Abstract
The review centers on the human gastrointestinal tract; focusing first on the bacterial stress responses needed to overcome the physiochemical defenses of the host, specifically how these stress survival strategies can be used as targets for alternative infection control strategies. The concluding section focuses on recent developments in molecular diagnostics; centring on the shifting paradigm from culture to molecular based diagnostics.
Introduction
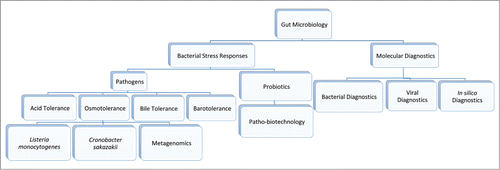
The unifying theme of my research centers on the microbiology of the gut (more specifically, the human gastrointestinal tract). My interests in this niche map to two main thematic areas (outlined in ). The first, deals with the elucidation of bacterial stress responses (i.e. the molecular mechanisms enabling bacterial gastrointestinal persistence), and the exploitation of these mechanisms in the design of novel biocontrol strategies. The second, molecular diagnostics, centers on the identification and characterization of new and emerging gastrointestinal pathogens. Herein, I review my contributions in this field.
Bacterial Stress Responses
My research career began with a focus on bacterial stress responses; determining how bacteria overcome the various stresses encountered in food production/storage and subsequently, following ingestion, within the gastrointestinal tract.
Pathogens
The Gram-positive foodborne bacterium Listeria monocytogenes, a physiologically robust and versatile gastrointestinal pathogen, proved to be the ideal model organism in the early years, which focused on four main stress resistance mechanisms:
Acid tolerance – The ability to resist the low pH of pickled foods and the acidic environment of the stomach (∼pH 2)
Osmotolerance – The ability to grow and survive in dried foods and to resist the low aw of the gastrointestinal tract (with an osmolarity equivalent to 0.3 M NaCl - approximately twice that of blood).
Bile tolerance – Resistance to the biological detergent bile, produced in the liver and stored inter-digestively in the gall bladder. As well as aiding the emulsification and digestion of fats, bile forms an important component of the innate defenses of the gut.
Barotolerance – the ability to overcome pressures in excess of 300 MPa, associated with high pressure processing of certain foods.
Acid tolerance
The first significant challenge encountered by gastrointestinal pathogens, following ingestion, is the low pH of the stomach. Gastric transit is thus an essential first step in establishing infection. In silico analysis of the L. monocytogenes genome identified lmo0292, a gene predicted to encode an HtrA-like serine protease.Citation1 Mutation analysis revealed a role for htrA in acid tolerance and virulence potential in L. monocytogenes. Furthermore, transcriptional analysis revealed that htrA is not only induced by low pH stress, but is also under the control of the two-component regulatory system LisRK.Citation2 Originally identified as an acid tolerance locus and listerial virulence factor, LisRK is also linked to β-lactam resistance in L. monocytogenes.Citation3 Moreover, we identified LisRK and HtrA as important osmotolerance loci.Citation2 Indeed, mutational analysis revealed that in addition to regulating HtrA, LisRK likely controls the transcription of one or more additional systems necessary for optimal listerial osmotolerance. Furthermore, it appears that LisRK may act at the initial stages of the listerial osmotic stress-response, with LisK possibly functioning as the primary sensor and LisR (and its genetic targets, e.g. htrA) as the primary respondent(s) to elevated osmolarity. LisRK may thus represent a new dimension in the osmosensing and osmoregulatory capabilities of L. monocytogenes, functioning independently of, but in tandem with, osmolyte uptake synthesis (described below).
