Abstract
We have recently characterized the degradation profiles of 2 human IgG1 monoclonal antibodies, the tumor-targeting mAb H10 and the anti-HIV mAb 2G12. Both mAbs were produced in plants either as stable transgenics or using a transient expression system based on leaf agroinfiltration. The purified antibodies were separated by 1DE and protein bands were characterized by N-terminal sequencing. The proteolytic cleavage sites identified in the heavy chain (HC) of both antibodies were localized in 3 inter-domain regions, suggesting that the number of proteolytic cleavage events taking place in plants is limited. One of the cleavage sites, close to the hinge region, was common to both antibodies.
In this work we report the characterization of mAb H10 degradation profile in plants by using 2-dimensional gel electrophoresis (2-DE) analysis, a technique that allowed us to separate protein species with higher resolution. By subsequent protein spots analysis we identified an additional cleavage site in the VH-CH1 inter-domain region of the HC. Interestingly, other cleavage sites in this region had been previously reported in the literature, supporting the hypothesis that different antibodies produced in plants share common degradation events, which appear to specifically occur in solvent accessible regions at domain interfaces.
Plants are ideal hosts for the production of heterologous proteins. With respect to traditional expression systems, based on bacterial and mammalian cells, plants offer several advantages, including reduced risk of contamination by human pathogens and low production costs.Citation1 A major obstacle hampering the production of recombinant proteins in plants is represented by degradation phenomena, which may take place either in the cell or during downstream processes and result in a dramatic reduction of the final yield of intact heterologous proteins. Moreover, both the quality and yield of the recombinant protein may be significantly influenced by the intrinsic stability of polypeptide chains expressed in heterologous cell environment.Citation2 Hundreds of genes encoding enzymes involved in proteolytic pathways have been identified in plants; as an example, about 800 genes directly or indirectly involved in the hydrolysis of peptide bonds are present in Arabidopsis thaliana.Citation3,4 In the case of antibodies, unintended proteolysis driven by this complex peptidase repertoire can affect the final yield of intact IgG, and may lead to almost complete degradation.Citation5 The presence of antibody fragments in plants has been ascribed to the following 3 phenomena: i) partial assembly intermediates; ii) extracellular peptidase activity after secretionCitation6; iii) activity of peptidases released during sample homogenization.Citation7 Additionally, fragments resulting from a first “opening cleavage” might be further processed into smaller fragments by plant proteases.Citation8 Recent studies have been focused on the identification of cleavage fragments resulting from in planta proteolysis by either mass spectrometry (MS)Citation5,9 or N-terminal sequencing by Edman degradation.Citation10 Mass analysis allowed 2 major degradation products to be identified, compatible with a cleavage presumably occurring close to the heavy chain (HC) hinge region of the H10 antibody.Citation5 Edman degradation analysis led to the identification of the N-terminal sequence of a fragment of the chimeric rat/human Lo-BM2 antibody localized in the hinge region of the HC.Citation10 In a recent study, Hehle and colleagues characterized by N-terminal sequencing the degradation profile of 2 human IgG1 monoclonal antibodies (mAbs) named 2G12 and H10 produced in tobacco plants. A limited number of proteolytic cleavage sites were identified in both the HC and light chain (LC) of the 2 mAbs, all of which are located within inter-domain regions.Citation11
Here we report the study of the degradation profile of the plant produced tumor-targeting mAb H10 determined by reducing 2-dimensional gel electrophoresis (2-DE) analysis. The mAb H10 was produced in Nicotiana benthamiana (N. benthamiana) plants by using a transient expression system based on agroinfiltration and subsequently purified by protein A affinity chromatography, as previously reported.Citation11 For 2-DE analysis, solubilized protein samples were supplemented with 350 μL of isoelectrofocusing (IEF) rehydration buffer and incubated with IPG-strips 3-11NL/18 cm (GE Healthcare, Uppsala,Sweden) O/N at room temperature essentially as described by Di Carli and colleagues.Citation12 Second dimension was run on 12.5 % (w/v) polyacrylamide gels using an Ettan DALT 12 unit (GE Healthcare, San Francisco, CA, USA) and gels were stained by Coomassie Blue as previously described.Citation12 As shown in , we focused our attention on protein spots of about 15–25 kDa with an experimental isoelectric point (pI) range of 3–7. In previous studies we demonstrated that the HC is specifically cleaved in plants yielding protein fragments of about 15 kDa and 25 kDa on reducing gel electrophoresis.Citation5,11 For this reason we expected that higher MW spots (∼50 kDa) corresponded to the complete HC and the selected spots could represent HC-derived fragments. Spots 1–6 have a molecular weight of ∼25 kDa, and are distributed within the pI range 4–7. In the pI range 3–4, 4 major spots are