Abstract
Azurin and Laz (lipidated azurin) are 2 bacterial proteins with anticancer, anti-viral and anti-parasitic activities. Azurin, isolated from the bacterium Pseudomonas aeruginosa, termed Paz, demonstrates anticancer activity against a range of cancers but not against brain tumors. In contrast, Laz is produced by members of Gonococci/Meningococci, including Neisseria meningitides which can cross the blood-brain barrier to infect brain meninges. It has been previously reported that Laz has an additional 39 amino acid moiety, called an H.8 epitope, in the N-terminal part of the azurin moiety that allows Laz to cross the entry barrier to brain tumors such as glioblastomas. Exactly, how the H.8 epitope helps the azurin moiety of Laz to cross the entry barriers to attack glioblastoma cells is unknown. In this paper, we describe the structural features of the H.8 moiety in Laz using X-ray crystallography and demonstrate that while the azurin moiety of Laz adopts a β-sandwich fold with 2 β-sheets arranged in the Greek key motif, the H.8 epitope was present as a disordered structure outside the Greek key motif. Structures of Paz and H.8 epitope-deficient Laz are well superimposed. The structural flexibility of the H.8 motif in Laz explains the extracellular location of Laz in Neisseria where it can bind the key components of brain tumor cells to disrupt their tight junctions and allow entry of Laz inside the tumors to exert cytotoxicity.
Introduction
Azurin, a blue copper-containing protein belonging to the cupredoxin superfamily, is localized in the periplasm of gram-negative bacteria and is involved in electron transport during respiration.Citation1 Although structural and functional characteristics of bacterial azurin as a redox protein have been well documented,Citation2 we found the following additional function of the protein: clinical isolates of Pseudomonas aeruginosa secrete azurin extracellularly in response to the presence of various human cancer cells (e.g., melanoma and breast cancer cells).Citation3 Azurin exhibits significant cytotoxicity against cancer cells, while little cytotoxicity is observed against normal cells.Citation4 The protein can preferentially enter cancer cells and bind to the tumor suppressor protein p53.Citation5 The complex formed by azurin and p53 as well as its electron transfer partner cytochrome c induces apoptosis in cancer cells through cell cycle arrest at the G1 phase or caspase-mediated mitochondrial cytochrome c release.Citation6,7 Azurin is therefore expected to be a potential drug for cancer therapy.Citation8,9
The precursor form of azurin (148 residues) produced by P. aeruginosa is converted to a mature form (Paz, 128 residues) through the release of a signal peptide (20 residues) across the inner membrane ().Citation10 Site-directed mutagenesis and deletion mutations indicate that the presence of a copper ion and redox activity was not essential for the cytotoxicity of Paz.Citation11 The internal sequence of Paz (residue nos. 50–77 in the mature form, termed p28) functions as a putative protein transduction domain responsible for azurin's penetration into cancer cellsCitation12 through endocytotic and nonendocytotic mechanisms.Citation13 Based on crystal structures, bacterial azurins show structural similarity to ephrinB2, a ligand for the receptor tyrosine kinase EphB2.Citation14,15 Cell signaling through Eph/ephrin is involved in cancer progression.Citation14,15 The chemically synthesized C-terminal domain of Paz (residue nos. 96–113 in the mature form), which is similar to ephrinB2 at the GH loop region responsible for receptor binding,Citation15 inhibits the growth of various cancer cells.Citation14 Indeed, it has been shown that azurin represses ephrinB2-mediated autophosphorlyation of the EphB2 tyrosine residue, thereby interfering with upstream cell signaling and contributing to cancer cell growth inhibition.Citation14
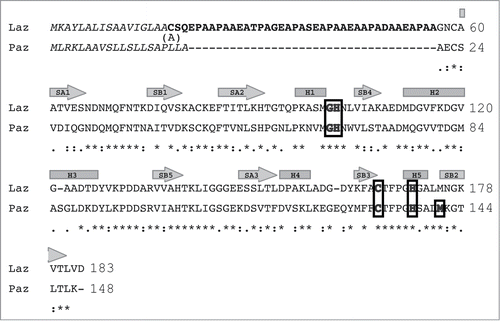
Figure 1. Amino acid sequence alignment of Laz and Paz using the CLUSTALW program (http://clustalw.genome.ad.jp/). Laz, Neisseria gonorrhoeae azurin; Paz, Pesudomonas aeruginosa azurin. Identical and similar amino acid residues in Laz and Paz are denoted by asterisks and dots, respectively. Letters in italic and bold indicate the signal peptide and H.8 epitope, respectively. The amino acid residues responsible for binding to the metal ion are boxed. The regions corresponding to cylinders and arrows represent α-helices and β-strands, respectively. The H.8 epitope is intrinsically disordered. Laz used in this study has Ala18 instead of Cys18.
