Abstract
The transcription factor NUCLEAR FACTOR Y (NF-Y) plays an essential role in many developmental and stress-responsive processes in plants. NF-Y composed of 3 subunits, NF-YA, NF-YB, and NF-YC, targets the CCAAT box, a common cis-element in eukaryotic promoters. We recently identified a gene TaNF-YA10–1 from the wheat salinity tolerant cultivar SR3 and found that recombinant TaNF-YA10–1 could successfully bind to the CCAAT motif in vitro. We also showed that the constitutive expression of TaNF-YA10–1 in Arabidopsis thaliana significantly increased the plant's sensitivity to salinity. Here, we further demonstrated that TaNF-YA10–1 -overexpressing plants conferred drought tolerance as judged from the relative root length and whole-plant growth under drought stress. These results suggest that TaNF-YA10–1 functions independently in salinity and drought stress. Our findings are helpful in understanding the distinct roles of NF-YA in plant stress responses.
Fine regulation of related genes at transcriptional level via different transcription factors is a vital part of plant defense responses in plant adaptive environmental stress response. Citation1-3 Currently, many kinds of transcription factors are known to be involved in plant life processes. Nuclear factor Y (NF-Y) is a type of typical multifunctional transcription factors, which is composed of 3 subunits: NF-YA, NF-YB and NF-YC. The three NF-Y subunits in the nucleus form an active heterotrimer that binds to the CCAAT box, a conserved motif in eukaryotic gene promoters, and thus regulate the transcription of downstream genes.Citation4-6 Each of the 3 NF-Y subunits in yeast and animals is encoded by a single gene, but in plants the NF-Y subunit proteins are present in the form of multigene families, which indicates a more complicated regulation of NF-Y proteins in plants than in other organisms.Citation7 For example, there are 36 NF-Y genes (10 NF-YA, 13 NF-YB and 13 NF-YC) in model plant Arabidopsis thaliana,Citation8 while in bread wheat (Triticum aestivum), the number of NF-Y genes is 80 (18 NF-YA, 34 NF-YB and 28 NF-YC).Citation9,10 By using the model plants (Arabidopsis and rice), it has been found that the members of NF-Y factors are involved in various processes of development and stress responses, including embryo development, flowering control, photosynthesis and the adaptive responses to drought stress, salt stress and nutrient deficiency.Citation9,11-14 Given the complex and large genome of wheat, the functions of NF-Ys in wheat have not been studied sufficiently. We recently isolated a wheat gene TaNF-YA10–1 from the salinity tolerant cultivar SR3 and showed that the constitutive expression of TaNF-YA10–1 in Arabidopsis significantly increased the plant's sensitivity to salinity.Citation15
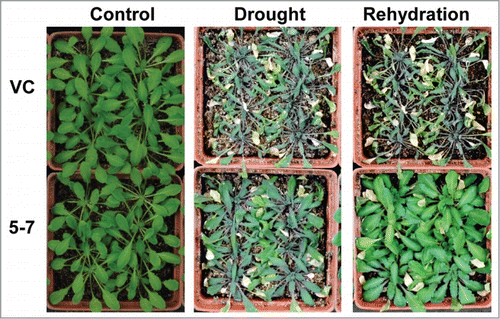
Moreover, NF-Ys have been identified as regulators of drought tolerance in different plant species. Transgenic Arabidopsis and maize (Zea mays) plants overexpressing AtNF-YA5, AtNF-YB1 and the maize ortholog ZmNF-YB2, respectively, have improved performance and survival under drought conditions.Citation11,16 TaNF-YB2, was reported to confer drought resistance and increase crop productivity under drought field tests.Citation10 To assess whether TaNF-YA10–1 also plays a role in drought stress response, we first examined the root elongation of the seedlings grown under medium containing a gradient concentration of mannitol. As shown in , the root length of TaNF-YA10–1 overexpression lines was longer than the wild type and vector control transgenic line when exposure to 300 or 350 mM mannitol. Assay of whole-plant drought tolerance confirmed that the seedlings of TaNF-YA10–1 overexpression line were more tolerant of the drought stress (). These interesting observations suggest that TaNF-YA10–1 is involved in different signaling pathways in response to different abiotic stresses.
Figure 1. The constitutive expression of TaNF-YA10–1 in A. thaliana increased the tolerance of transgenic plants to osmotic stress. Comparison of root length between wild type (Col-0), the empty vector control (VC) and TaNF-YA10–1 overexpression lines (3–6 and 5–7) under normal conditions (A), 300 mM mannitol (B) and 350 mM mannitol (C). (D) Total root length of above seedlings. Man: mannitol. All data are given as mean ± SD from 3 independent experiments. The asterisks and double asterisks represent significant difference determined by the Student's t-test at P < 0.05 and P < 0.01 respectively.
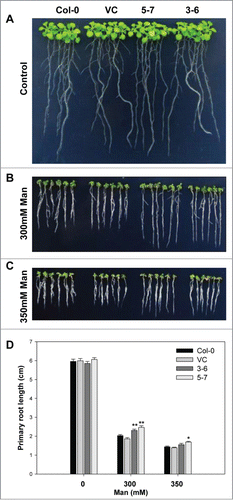
Figure 2. The phenotype of TaNF-YA10–1 overexpression line under drought stress conditions. VC: the empty vector control; 5–7: TaNF-YA10–1 overexpression line. For the drought stress treatment, watering was withheld from 3-week-old plants for 3 weeks before the photograph was taken. For the rehydration treatment, the photograph was taken 3 d after rewatering.
