents on inulinase activity were determined in McIlvaine buffer (pH 5.3) at 45°C. We added 1.0 and 10.0 mM (final concentration) CaCl2, NiSO4, CoCl2, MgSO4, KCl, ZnSO4, FeCl3, Pb(CH3COO)2, MnSO4, FeSO4, HgCl2, EDTA, β-mercaptoethanol, or SDS, or 3.0–30.0% (w/v) NaCl to the reaction solution. The reaction without metal ions or organic reagents was used as a control.
Hydrolysis products
Jerusalem artichoke tubers (purchased from a local market) were sliced, sun-dried, and then ground into powder form. The hydrolysis of inulin (0.5%, w/v; pH 5.3), levan (0.5%, w/v; pH 5.3), and artichoke powder (10.0%, w/v; pH 5.3) was performed with the reaction system of 1.0 U/mL rInuAHJ7 at 45°C for 3 h. The hydrolysis products were analyzed by thin layer chromatography (TLC) as previously described.Citation5 Glucose, fructose, and fructo-oligosaccharide mixture (kestose, nystose, and fructofuranosylnystose) were used as standards. Inulin, levan, or artichoke powder with the inactivated rInuAJB13 (90°C for 5 min) was used as a control.
Nucleotide sequence accession numbers
The nucleotide sequences for the Arthrobacter sp HJ7 16S rDNA and exo-inulinase gene (inuAHJ7) were deposited in GenBank under the accession numbers JQ863112 and JQ863113, respectively.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest were disclosed.
Figure_S1.pdf
Download PDF (161.5 KB)Funding
This work was supported by the Key Technologies Research and Development Program of China (2013BAD10B01), the National Nature Science Foundation of China (31260215), and the Science Research Foundation of Yunnan Provincial Education Committee (2013Y432).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Kango N, Jain SC. Production and properties of microbial inulinases: recent advances. Food Biotechnol 2011; 25:165-212; PMID:24793124; http://dx.doi.org/10.1080/08905436.2011.590763
- Vijayaraghavan K, Yamini D, Ambika V, Sowdamini NS. Trends in inulinase production-a review. Crit Rev Biotechnol 2009; 29:67-77; PMID:19514896; http://dx.doi.org/10.1080/07388550802685389
- Chi ZM, Zhang T, Cao TS, Liu XY, Cui W, Zhao CH. Biotechnological potential of inulin for bioprocesses. Bioresource Technol 2011; 102:4295-303; PMID:21247760; http://dx.doi.org/10.1016/j.biortech.2010.12.086
- Lima DM, Fernandes P, Nascimento DS, Ribeiro R, de Assis SA. Fructose syrup: a biotechnology asset. Food Technol Biotech 2011; 49:424-34
- Zhou JP, Lu Q, Peng MZ, Zhang R, Mo MH, Tang XH, Li JJ, Xu B, Ding JM, Huang ZX. Cold-active and NaCl-tolerant exo-inulinase from a cold-adapted Arthrobacter sp. MN8 and its potential for use in the production of fructose at low temperatures. J Biosci Bioeng 2015; 119:267-74; PMID:25266375; http://dx.doi.org/10.1016/j.jbiosc.2014.08.003
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 2009; 37:D233-8; PMID:18838391; http://dx.doi.org/10.1093/nar/gkn663
- Zhou JP, Gao YJ, Dong YY, Tang XH, Li JJ, Xu B, Mu YL, Wu Q, Huang ZX. A novel xylanase with tolerance to ethanol, salt, protease, SDS, heat, and alkali from actinomycete Lechevalieria sp. HJ3. J Ind Microbiol Biot 2012; 39:965-75; PMID:22430498; http://dx.doi.org/10.1007/s10295-012-1113-1
- Zhou JP, Wu Q, Zhang R, Mo MH, Tang XH, Li JJ, Xu B, Ding JM, Lu Q, Huang ZX. A thermo-halo-tolerant and proteinase-resistant endoxylanase from Bacillus sp. HJ14. Folia Microbiol 2014; 59:423-31; PMID:24728834; http://dx.doi.org/10.1007/s12223-014-0316-4
- Margesin R, Schinner F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 2001; 5:73-83; PMID:11354458; http://dx.doi.org/10.1007/s007920100184
- Nagem RAP, Rojas AL, Golubev AM, Korneeva OS, Eneyskaya EV, Kulminskaya AA, Neustroev KN, Polikarpov I. Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition. J Mol Biol 2004; 344:471-80; PMID:15522299; http://dx.doi.org/10.1016/j.jmb.2004.09.024
- Norman JM, Bunny KL, Giffard PM. Characterization of levJ, a sucrase/fructanase-encoding gene from Actinomyces naeslundii T14V, and comparison of its product with other sucrose-cleaving enzymes. Gene 1995; 152:93-8; PMID:7828936; http://dx.doi.org/10.1016/0378-1119(94)00695-o
- Gill PK, Manhas RK, Singh J, Singh P. Purification and characterization of an exoinulinase from Aspergillus fumigatus. Appl Biochem Biotech 2004; 117:19-32; PMID:15126701; http://dx.doi.org/10.1385/abab:117:1:19
- Kushi RT, Monti R, Contiero J. Production, purification and characterization of an extracellular inulinase from Kluyveromyces marxianus var. bulgaricus. J Ind Microbiol Biot 2000; 25:63-9; http://dx.doi.org/10.1038/sj.jim.7000032