In addition to azurin, rusticyanin, another bacterial cupredoxin, causes apoptosis in human cancer cells.Citation16 Neisserial azurin-like protein, a “lipobox”-dependent lipid-modified azurin (Laz) bound to the outer membrane,Citation17 also enters brain tumors such as glioblastomas and exhibits significant toxicity against the cancer cells,Citation18 suggesting that Laz must cross the entry barrier to reach brain tumor cells. Laz includes a 39-amino acid residue region called the H.8 epitope at the N terminus prior to the azurin domain. In our previous studies with Laz, the H.8 epitope of the protein, but not its lipidation, was demonstrated to be essential for disrupting the entry barrier to provide access to brain tumor cells.Citation18 To obtain clues on the entry of Laz into brain tumor cells, we determined the structure of Laz by X-ray crystallography and our findings are described herein.
Results
Comparison of primary structure between Laz and Paz
Laz consists of 183 amino acid residues with a molecular weight of 18532 ().Citation17 Similar to other bacterial outer membrane-bound lipoproteins, Laz includes a conserved 4-amino acid sequence termed lipobox [LVI][ASTVI][GAS]C at the N terminus.Citation19,20 The thiol group of the cysteine residue in the lipobox accepts diacylglycerol from phosphatidylglycerol through the action of transferase Lgt, and the resultant lipid-modified proteins are processed by signal peptidase II. The N-terminal 17 amino acid residues are therefore considered to function as a sorting signal to the outer membrane. On the other hand, Paz is known to be a protein secreted through processing of a 20-residue signal peptide by signal peptidase I.
After processing by signal peptidase II, mature Laz consists of 166 residues with the N-terminal 39-residue sequence designated as the H.8 epitope (). This epitope includes a repeated sequence of 5 amino acid residues and exhibits a significantly low isoelectric point (pI 3.4). A substantial sequence identity (56%) exists in the azurin domain between Laz and Paz. Although the crystal structure of Paz has already been solved, no information is available on the structure of Laz, especially of the H.8 epitope. Thus, X-ray crystallography of Laz was performed to clarify the structure–function relationship of the H.8 epitope involved in disrupting the entry barrier to brain tumor cells.
Purification and characterization of Laz
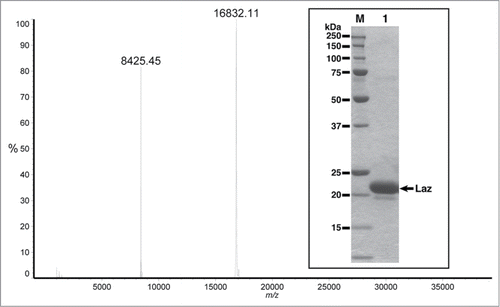
Lipidation is known to hinder crystallization of lipoproteins. Thus, the Laz mutant with a pectate lyase B (pelB) signal peptide instead of the native lipobox peptide was purified and characterized because this mutant can also enter glioblastoma cells and exhibit cytotoxicity.Citation18 In the mutant, the signal peptide was fused to the H.8 epitope with the N-terminal cysteine residue (Cys18) substituted with alanine (Ala18) to avoid lipidation (). Laz purified from the recombinant E. coli cells showed a single protein band of 22 kDa on the SDS-PAGE gel (, inset). Positive-mode matrix-assisted laser desorption/ionization time-of-flight mass spectrometry demonstrated that the molecular weight of the purified protein was 16832 (). The signal at m/z 8425 was considered to be derived from the sample in the divalent ion form. The N-terminal amino acid sequence of the purified Laz was determined to be NH2–ASQEP corresponding toCitation18ASQEP,Citation22 indicating that the mature protein, formed after cleavage of the signal peptide, consists of 166 amino acid residues with a theoretical molecular weight of 16841 which is in good agreement with that determined by mass spectrometry (16832). The difference in molecular weight between SDS-PAGE and mass spectrometry is possibly due to the significantly low pI of the H.8 epitope. Inductively coupled plasma atomic emission spectrometry analysis revealed that the protein contained metal ions such as zinc and copper. The content of zinc ion was fold10- higher than that of copper ion.
Structure determination
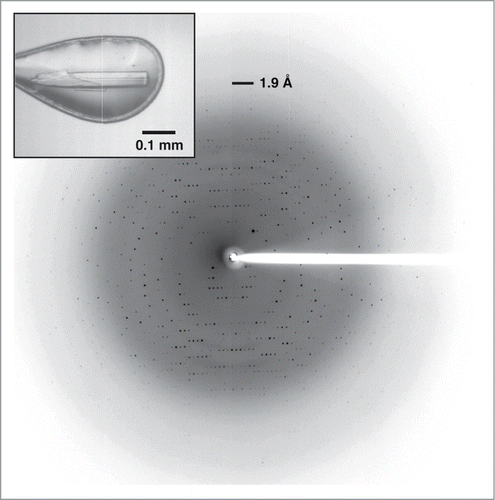
Stick-shaped crystals of Laz were found in a droplet consisting of 1.5 M ammonium sulfate, 0.1 M Tris-HCl (pH 8.5), and 20% glycerol (, inset). Diffraction images of the crystal were collected at up to 1.90 Å resolution (). The structure of Laz was solved by molecular replacement, using Paz as a reference model. Statistics for data collection and structure refinement are shown in. The N-terminal sequence (40 amino acid residues, Ala18–Gly57) closely corresponding to the H.8 epitope could not be assigned in the 2Fo-Fc map. The refined model in an asymmetric unit consisted of 2 identical monomers (126 amino acid residues, Asn58–Asp183 × 2) termed molecules A and B with 161 water molecules, 2 metal ions, 2 sulfate ions, and 2 glycerol molecules; root-mean-square deviation (rmsd) between molecules A and B was calculated as 0.191 Å for all residues (126 Cα atoms), indicating that the structures of molecules A and B are basically identical to each other. On the basis of theoretical curves in the plot calculated according to Luzzati,Citation21 the absolute positional error was estimated to be 0.214 Å at 1.90 Å resolution. Ramachandran plot analysisCitation22 in which the stereochemical correctness of the backbone structure is indicated by (ϕ, ψ) torsion angles showed that 93.1% of nonglycine residues lie within the most favored regions and 6.9% of nonglycine residues lie in the additionally allowed regions.