visible: spots 7 and 8, whose molecular weight is ∼23 kDa and ∼18 kDa, respectively; spots 9 and 10, whose molecular weight is ∼20 kDa. To identify the molecular species associated with each spot, Western blot analysis was performed using anti-LC and anti-HC specific antibodies. Briefly, proteins were separated by 2-DE as reported above, blotted on a PVDF membrane (Millipore, Bedford MA) and incubated with either anti-human γ chain (8419; Sigma Aldrich) or anti-human λ chain (A5175; Sigma Aldrich) horseradish peroxidase labeled antibodies for 1h at room temperature in 2% (w/v) non-fat milk in PBS. Detection was performed using ECL Plus Western blotting reagent (GE; Healthcare). In the anti-LC Western blot analysis () major signals at 25 kDa corresponded to spots 1 to 6 observed in the Coomassie stained gel (spot 5 representing the most abundant one). Their molecular weight (∼25 kDa) is in agreement with the presence of the intact form of the LC confirming previous results that showed no appreciable degradation of LC in plantsCitation11 and their different pI values are probably related to different post-translational modifications. Only a faint spot with lower molecular weight and acidic pI was observed, probably corresponding to spot 10 in . The anti-HC Western blot analysis () revealed the presence of 2 spots, an intense one at higher molecular weight (∼23 kDa) which, based on pI and MW values, corresponded to spot 7 on Coomassie gel and a less intense one at lower molecular weight (∼20 kDa) possibly matching to spot 10 of . Based on these results spots 7 and 10 were selected for N-terminal sequencing analysis. This involves a series of chemical reactions to derivatize and remove one amino acid at the time from the N-terminus of purified peptides, enabling the sequential identification of N-terminal residues. N-terminal protein analysis was performed by Dr. Mike Weldon, University of Cambridge, using an ABI Procise 494HT Protein Sequencer. This analysis was limited to 5 residues, which is the minimum number of amino acids required to unequivocally identify a proteolytic cleavage site. N-terminal sequences of spot 7 (pI 4, ∼23 kDa) and spot 10 (pI 3, ∼20 kDa) are shown in . The amino acid sequence of spot 10 matched to the intact N-terminus of the LC while, differently to what observed in the anti-HC Western blot analysis, no heavy chain sequences were detected. A possible explanation for this result is that spot 10 contains a low abundance of HC fragments (a faint signal was observed in the anti-HC Western blot analysis) or that their N-terminus could be blocked either naturally or as a result of sample processing. In fact, it has been demonstrated that many proteins cannot be directly sequenced by Edman degradation because they have a blocked N-terminal residue.Citation13 In the case of spot 7 we found the N-terminal sequence STKGP corresponding to position 119–123 of the HC, localized at the beginning of the CH1 domain. Most interestingly, cleavage sites at the interface between VH and CH1, very close to the STKGP sequence, were previously identified in mAb 2G12 by 2 other research groupsCitation11,14 (), suggesting that this antibody region might be particularly susceptible to proteolytic cleavage. The molecular weight of the species in spot 7 (∼23 kDa) indicates that the C-terminal sequence is likely to be localized in the inter-domain region CH2-CH3 (a schematic illustration is found in ), the same region where cleavage sequences had been previously identified by Hehle and colleagues.Citation11
Figure 1. Two-dimensional gel electrophoresis (2-DE) analysis of mAb H10. (A) Coomassie stained 2-DE of mAb H10 produced in agroinfiltrated N. benthamiana leaves. 800μl of protein A purified antibody (at 1mg/ml concentration) was separated by isoelectrofocusing (IEF) at pH 3-11 Non Linear (NL), followed by SDS-12.5% (w/v) PAGE. The major visible spots are indicated by numbers. Spots 7 and 10 (bold) were further characterized by N-terminal sequencing analysis. Western blot analysis of purified mAb H10 separated by 2DE using anti-LC (B) or anti-HC (C) specific antibodies. Spot numbers in brackets indicate the correspondence (based on observed pI and molecular weight values) to the spots on Coomassie stained gel of Figure 1A.
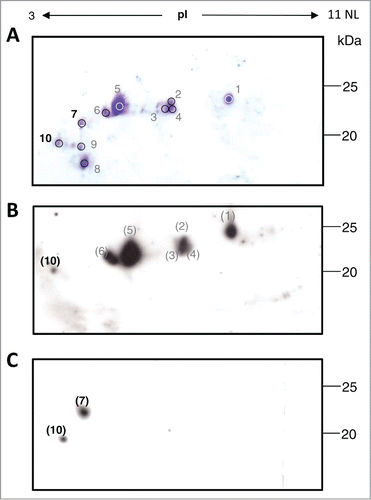
Figure 2. N-terminal sequence analysis of spots 7 and 10 and Heavy chain (HC) cleavage sites of mAbs 2G12 and H10. (A) Summary of results obtained by N-terminal sequence analysis of spots 7 and 10. Top row: domain architecture of complete LC and HC antibody sequences. Middle and bottom rows: the N-terminal sequences of mAb H10 fragments in spots 7 and 10 is reported, together with the deduced molecular species. n.d.: N-terminal sequences in that region were not detected. (B) Alignment of mAbs H10 and 2G12 HC amino acid sequences. The variable (VH) and constant (CH1, CH2 and CH3) domains and hinge region of the heavy chain are indicated and separated by black vertical bars. Open arrows indicate cleavage sites of mAbs 2G12 and H10 previously identified by Hehle and colleagues,Citation11 filled arrows indicate mAb 2G12 cleavage sites identified by Niemer and colleagues.Citation14 The newly identified cleavage site in mAb H10, at the VH-CH1 interface, is indicated by the black diamond-arrow sign and the STKGP N-terminal sequence deriving from it is underlined.