- Madern D, Ebel C, Zaccai G. Halophilic adaptation of enzymes. Extremophiles 2000; 4:91-8; PMID:10805563
- Tsujimoto Y, Watanabe A, Nakano K, Watanabe K, Matsui H, Tsuji K, Tsukihara T, Suzuki Y. Gene cloning, expression, and crystallization of a thermostable exo-inulinase from Geobacillus stearothermophilus KP1289. Appl Microbiol Biot 2003; 62:180-5; PMID:12883863; http://dx.doi.org/10.1007/s00253-003-1261-3
- Kwon YM, Kim HY, Choi YJ. Cloning and characterization of Pseudomonas mucidolens exoinulinase. J Microbiol Biotechn 2000; 10:238-43
- Liebl W, Brem D, Gotschlich A. Analysis of the gene for β-fructosidase (invertase, inulinase) of the hyperthermophilic bacterium Thermotoga maritima, and characterisation of the enzyme expressed in Escherichia coli. Appl Microbiol Biot 1998; 50:55-64; PMID:9720201; http://dx.doi.org/10.1007/s002530051256
- Liu GL, Fu GY, Chi Z, Chi ZM. Enhanced expression of the codon-optimized exo-inulinase gene from the yeast Meyerozyma guilliermondii in Saccharomyces sp. W0 and bioethanol production from inulin. Appl Microbiol Biot 2014; 98:9129-38; PMID:25236803; http://dx.doi.org/10.1007/s00253-014-6079-7
- Zhang LH, Zhao CX, Ohta WY, Wang YJ. Inhibition of glucose on an exoinulinase from Kluyveromyces marxianus expressed in Pichia pastoris. Process Biochem 2005; 40:1541-5; http://dx.doi.org/10.1016/j.procbio.2004.01.057
- Kwon HJ, Jeon SJ, You DJ, Kim KH, Jeong YK, Kim YH, Kim YM, Kim BW. Cloning and characterization of an exoinulinase from Bacillus polymyxa. Biotechnol Lett 2003; 25:155-9; PMID:12882292; http://dx.doi.org/10.1023/a:1021987923630
- Gao J, Xu YY, Yang HM, Xu H, Xue F, Li S, Feng XH. Gene cloning, expression, and characterization of an exo-inulinase from Paenibacillus polymyxa ZJ-9. Appl Biochem Biotech 2014; 173:1419-30; PMID:24807534; http://dx.doi.org/10.1007/s12010-014-0950-y
- Moriyama S, Akimoto H, Suetsugu N, Kawasaki S, Nakamura T, Ohta K. Purification and properties of an extracellular exoinulinase from Penicillium sp. strain TN-88 and sequence analysis of the encoding gene. Biosci Biotech Bioch 2002; 66:1887-96; PMID:12400688; http://dx.doi.org/10.1271/bbb.66.1887
- Sheng J, Chi ZM, Gong F, Li J. Purification and characterization of extracellular inulinase from a marine yeast Cryptococcus aureus G7a and inulin hydrolysis by the purified inulinase. Appl Biochem Biotech 2008; 144:111-21; PMID:18456943; http://dx.doi.org/10.1007/s12010-007-8025-y
- Wang L, Huang Y, Long X, Meng X, Liu Z. Cloning of exoinulinase gene from Penicillium janthinellum strain B01 and its high-level expression in Pichia pastoris. J Appl Microbiol 2011; 111:1371-80; PMID:21895898; http://dx.doi.org/10.1111/j.1365-2672.2011.05145.x
- Wanker E, Huber A, Schwab H. Purification and characterization of the Bacillus subtilis levanase produced in Escherichia coli. Appl Environ Microb 1995; 61:1953-8; PMID:7646030
- Kuzuwa S, Yokoi K, Kondo M, Kimoto H, Yamakawa A, Taketo A, Kodaira KI. Properties of the inulinase gene levH1of Lactobacillus casei IAM 1045; cloning, mutational and biochemical characterization. Gene 2012; 495:154-62; PMID:22197658; http://dx.doi.org/10.1016/j.gene.2011.12.004
- Kobayashi T, Uchimura K, Deguchi S, Horikoshi K. Cloning and sequencing of inulinase and β-fructofuranosidase genes of a deep-sea microbulbifer species and properties of recombinant enzymes. Appl Environ Microb 2012; 78:2493-5; PMID:22286980; http://dx.doi.org/10.1128/aem.07442-11
- Kim KY, Koo BS, Jo D, Kim SI. Cloning, expression, and purification of exoinulinase from Bacillus sp. snu-7. J Microbiol Biotechn 2004; 14:344-9
- Gill PK, Manhas RK, Singh P. Purification and properties of a heat-stable exoinulinase isoform from Aspergillus fumigatus. Bioresource Technol 2006; 97:894-902; PMID:15964186; http://dx.doi.org/10.1016/j.biortech.2005.04.034
- Zhou JP, Huang HQ, Meng K, Shi PJ, Wang YR, Luo HY, Yang PL, Bai YG, Yao B. Cloning of a new xylanase gene from Streptomyces sp. TN119 using a modified thermal asymmetric interlaced-PCR specific for GC-rich genes and biochemical characterization. Appl Biochem Biotech 2010; 160:1277-92; PMID:19412576; http://dx.doi.org/10.1007/s12010-009-8642-8
- Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expres Purif 2005; 41:207-34; PMID:15915565; http://dx.doi.org/10.1016/j.pep.2005.01.016