Table 1. Data collection and refinement statistics
Overall structure
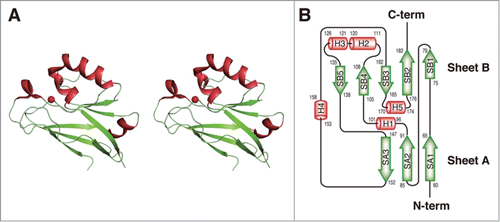
The overall structure () and topology of secondary structure elements () indicate that the H.8 epitope-deficient Laz (C-terminal domain of Laz, Laz-C) adopts a β-sandwich fold as a basic scaffold. Five additional α-helices (H1, residues 96–101; H2, 111–120; H3, 121–126; H4, 153–158; and H5, 170–174) are included in the domain. The β-sandwich fold consists of 2 β-sheets arranged in the Greek key motif. One sheet (SA) is composed of 3 strands (SA1, 60–65; SA2, 85–91; and SA3, 147–152) and the other (SB) has 5 strands (SB1, 75–79; SB2, 176–182; SB3, 162–165; SB4, 105–108; and SB5, 135–138).
Figure 4. Structure of Laz. (A) Overall structure (stereo diagram). (B) Topology diagram. α-Helices are shown as orange cylinders and β-strands as green arrows. The metal ion is depicted by a red ball.
Structural homologs of Laz-C were searched in Protein Data Bank (PDB) using the DALI program.Citation23 Bacterial azurin proteins were found to exhibit significant structural homology with Laz-C (). The overall structure of Laz-C is most similar to that of Alcaligenes xylosoxidans azurin (PDB code, 1RKR; Z = 24.7) with rmsd of 1.0 Å for 126 Cα. Laz-C is also structurally similar to Paz (Z = 24.5). Superimposition of the crystal structures of Laz-C and Paz (PDB code, 1AG0) is shown in . Because a large number of bacterial azurins and plant plastocyanins with the Greek key motif are categorized into the plastocyanin/azurin-like family in the cupredoxin superfamily on the SCOP database (http://scop.mrc-lmb.cam.ac.uk/scop/), Laz-C is considered to be structurally classified into this family.
Table 2. Structure-based homology with Laz
Figure 5. Structural comparison. (A) Superimposition of the overall structures of Laz-C (green) and Paz (blue) (PDB code, 3AZU). (B) Metal-binding site. Left, Laz (carbon, green; nitrogen, blue; oxygen, red; and sulfur, yellow); right, superimposition of copper (blue)- and zinc (purple)-binding Paz proteins. Balls colored red and orange indicate the zinc and copper ions, respectively.
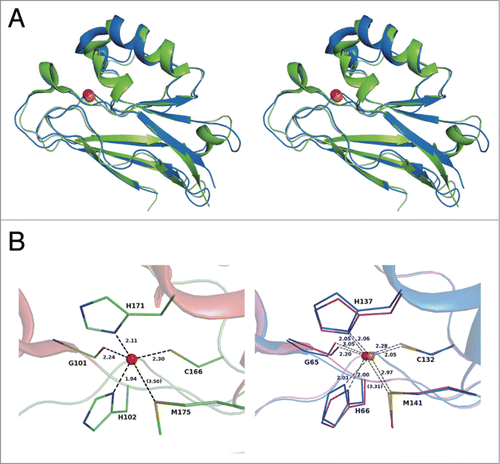
Metal-binding site
A density map for metal ions was observed in each protein molecule, suggesting that Laz contained one molecule of metal ion per protein molecule. Although inductively coupled plasma atomic emission spectrometry showed that 2 ions such as zinc and copper are included in Laz, the protein was suggested to contain a zinc or copper ion. Based on the content of metal ions, Laz used in this study is considered to incorporate a zinc rather than a copper ion. This is possibly due to an insufficient supply of copper ions during culturing of the bacteria, purification of the protein, or both. The four atoms, O of Gly101, Nδ of His102, Sγ of Cys166, and Nδ of His171, are coordinated to the zinc ion, and the coordination geometry comprises a distorted tetrahedron (, left). The distance between the zinc ion and these atoms ranges from 1.94 to 2.30 Å (average, 2.15 Å).
