Abstract
Creatinase (creatine amidinohydrolase), an important medical enzyme, has been used for clinical diagnosis of renal function because of its high substrate specificity. Recently, we successfully cloned a NaN3-resistant creatinase encoding gene from Arthrobacter nicotianae. By optimizing the cultivation process, we realized its high-level expression in Escherichia coli. In this addendum, production of this NaN3-resistant creatinase in E. coli and future research were further discussed.
In recent years, although the Jaffe's reaction has been widely used for clinical diagnosis of renal function,Citation1,2 the low substrate specificity cannot meet the requirement of precision measurement.Citation3 As an alternative approach, the enzymatic method applying creatinase (EC 3.5.3.3) and the other 2 enzymes (creatininase, EC 3.5.2.10; sarcosine oxidase, EC 1.5.3.1) has been received intensive attention. In addition, the preservatives and surfactants such as NaN3, EDTA, Tween 20, Tween 80 and Triton X-100 were usually supplemented during storage. Thus, achievement of high-level production of creatinase with desirable resistance to these reagents (especially sodium azide) will be advantageous for clinical applications.
To date, many different creatinase proteins from Pseudomonas,Citation4 Alcaligenes sp.,Citation5 and Flavobacterium spCitation6 have been extracted and studied. In particular, the creatinase from Pseudomonas putida has been successfully purified and crystallized.Citation7 Nevertheless, novel creatinases with good properties (such as high resistance to sodium azide) are much more attractive. From this perspective, Zhi et al. recently purified and characterized a NaN3-resistant creatinase from an isolated strain Arthrobacter nicotianae 02181.Citation8 To accumulate this creatinase, the expensive inducer creation has to be supplemented during cultivation which impairs its potential application.Citation9 Moreover, the low expression level and complicated long cultivation process will also restrict its practical applications in creatinase production. Therefore, identification of novel creatinase encoding genes and construction of robust recombinant cell factories for creatinase production is imperative.
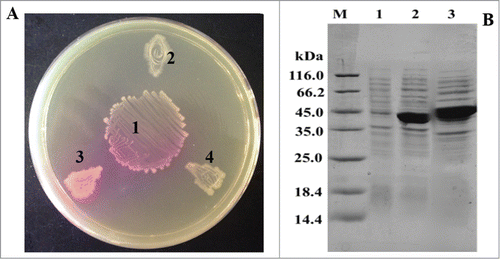
More recently, we cloned a novel creatinase encoding gene from A. nicotianae 23710 and successfully achieved its active expression in Escherichia coli.Citation10 By co-cultivating the recombinant Escherichia coli with an acid urease producing strain Lactobacillus reuteri CICC6124Citation11 on an indicator plate (creatine and phenol red were added), a clear transparent zone could be visually observed. At the same time, it could also be found that compared with the creatinase from P. putida, this novel creatinase showed higher enzymatic activities (). In parallel, SDS-PAGE analysis results also confirmed that this creatinase gene from A. nicotianae 23710 obtained higher-level expression in E. coli than that from P. putida (). Furthermore, enzymatic property analysis results demonstrated that this recombinant creatinase has excellent resistance to NaN3 (20 mM), EDTA, Tween20, Tween80 and Triton X-100 which is of great importance for commercial applications.
Figure 1. Functional expression of the recombinant creatinase in Escherichia coli. (A) Visualized analysis of the active recombinant creatinases with an indicator plate that containing creatine and phenol red. One, Lactobacillus reuteri CICC6124 (an acid urease producing strain)Citation11; 2, E. coli BL21 (pET20b) (control); 3, E. coli BL21A (containing the creatinase gene from Arthrobacter nicotianae 23710); 4, E. coli BL21P (containing the creatinase gene from P. putida KT2440). (B) SDS-PAGE of the recombinant creatinases. Lanes 1, 2 and 3 are the intracellular supernatants of the recombinant strains E. coli BL21 (pET20), E. coli BL21P and E. coli BL21A10, respectively.