In transgenic A. thaliana constitutively expressing TaNF-YA10–1, the ABA dependent pathway genes AtRAB18, AtRD29B and AtABI5, as well as the ABA-independent pathway genes AtCBF1 and AtCBF3, were all down-regulated when plants were exposed to salinity stress.Citation15 The NF-Y transcription factors have mainly been considered as activators of transcription; however, recent evidence suggests their involvement in gene repression.Citation17,18 Thus, TaNF-YA10–1 might function as repressor of transcription under salt stress conditions. It is suggested that NF-YAs could act as transcriptional activators of a subset of genes that contain the CCAAT-box and as repressors of genes whose promoters lack the CCAAT-box.Citation18 However, clear CCAAT motif enrichment was detected in the promoters of AtCBF1, AtCBF3 and AtRAB18. Transcriptional activation or repression using protoplasts or yeast one-hybrid system will be required to provide direct evidences. Here, we further demonstrated that TaNF-YA10–1-overexpressing plants conferred drought tolerance, suggesting that TaNF-YA10–1 functions independently in salinity and drought stress. Transcriptional behavior of stress responsive genes in TaNF-YA10–1 overexpression lines under osmotic stress conditions remains to be elucidated.
The expansion of NF-Y families in plants, combined with their heterotrimeric nature, means that many possible NF-Y complexes can form. This leads to the formation of a flexible, combinatorial system of transcription factors that may allow subtle adjustments to many different environmental conditions.Citation19 These different functions of TaNF-YA10–1 might be explained by the diverse combination of NF-YA with other NF-YB/NF-YC factors, which thus affected different regulating pathways. We propose that there exists dual transcriptional control of TaNF-YA10–1 through the interaction with different NF-YB/NF-YC heterodimers via its role in activating target drought-responsive genes or repressing target salt-responsive genes. Alternatively, other proteins associating with TaNF-YA10–1 subunit of NF-Y complexes may be different in the presence of salinity and drought stresses. Further study on protein-protein interactions will be needed to elucidate the inner mechanisms.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Funding
This research was financially supported by grants from the Shandong Province Scientific Research Award Foundation for Excellent Young Scientists (2012BSE27117).
References
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A 2006; 103:12987-92; PMID:16924117; http://dx.doi.org/10.1073/pnas.0604882103
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998; 280:104-6; PMID:9525853; http://dx.doi.org/10.1126/science.280.5360.104
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 1999; 17:287-91; PMID:10096298; http://dx.doi.org/10.1038/7036
- Frontini M, Imbriano C, Manni I, Mantovani R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle 2004; 3:217-22; PMID:14712092; http://dx.doi.org/10.4161/cc.3.2.654
- Kahle J, Baake M, Doenecke D, Albig W. Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol Cell Biol 2005; 25:5339-54; PMID:15964792; http://dx.doi.org/10.1128/MCB.25.13.5339-5354.2005
- Steidl S, Tuncher A, Goda H, Guder C, Papadopoulou N, Kobayashi T, Tsukagoshi N, Kato M, Brakhage AA. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J Mol Biol 2004; 342:515-24; PMID:15327951; http://dx.doi.org/10.1016/j.jmb.2004.07.011
- Edwards D, Murray JA, Smith AG. Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol 1998; 117:1015-22; PMID:9662544; http://dx.doi.org/10.1104/pp.117.3.1015
- Gusmaroli G, Tonelli C, Mantovani R. Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 2002; 283:41-8; PMID:11867211; http://dx.doi.org/10.1016/S0378-1119(01)00833-2
- Qu B, He X, Wang J, Zhao Y, Teng W, Shao A, Zhao X, Ma W, Li B, Li Z, Tong Y. A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol 2014; 167:411-23; PMID:25489021; http://dx.doi.org/10.1104/pp.114.246959
- Stephenson TJ, McIntyre CL, Collet C, Xue GP. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol Biol 2007; 65:77-92; PMID:17598077; http://dx.doi.org/10.1007/s11103-007-9200-9
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008; 20:2238-51; PMID:18682547; http://dx.doi.org/10.1105/tpc.108.059444
- Li YJ, Fang Y, Fu YR, Huang JG, Wu CA, Zheng CC. NFYA1 is involved in regulation of postgermination growth arrest under salt stress in Arabidopsis. PloS One 2013; 8:e61289; PMID:23637805
- Mu J, Tan H, Hong S, Liang Y, Zuo J. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant 2013; 6:188-201; PMID:22933713; http://dx.doi.org/10.1093/mp/sss061
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006; 18:2971-84; PMID:17138697; http://dx.doi.org/10.1105/tpc.106.043299
- Ma X, Zhu X, Li C, Song Y, Zhang W, Xia G, Wang M. Overexpression of wheat NF-YA10 gene regulates the salinity stress response in Arabidopsis thaliana. Plant Physiol Biochem 2015; 86:34-43; PMID:25461698; http://dx.doi.org/10.1016/j.plaphy.2014.11.011
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci U S A 2007; 104:16450-5; PMID:17923671; http://dx.doi.org/10.1073/pnas.0707193104
- Ceribelli M, Dolfini D, Merico D, Gatta R, Vigano AM, Pavesi G, Mantovani R. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol 2008; 28:2047-58; PMID:18212061; http://dx.doi.org/10.1128/MCB.01861-07
- Leyva-Gonzalez MA, Ibarra-Laclette E, Cruz-Ramirez A, Herrera-Estrella L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PloS One 2012; 7:e48138; PMID:23118940
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF, 3rd, Mantovani R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012; 24:4777-92; PMID:23275578; http://dx.doi.org/10.1105/tpc.112.105734