The copper ion in Paz (Cu-Paz) is known to be bound to 5 atoms, O of Gly65, Nδ of His66, Sγ of Cys132, Nδ of His137, and Sδ of Met141 (, right, purple).Citation24 These 5 residues (G/H/C/H/M) involved in binding to the copper ion are also completely conserved in Laz (), although Met175 in Laz-C is located slightly farther from the zinc ion (3.50 Å) (, left). Among a large number of azurin-like crystal structures determined to date, a zinc ion-bound Paz (Zn-Paz) expressed in recombinant E. coli cells has been isolated and structurally characterized.Citation25 In the crystal structure of Zn-Paz (PDB code, 1E67), the distance between the zinc ion and Sδ of Met141 is 3.31 Å (, right, blue), while the corresponding distance is 2.97 Å in Cu-Paz (PDB code, 3AZU) (, right, purple), indicating that Laz-C is structurally similar to Zn-Paz.
Discussion
Because of disorder, the structure of the H.8 epitope could not be determined. Recently, natively unfolded or intrinsically disordered proteins (IDPs) or regions have been found mainly in eukaryotic cells.Citation26,27 In addition, the sigma factor inhibitor FlgM in bacteria has been identified as an IDP.Citation28 A large number of IDPs are classified as a superfamily of macromolecule (DNA, RNA, or protein)-associated proteins such as transcriptional factors, RNA-binding proteins, and cell cycle regulators.Citation27 A typical IDP is the transcriptional factor leucine zipper protein GCN4.Citation29 The basic DNA-binding region in GCN4 is unfolded in the absence of DNA, while a helical structure is inducibly formed in the region of DNA binding. This is called the coupled binding and folding process.Citation26
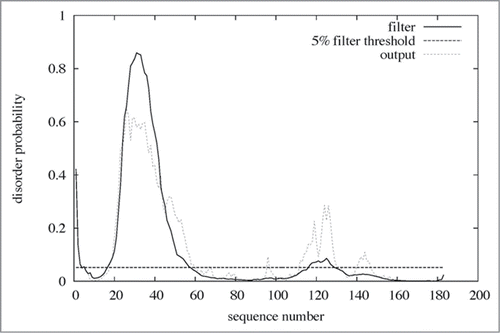
Through bioinformatics approaches, 5 common features of IDPs are postulated:Citation26,27 (i) Short sequences are repeated in IDPs. (ii) Little sequence homology is observed in disordered regions among IDPs. (iii) Disordered regions are located at the N or C terminus of IDPs. (iv) IDPs are primarily produced by eukaryotes. (v) IDPs are frequently localized in the nucleus and rarely in mitochondria. In case of Laz, the N-terminal H.8 epitope with a 5 residue-repeated sequence (AAEAP, a typical but somewhat variable sequence) is attached to the azurin domain (). No proteins or peptides were found to be homologous to the H.8 epitope in the protein databases. Several programs for identification of intrinsically disordered regions in IDPs are available on the web. The H.8 epitope of Laz was predicted to be intrinsically disordered by all the typical programs, PONDR-FIT,Citation30 DISOPRED 2,Citation31 and DisEMBLCitation32 ( and ). Although the target molecules that bind to Laz have not been identified in the entry barrier to brain tumors, the intrinsically disordered H.8 epitope may inducibly fold into a rigid structure by binding to target molecules.
Figure 6. Disordered region in Laz. The amino acid sequence of Laz (183 residues) was analyzed by the disorder prediction program DISOPRED 2 (http://bioinf.cs.ucl.ac.uk/disopred/). Residues (20–58) corresponding to the H.8 epitope were identified as a disordered region.
Why is the H.8 epitope of Laz intrinsically disordered rather than being integrated with the folded region of Laz-C? It should be emphasized that the H.8 epitope of Laz seems to have 2 major functions in which a loosely associated H.8 epitope may be required. First, the H.8 epitope has been shown to be involved in facilitating the entry of Laz-C (the 127 amino acid azurin moiety) into glioblastomasCitation18 as well as its passage through the blood–brain barrier.Citation33 Thus, the presence of the H.8 epitope allows the Laz-C moiety to enter glioblastoma or other brain tumors when Neisseria such as N. meningitides are present in the brain meninges. Because P. aeruginosa does not normally cause infections in the brain, it has no particular need to equip its anticancer weapon Paz with a blood–brain barrier-crossing capability. The other hypothetical function of the H.8 epitope is to allow N. meningitides to protect its weapon Laz-C from being attacked by the immune system following the release of Laz-C from the bacterial cells for entry into the tumor. Interestingly, the H.8 epitope has been shownCitation34 to induce formation of blocking antibodies that bind complements or other antibodies generated by the host to remove the invading bacteria thereby inactivating the host-generated antibodies. Such induction of blocking antibodies by the H.8 epitope is believed to prevent complement-mediated bactericidal action and provide a survival advantage to group B meningococci.Citation34 It is, however, equally possible that such blocking antibodies protect Laz from immune attack when released for entry into brain tumors. Normally, azurin (Paz) demonstrates low immunogenicity because of its structural similarity with immunoglobulins. Both azurin and immunoglobulins, while having low sequence identity, demonstrate structural features with β-sandwich packing.Citation8 Both these proteins exhibit tyrosine residues in a motif called the “tyrosine corner,” which plays a significant role in the structural stability and folding of the Greek key β-barrel structure exhibited by azurin and immunoglobulins.Citation8 This low immunogenicity of azurin (Laz-C) is further enhanced by the presence of the intrinsically disordered H.8 epitope that elicits blocking antibodies to protect Laz from immune attack.