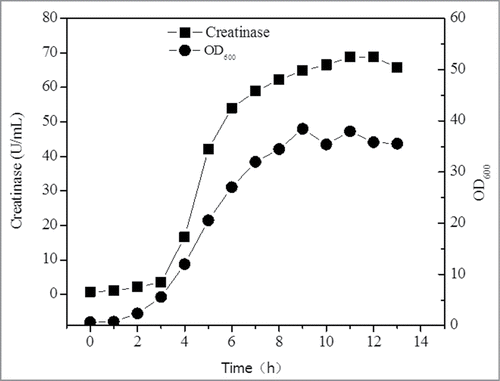
To realize its high level expression and pave the way for industrial production, the cultivation process was systematically optimized. First, the commonly used LB and TB media were compared, and the results showed that more biomass and creatinase were obtained in TB medium. At 7h, the creatinase titer in TB medium was increased to 21.5 U/mL with a biomass value of 13.5 OD600 () while merely 6.95 U/mL creatinase was accumulated when cultivating in LB medium (), confirming TB medium is much more applicable for creatinase expression. Next, the culture pH, temperature and inducer concentration were identified as the crucial fermentation process parameters and optimized to enhance the expression of creatinase. After optimization (the pH, temperature and concentration of IPTG were 7.0, 30 °C and 0.6 mM, respectively), the creatinase production was improved to 33.3 U/mL. In consideration of cell growth and creatinase expression, a 2-stage temperature control strategy was further applied in batch fermentation. Specifically, the temperature was switched from 37 °C to 30 °C for creatinase expression. As expected, the titer of the recombinant creatinase was significantly increased to 61.3 U/mL at 10 h with a productivity of 6.1 U/mL/h. Ten By comparing cell growth curve and the titer of creatinase, it could be found that expression of creatinase was coupled with cell growth. In view of this result, glycerol as carbon source was supplemented during fed-batch fermentation. As shown in , the production of the recombinant creatinase was improved to 68.8 U/mL. To the best of our knowledge, this was the highest reported value until now.
Figure 2. Flask cultivation results in the media of TB and LB. (A) TB medium; (B) LB medium. Specifically, one enzyme unit was defined as the amount of enzyme forming 1 μmol urea per minute under the conditions described before.Citation10
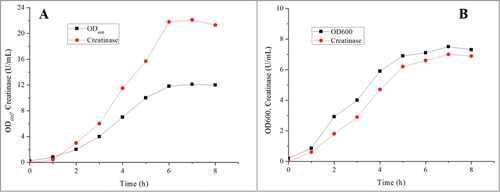
Figure 3. Fed-batch fermentation in 3L fermentor. Glycerol was supplied at 7h with a concentration of 5g/L.
To further reduce the fermentation cost, other expression systems without addition of inducers should be applied for creatinase expression. For instance, Kang et al. developed a stress-induced system in E. coli and successfully applied it for high-yield production of polyhydroxyalkanoates.Citation12 In addition, due to many advantages such as powerful capacity for protein secretion, high amenability for genetic engineering and large-scale fermentation, Bacillus subtilis has been intensively modified for the secretory production of pharmaceutical proteins and industrial enzymes.Citation13 As a result, it will be much more attractive to achieve large scale secretory production of creatinase in B. subtilis by engineering its secretory machinery.Citation14 To expedite its practical applications, further optimization of the enzymatic properties (such as improvement of the thermostability) with directed evolution strategies and synthetic biology tools should be carried out in near future.Citation15
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was financially supported by the National High Technology Research and Development Program of China (863 Program, 2011AA100905), Program for Changjiang Scholars and Innovative Research Team in University (no. IRT1135), the National Science Foundation for Post-doctoral Scientists of China (2013M540414), the Jiangsu Planned Projects for Postdoctoral Research Funds (1301010B) and the 111 Project.