In conclusion, 2-DE analysis coupled with N-terminal sequencing proved to be a powerful method to characterize the products of antibody degradation in plants and identify putative sequence features responsible for it. In this work, we identified a novel mAb H10 proteolytic cleavage site within the antibody HC region, which is located at the interface between VH and CH1 domains and is largely exposed to the solvent. This result strengthens the hypothesis that different antibodies produced in plants share common cleavage events, and that these events preferably take place in solvent accessible, inter-domain regions of antibody molecules.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Paul M, Ma JK-C. Plant-made pharmaceuticals: leading products and production platforms. Biotechnol Appl Biochem 58:58-67; PMID:21446960; http://dx.doi.org/10.1002/bab.6
- Faye L, Boulaflous A, Benchabane M, Gomord V, Michaud D. Protein modifications in the plant secretory pathway: current status and practical implications in molecular pharming. Vaccine 2005; 23:1770-8; PMID:15734039; http://dx.doi.org/10.1016/j.vaccine.2004.11.003
- Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res 2008; 36:D320-5; PMID:17991683; http://dx.doi.org/10.1093/nar/gkm954
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 2004; 55:555-90; PMID:15377232; http://dx.doi.org/10.1146/annurev.arplant.55.031903.141801
- Villani ME, Morgun B, Brunetti P, Marusic C, Lombardi R, Pisoni I, Bacci C, Desiderio A, Benvenuto E, Donini M. Plant pharming of a full-sized, tumour-targeting antibody using different expression strategies. Plant Biotechnol J 2009; 7:59-72; PMID:18793269; http://dx.doi.org/10.1111/j.1467-7652.2008.00371.x
- Hehle VK, Paul MJ, Drake PM, Ma JKC, van Dolleweerd CJ. Antibody degradation in tobacco plants: a predominantly apoplastic process. BMC Biotechnol 2011; 11:128; PMID:22208820; http://dx.doi.org/10.1186/1472-6750-11-128
- Sharp JM, Doran PM. Characterization of monoclonal antibody fragments produced by plant cells. Biotechnol Bioeng 2001; 73:338-46; PMID:11320504; http://dx.doi.org/10.1002/bit.1067
- De Muynck B, Navarre C, Boutry M. Production of antibodies in plants: status after twenty years. Plant Biotechnol J 2010; 8:529-63; PMID:20132515; http://dx.doi.org/10.1111/j.1467-7652.2009.00494.x
- Ramessar K, Rademacher T, Sack M, Stadlmann J, Platis D, Stiegler G, Labrou N, Altmann F, Ma J, Stöger E, et al. Cost-effective production of a vaginal protein microbicide to prevent HIV transmission. Proc Natl Acad Sci U S A 2008; 105:3727-32; PMID:18316741; http://dx.doi.org/10.1073/pnas.0708841104
- De Muynck B, Navarre C, Nizet Y, Stadlmann J, Boutry M. Different subcellular localization and glycosylation for a functional antibody expressed in Nicotiana tabacum plants and suspension cells. Transgenic Res 2009; 18:467-82; PMID:19140023; http://dx.doi.org/10.1007/s11248-008-9240-1
- Hehle VK, Lombardi R, van Dolleweerd CJ, Paul MJ, Di Micco P, Morea V, Benvenuto E, Donini M, Ma JK-C. Site-specific proteolytic degradation of IgG monoclonal antibodies expressed in tobacco plants. Plant Biotechnol J 2015; 13:235-45; PMID:25283551; http://dx.doi.org/10.1111/pbi.12266
- Di Carli M, Tanno B, Capodicasa C, Villani ME, Salzano AM, Scaloni A, Raschellà G, Benvenuto E, Donini M. Proteome changes induced by c-myb silencing in human chronic myeloid leukemia cells suggest molecular mechanisms and putative biomarkers of hematopoietic malignancies. J Proteomics 2014; 96:200-22; PMID:24220303; http://dx.doi.org/10.1016/j.jprot.2013.10.040
- Leone JW, Hampton B, Fowler E, Moyer M, Krishna RG, Chin CCQ. Removal of N-terminal blocking groups from proteins. Curr Protoc Protein Sci 2011; Chapter 11:Unit11.7; PMID:21400688
- Niemer M, Mehofer U, Torres Acosta JA, Verdianz M, Henkel T, Loos A, Strasser R, Maresch D, Rademacher T, Steinkellner H, et al. The human anti-HIV antibodies 2F5, 2G12, and PG9 differ in their susceptibility to proteolytic degradation: down-regulation of endogenous serine and cysteine proteinase activities could improve antibody production in plant-based expression platforms. Biotechnol J 2014; 9:493-500; PMID:24478053; http://dx.doi.org/10.1002/biot.201300207