In conclusion, this is the first report on the structure of Neisserial azurin (Laz), which is cytotoxic to brain and other tumor cells. Laz is composed of an N-terminal intrinsically disordered H.8 epitope and a rigid azurin domain with the Greek key motif. In contrast, the P. aeruginosa azurin Paz, which lacks the H.8 epitope, is deficient in the ability to enter brain tumor cells and consequently demonstrates significantly lower cytotoxicity against such cells.Citation18,33 It should be noted, however, that as mentioned earlier, Paz, unlike Laz, is deficient in entry in glioblastoma cells, while H.8-Paz, where the H.8 epitope was cloned in the N-terminal of Paz, had significant cytotoxicity against glioblastoma cells, confirming the presence of the H.8 epitope as critical for entry in brain tumors. It should also be noted that Paz, Laz and H.8-Paz had not only anticancer activity, but strong anti-viral activity against the AIDS-causing HIV-1 virus and strong anti-parasitic activity against parasites such as the malarial parasite Plasmodium falciparumCitation35 and the toxoplasmosis-causing parasite Toxoplasma gondii.Citation36 The potential clinical importance of such bacterial proteins as Paz and Laz is reflected in the fact that a 28 amino acid peptide fragment from azurin (azurin 50–77), termed p28, has shown very little toxicity in 15 stage IV cancer patients in a phase I clinical trial, but significant beneficial effect including partial and complete regression of the drug-resistant tumors in several patients.Citation37 Another phase I trial with p28, currently in progress in pediatric brain tumor patients in 11 hospitals in the US (http://clinicaltrials.gov/ct2/show/NCT01975116) for more than a year and 3 months appears to indicate that p28, even lacking the H.8 epitope but with a smaller size than azurin, may be able to enter the brain tumors to allow their regression.
Materials and Methods
Microorganisms and culture conditions
Escherichia coli strain BL21(DE3) (Novagen) was used as a host for expression of Neisseria gonorrhoeae Laz. For expression in E. coli, cells were aerobically precultured at 30°C in Luria–Bertani mediumCitation38 supplemented with sodium ampicillin (0.1 mg/ml). When the turbidity reached approximately 0.5 at 600 nm, isopropyl-β-D-thiogalactopyranoside was added to the culture (0.1 mM) and the cells were further cultured for 44 h at 16°C.
Purification
Unless otherwise specified, all operations were performed at 0–4°C. E. coli cells harboring pET25b-lcaCitation18 were grown in 6 l of LB medium (1.5 l/flask), collected by centrifugation (6000 ×g for 5 min at 4°C), washed with 20 mM Tris-HCl (pH 7.5), and subsequently resuspended in the same buffer. The cells were ultrasonically disrupted (Insonator Model 201M; Kubota) at 9 kHz for 20 min at 0°C, and the clear solution obtained on centrifugation at 20000 ×g and 4°C for 20 min was used as the cell extract. The extract was applied to a DEAE-Toyopearl 650 M (Tosoh) column (2.6 cm × 10 cm) previously equilibrated with 20 mM Tris-HCl (pH 7.5). The absorbed proteins were eluted with a linear gradient of NaCl (0–1 M) in 20 mM Tris-HCl (pH 7.5, 300 ml), and a 6.6-ml fraction was collected every 7 min. The fractions containing Laz, eluted with 0.2–0.3 M NaCl, were combined and saturated with ammonium sulfate (30%). The Laz solution was then applied to a Butyl-Toyopearl 650M (Tosoh) column (2.6 cm × 10 cm) previously equilibrated with 20 mM Tris-HCl (pH 7.5) containing 30% saturated ammonium sulfate. The absorbed proteins were eluted with a linear gradient of saturated ammonium sulfate (30–0%, 300 ml), and a 6.6-ml fraction was collected every 10 min. The Laz fractions, eluted with 20–10% saturated ammonium sulfate, were combined and dialyzed for 3 h against 20 mM Tris-HCl (pH 7.5). The dialysate was applied to a SuperQ-Toyopearl 650S (Tosoh) column (1 cm × 10 cm) previously equilibrated with 20 mM Tris-HCl (pH 7.5). The absorbed proteins were eluted with a linear gradient of NaCl (0–0.5 M) in 20 mM Tris-HCl (pH 7.5, 30 ml), and a 1-ml fraction was collected every 1 min. The Laz fractions, eluted with 0.2–0.25 M NaCl, were combined and applied to a HiLoad 16/60 Superdex 75 pg (GE healthcare) column (1.6 cm × 60 cm) previously equilibrated with 20 mM Tris-HCl (pH 7.5) containing 0.15 M NaCl. Laz was eluted with the same buffer (120 ml), and a 2-ml fraction was collected every 2 min. The Laz fractions were combined and dialyzed for 3 h against 20 mM Tris-HCl (pH 7.5). The dialysate was used as the purified Laz. The homogeneity of the purified protein was confirmed by SDS-PAGE.Citation39 The protein content was determined by measuring the absorbance at 280 nm using a cuvette with a path length of 1 cm, assuming that E280 = 0.177 (Laz) corresponds to 1 mg/ml.