References
- Kim EJ, Haruyama T, Yanagida Y, Kobatake E, Aizawa M. Disposable creatinine sensor based on thick-film hydrogen peroxide electrode system. Analytica Chimica Acta 1999; 394:225-31; http://doi.org/doi:10.1016/S0003-2670(99)00308-6
- Lad U, Khokhar S, Kale GM. Electrochemical creatinine biosensors. Anal Chem 2008; 80: 7910-7. PMID:18975861; http://doi.org/10.1021/ac801500t
- Nilsson D, Hansson LE, Johansson K, Nyström C, Paalzow L, Aquilonius SM. Long-term intraduodenal infusion of a water based levodopa-carbidopa dispersion in very advanced Parkinson disease. Acta Neurol Scand 1998; 97: 175-83; PMID:9531434
- Roche J, Lacombe G, Girard H. On the specificity of certain bacterial deguanidases generating urea and on arginindihydrolase. Biochim Biophys Acta 1950; 6: 210-6. PMID:14791411
- Matsuda Y, Wakamatsu N, Inouye Y, Uede S, Hashimoto Y, Asano K, Nakamura S. Purification and characterization of creatine amidinohydrolase of Alcaligenes origin. Chem Pharm Bull 1986; 34:2155-60. PMID:3742654
- Koyama Y, Kitao S, Yamamoto-Otake H, Suzuki M, Nakano E. Cloning and expression of the creatinase gene from Flavobacterium sp. U-188 in Escherichia coli. Agric Biol Chem 1990; 54:1453-7; PMID: 1368564. http://doi.org/10.1271/bbb1961.54.1453
- Yoshimoto T, Oka I, Tsuru D. Purification, crystallization, and some properties of creatine amidinohydrolase from Pseudomonas putida. J Biochem 1976; 79:1381-3; PMID: 8443
- Zhi Q, Kong P, Zang J, Cui Y, Li S, Li P. Biochemical and molecular characterization of a novel high activity creatine amidinohydrolase from Arthrobacter nicotianae strain 02181. Process Biochem 2009; 44:460-65; http://dx.doi.org/10.1016/j.procbio.2008.12.014
- Chang MC, Chang CC, Chang JC. Cloning of a creatinase gene from Pseudomonas putida in Escherichia coli by using an indicator plate. Appl Environ Microbiol 1992; 58: 3437-40; PMID:1444379. http://dx.doi.org/0099-2240/92/103437-04$02.00/0
- Dai J, Zhang L, Kang Z, Chen J, Du G. High-level Production of Creatine Amidinohydrolase from Arthrobacter nicotianae 23710 in Escherichia coli. Appl Biochem Biotechnol 2015; 175:2564-73; PMID: 25536878. http://dx.doi.org/10.1007/s12010-014-1460-7
- Yang Y, Kang Z, Zhou J, Chen J, Du G. High-level expression and characterization of recombinant acid urease for enzymatic degradation of urea in rice wine. Appl Microbiol Biotechnol 2015; 99:301-8; PMID:25027572; http://dx.doi.org/10.1007/s00253-014-5916-z
- Kang Z, Wang Q, Zhang H, Qi Q. Construction of a stress-induced system in Escherichia coli for efficient polyhydroxyalkanoates production. Appl Microbiol Biotechnol 2008; 79:203-8; PMID:18347791; http://dx.doi.org/10.1007/s00253-008-1428-z
- Kang Z, Yang S, Du G, Chen J. Molecular engineering of secretory machinery components for high-level secretion of proteins in Bacillus species. J Ind Microbiol Biotechnol 2014; 41:1599-607; PMID:25212246; http://dx.doi.org/10.1007/s10295-014-1506-4
- Mulder KC, Bandola J, Schumann W. Construction of an artificial secYEG operon allowing high level secretion of α-amylase. Protein Expr Purif 2013; 89:92-6; PMID:23473827; http://dx.doi.org/10.1016/j.pep.2013.02.008
- Kang Z, Zhang J, Jin P, Yang S. Directed evolution combined with synthetic biology strategies expedite semi-rational engineering of genes and genomes. Bioengineered 2015; 6:136-40; PMID:25621864; http://dx.doi.org/10.1080/21655979.2015.1011029