Analytical methods
The N-terminal amino acid sequence of Laz was determined by the Edman degradation method using a Procise 492 protein sequencer (Applied Biosystems). The molecular weight of Laz was determined using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (AXIMA Performance; Shimadzu); sinapinic acid was used as the matrix. Metal ions included in Laz were examined using an inductively coupled plasma atomic emission spectrometer (ICPS-8100; Shimadzu).
Crystallization and X-ray diffraction
Laz (98 mg/ml) was crystallized at 20°C using the sitting drop vapor diffusion method. The mother liquor consisting of 1.5 M ammonium sulfate, 0.1 M Tris-HCl (pH 8.5), and 20% glycerol (100 μl) was used as a reservoir solution, and 1 μl of the Laz solution was mixed with 1 μl of the reservoir solution to form the drop. The Laz crystal was removed from the drop solution using a mounted nylon loop (Hampton Research) and then placed directly into a cold nitrogen gas stream at −173°C. X-ray diffraction images of the crystal were collected at −173°C under a nitrogen gas stream using a Jupiter 210 charge-coupled device detector and synchrotron radiation at the BL-38B1 station of SPring-8 (Japan). The diffraction data for the native crystal were collected at 1.90 Å resolution and processed using the HKL2000 program package.Citation40 Data collection statistics from the crystal are summarized in .
Structure determination and refinement
The crystal structure of Laz was solved by molecular replacement using the Molrep programCitation41 in the CCP4 program package;Citation42 the Paz structure (PDB code, 1AG0) was used as a reference model. The Coot programCitation43 was used for manual modification of the initial model. Initial rigid body refinement and several rounds of restrained refinement against the dataset were performed using the Refmac5 program.Citation44 Water molecules were incorporated where the difference in density exceeded 3.0σ above the mean and the 2Fo-Fc map showed a density of over 1.0σ. At this stage, metal ions were included in the calculation and refinement continued until convergence at maximum resolution. Protein models were superimposed and their rmsd was determined using the LSQKAB program,Citation45 a part of CCP4. Final model quality was determined using the PROCHECK program.Citation46 Ribbon plots were prepared using the PyMOL program.Citation47 Coordinates used in this study were taken from PDB, Research Collaboratory for Structural Bioinformatics.Citation48
Protein structure accession number
The atomic coordinates and structure factors (PDB code, 3AY2) of Laz have been deposited in PDB, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs. S Baba and N Mizuno of the Japan Synchrotron Radiation Research Institute (JASRI) for their kind help in data collection. Diffraction data for crystals were collected at the BL-38B1 station of SPring-8 (Hyogo, Japan) with the approval of JASRI.
References
- Lancaster KM, Farver O, Wherland S, Crane EJ, Richards JH, Pecht I, Gray HB. Electron transfer reactivity of type zero Pseudomonas aeruginosa azurin. J Am Chem Soc 2011; 133: 4865-73; PMID:21405124; http://dx.doi.org/10.1021/ja1093919
- Savelieff MG, Lu Y. Cu(A) centers and their biosynthetic models in azurin. J Biol Inorg Chem 2010; 15: 461-83; PMID:20169379; http://dx.doi.org/10.1007/s00775-010-0625-2
- Mahfouz M, Hashimoto W, Das Gupta TK, Chakrabarty AM. Bacterial proteins and CpG-rich extrachromosomal DNA in potential cancer therapy. Plasmid 2007; 57:4-17; PMID:17166586; http://dx.doi.org/10.1016/j.plasmid.2006.11.001
- Punj V, Bhattacharyya S, Saint-Dic D, Vasu C, Cunningham EA, Graves J, Yamada T, Constantinou AI, Christov K, White B, et al. Bacterial cupredoxin azurin as an inducer of apoptosis and regression in human breast cancer. Oncogene 2004; 23: 2367-78; PMID:14981543; http://dx.doi.org/10.1038/sj.onc.1207376
- Yamada T, Goto M, Punj V, Zaborina O, Kimbara K, Das Gupta TK, Chakrabarty AM. The bacterial redox protein azurin induces apoptosis in J774 macrophages through complex formation and stabilization of the tumor suppressor protein p53. Infect Immun 2002; 70: 7054-62; PMID:12438386; http://dx.doi.org/10.1128/IAI.70.12.7054-7062.2002
- Hiraoka Y, Yamada T, Goto M, Das Gupta TK, Chakrabarty AM. Modulation of mammalian cell growth and death by prokaryotic and eukaryotic cytochrome c. Proc Natl Acad Sci USA 2004; 101:6427-32; PMID:15082831; http://dx.doi.org/10.1073/pnas.0401631101
- Yamada T, Hiraoka Y, Ikehata M, Kimbara K, Avner BS, Das Gupta TK, Chakrabarty AM. Apoptosis or growth arrest: modulation of tumor suppressor p53s specificity by bacterial redox protein azurin. Proc Natl Acad Sci USA 2004; 101: 4770-5; PMID:15044691; http://dx.doi.org/10.1073/pnas.0400899101
- Fialho AM, Stevens FJ, Das Gupta TK, Chakrabarty AM. Beyond host-pathogen interactions: microbial defense strategy in the host environment. Curr Opin Biotechnol 2007; 18:279-86; PMID:17451932; http://dx.doi.org/10.1016/j.copbio.2007.04.001
- Chakrabarty AM, Bernardes N, Fialho AM. Bacterial proteins and peptides in cancer therapy: today and tomorrow. Bioengineered 2014; 5: 234-42; PMID:24875003; http://dx.doi.org/10.4161/bioe.29266
- Arvidsson RH, Nordling M, Lundberg LG. The azurin gene from Pseudomonas aeruginosa. cloning and characterization. Eur J Biochem 1989; 179: 195-200; PMID:2537198; http://dx.doi.org/10.1111/j.1432-1033.1989.tb14540.x
- Goto M, Yamada T, Kimbara K, Horner J, Newcomb M, Das Gupta TK, Chakrabarty AM. Induction of apoptosis in macrophages by Pseudomonas aeruginosa azurin: tumour-suppressor protein p53 and reactive oxygen species, but not redox activity, as critical elements in cytotoxicity. Mol Microbiol 2003; 47: 549-59; PMID:12519204; http://dx.doi.org/10.1046/j.1365-2958.2003.03317.x
- Yamada T, Fialho AM, Punj V, Bratescu L, Das Gupta TK, Chakrabarty AM. Internalization of bacterial redox protein azurin in mammalian cells: entry domain and specificity. Cell Microbiol 2005; 7: 1418-31; PMID:16153242; http://dx.doi.org/10.1111/j.1462-5822.2005.00567.x
- Taylor BN, Mehta RR, Yamada T, Lekmine F, Christov K, Chakrabarty AM, Green A, Bratescu L, Shilkaitis A, Beattie CW, et al. Noncationic peptides obtained from azurin preferentially enter cancer cells. Cancer Res 2009; 69: 537-46; PMID:19147567; http://dx.doi.org/10.1158/0008-5472.CAN-08-2932
- Chaudhari A, Mahfouz M, Fialho AM, Yamada T, Granja AT, Zhu Y, Hashimoto W, Schlarb-Ridley B, Cho W, Das Gupta TK, et al. Cupredoxin-cancer interrelationship: azurin binding with EphB2, interference in EphB2 tyrosine phosphorylation, and inhibition of cancer growth. Biochemistry 2007; 46: 1799-810; PMID:17249693; http://dx.doi.org/10.1021/bi061661x
- Chrencik JE, Brooun A, Recht MI, Nicola G, Davis LK, Abagyan R, Widmer H, Pasquale EB, Kuhn P. Three-dimensional structure of the EphB2 receptor in complex with an antagonistic peptide reveals a novel mode of inhibition. J Biol Chem 2007; 282: 36505-13; PMID:17897949; http://dx.doi.org/10.1074/jbc.M706340200
- Yamada T, Hiraoka Y, Das Gupta TK, Chakrabarty AM. Rusticyanin, a bacterial electron transfer protein, causes G1 arrest in J774 and apoptosis in human cancer cells. Cell Cycle 2004; 3: 1182-7; PMID:15467448
- Cannon JG. Conserved lipoproteins of pathogenic neisseria species bearing the H.8 epitope: lipid-modified azurin and H.8 outer membrane protein. Clin Microbiol Rev 1989; 2: S1-4; PMID:2470496
- Hong CS, Yamada T, Hashimoto W, Fialho AM, Das Gupta TK, Chakrabarty AM. Disrupting the entry barrier and attacking brain tumors: the role of the neisseria H.8 epitope and the Laz protein. Cell Cycle 2006; 5: 1633-41; PMID:16861907; http://dx.doi.org/10.4161/cc.5.15.2991
- Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol 2006; 188: 2761-73; PMID:16585737; http://dx.doi.org/10.1128/JB.188.8.2761-2773.2006
- Narita S, Tokuda H. An ABC transporter mediating the membrane detachment of bacterial lipoproteins depending on their sorting signals. FEBS Lett 2006; 580: 1164-70; PMID:16288742; http://dx.doi.org/10.1016/j.febslet.2005.10.038
- Luzzati V. Traitement statistique des erreurs dans la determination des structures cristallines. Acta Crystallogr 1952; 5: 802-10; http://dx.doi.org/10.1107/S0365110X52002161
- Ramachandran GN, Sasisekharan V. Conformations of polypeptides and proteins. Adv Protein Chem 1968; 23: 283-438; PMID:4882249; http://dx.doi.org/10.1016/S0065-3233(08)60402-7
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol 1993; 233:123-38; PMID:8377180; http://dx.doi.org/10.1006/jmbi.1993.1489
- Nar H, Messerschmidt A, Huber R, van de Kamp M, Canters GW. X-ray crystal structure of the two site-specific mutants His35Gln and His35Leu of azurin from Pseudomonas aeruginosa. J Mol Biol 1991; 218: 427-47; PMID:1901363; http://dx.doi.org/10.1016/0022-2836(91)90723-J
- Nar H, Huber R, Messerschmidt A, Filippou AC, Barth M, Jaquinod M, van de Kamp M, Canters GW. Characterization and crystal structure of zinc azurin, a by-product of heterologous expression in Escherichia coli of Pseudomonas aeruginosa copper azurin. Eur J Biochem 1992; 205: 1123-9; PMID:1576995; http://dx.doi.org/10.1111/j.1432-1033.1992.tb16881.x
- Nishikawa K. An overview on natively unfolded proteins. Seibutsu Butsuri 2009; 49: 4-10; http://dx.doi.org/10.2142/biophys.49.004
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 1999; 293: 321-31; PMID:10550212; http://dx.doi.org/10.1006/jmbi.1999.3110
- Daughdrill GW, Hanely LJ, Dahlquist FW. The C-terminal half of the anti-sigma factor FlgM contains a dynamic equilibrium solution structure favoring helical conformations. Biochemistry 1998; 37:1076-82; PMID:9454599; http://dx.doi.org/10.1021/bi971952t
- Weiss MA, Ellenberger T, Wobbe CR, Lee JP, Harrison SC, Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature 1990; 347:575-8; PMID:2145515; http://dx.doi.org/10.1038/347575a0
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 2010; 1804: 996-1010; PMID:20100603; http://dx.doi.org/10.1016/j.bbapap.2010.01.011
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 2004; 337: 635-45; PMID:15019783; http://dx.doi.org/10.1016/j.jmb.2004.02.002
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure 2003; 11: 1453-9; PMID:14604535; http://dx.doi.org/10.1016/j.str.2003.10.002
- Hong CS, Yamada T, Fialho AM, Das Gupta TK, Chakrabarty AM. Transport agents for crossing the blood-brain barrier and into brain cancer cells, and methods of use thereof. US patent 2010; 7807183
- Dutta Ray T, Lewis LA, Gulati S, Rice PA, Ram S. Novel blocking human IgG directed against the pentapeptide repeat motifs of Neisseria meningitides Lip/H.8 and Laz lipoproteins. J Immunol 2011; 186: 4881-94; PMID:21402895; http://dx.doi.org/10.4049/jimmunol.1003623
- Chaudhari A, Fialho AM, Ratner D, Gupta P, Hong CS, Kahali S, Yamada T, Haldar K, Murphy S, Cho W, et al. Azurin, Plasmodium falciparum malaria and HIV/AIDS: inhibition of parasitic and viral growth by azurin. Cell Cycle 2006; 5: 1642-8; PMID:16861897; http://dx.doi.org/10.4161/cc.5.15.2992
- Naguleswaran A, Fialho AM, Chaudhari A, Hong CS, Chakrabarty AM, Sullivan WJ, Jr. Azurin-like protein blocks invasion of Toxoplasma gondii through potential interactions with parasite surface antigen SAG1. Antimicrob Agents Chemother 2008; 52: 402-8; PMID:18070964; http://dx.doi.org/10.1128/AAC.01005-07
- Warso MA, Richards JM, Mehta D, Christov K, Schaeffer C, Rae Bressler L, Yamada T, Majumdar D, Kennedy SA, Beattie CW, et al. A first-in-class, first-in-human phase I trial of p28, a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in patients with advanced solid tumours. Br J Cancer 2013; 108: 2061-70; PMID:23449360; http://dx.doi.org/10.1038/bjc.2013.74
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A Laboratory Manual, second ed., Taylor & Francis Harbor, NY. 1989.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227: 680-5; PMID:5432063; http://dx.doi.org/10.1038/227680a0
- Otwinoski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276: 307-26; http://dx.doi.org/10.1016/S0076-6879(97)76066-X
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr Sect D Biol Crystallogr 2010; 66: 22-5; PMID:20057045; http://dx.doi.org/10.1107/S0907444909042589
- Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr 1994; 50: 760-3; PMID:15299374; http://dx.doi.org/10.1107/S0907444994003112
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr 2004; 60: 2126-32; PMID:15572765; http://dx.doi.org/10.1107/S0907444904019158
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D Biol Crystallogr 1997; 53: 240-55; PMID:15299926; http://dx.doi.org/10.1107/S0907444996012255
- Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr Sect A 1976; 32: 922-3; http://dx.doi.org/10.1107/S0567739476001873
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 1993; 26: 283-91; http://dx.doi.org/10.1107/S0021889892009944
- DeLano WL. The PyMOL Molecular Graphics System. Taylor & Francis, San Carlos, CA. 2004
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res 2000; 28: 235-42; PMID:10592235; http://dx.doi.org/10.1093/nar/28.1.235