Osmotolerance
L. monocytogenes
In the late 1990s, knowledge of the listerial osmotolerance response was limited; confined mainly to physiological investigations. My strategy was to apply genetic approaches to gain fresh insights into the mechanisms of bacterial osmoadaptation. Indeed, my first published paperCitation4 described the identification and characterization of BetL, a membrane protein which protects Listeria at elevated osmolarities, by transporting the osmoprotective compound betaine. Representing the first genetic analysis of osmotolerance in Listeria, this study also identified a putative σB-dependent promoter binding site upstream of betL, suggesting that betaine uptake in Listeria might be regulated, at least in part, at the level of transcription. Indeed, we later proved the hypothesis, recording a 1.6-fold increase in betL transcript levels following exposure to elevated osmolarity.Citation5 Later, while on sabbatical at the laboratory of Prof. Janet Wood at the University of Guelph, Ontario, Canada, I employed a nisin-controlled expression (NICE) system to decouple transcriptional control of betL, revealing that the BetL protein is itself activated (post-translationally) in response to changes in osmolarity. Rapid activation of pre-existing BetL, in response to relatively low NaCl concentrations, suggested that the protein is one of the primary respondents to rapid fluxes in osmolarity.Citation6 Furthermore, in addition to transcriptional and post-translational control, in silico analysis strongly suggests a role for translational control, not only of betL but of listerial osmoregulation in general.Citation7
However, despite its obvious role in listerial osmotolerance, deleting betL did not significantly impair growth of the pathogen at elevated osmolarity; suggesting the existence of multiple osmolyte uptake systems. In support of this, we described the identification and characterization of OpuC, a carnitine uptake system which contributes to osmotolerance, chill stress and virulence potential of L. monocytogenes.Citation8,9 Indeed, Sleator et al.,Citation8 provided the first direct link between osmotolerance and virulence in Listeria, a link which we ascribed to the accumulation of carnitine, a protective compound similar to betaine but present at much higher concentrations in animal tissues. Following the discovery of Gbu, a third osmolyte uptake system, by Gary Smith's group at the University of California, I undertook a systematic approach to knocking out each of the three uptake systems; BetL, OpuC and Gbu, creating a bank of single, double and triple mutants. This bank allowed me to elucidate, for the first time, the exact role of each transporter against its native background; plotting a hierarchy of osmolyte and transporter importance under osmoticCitation10 chill stressCitation11 and desiccation conditionsCitation12 We subsequently pushed the analysis further;Citation13 establishing the relative importance of each transporter in food and during infection. Interestingly, while betaine appeared to confer most protection in food, the hierarchy of transporter importance differs depending on the food type. While in the animal model, OpuC appears to play the dominant role, with the remaining systems contributing little to the infection process. These findings have had a significant impact on the development of effective control strategies for L. monocytogenes in foods and during infection.
In addition to osmolyte uptake, we also identified the first osmolyte synthesis system in L. monocytogenes, proBA Citation14 While inactivation of proBA had no appreciable effect on virulence, growth at elevated osmolarity was significantly reduced in the absence of efficient proline synthesis. Thus, while proline biosynthesis plays little, if any, role in the intracellular life cycle and infectious nature of L. monocytogenes, it is required for survival in osmolyte-depleted environments of elevated osmolarity. Furthermore, we showed that proline synthesis in Listeria is regulated by feedback inhibition of γ-glutamyl kinase, the first enzyme of the proline biosynthesis pathway, encoded by the proB gene. Random saturation mutagenesis of the proBA operon generated 3 independent mutations in proB, leading to proline overproduction.Citation15 However, as observed for proline auxotrophy, proline hyperproduction has no apparent impact on listerial osmotolerance or indeed virulence potential. Later, using the same random saturation mutagenesis strategy to target betL, we identified a single point mutation in a putative promoter region upstream of betL which led to a dramatic increase in transcript levels under osmo- and chill-stress conditions, and a concomitant increase in stress tolerance.Citation16 Furthermore, the mutation appears to counter a previously unreported “twisted” cell morphology observed for L. monocytogenes grown at elevated osmolarity.
By 2001, my work, and that of others, had fully elucidated the listerial osmotolerance response.Citation17 In less than 4 years, all of the principal osmolyte uptake and synthesis mechanisms had been identified and characterized. This culminated with the publication of the L. monocyogenes genome sequence which we used to perform a complete in silico analysis of osmotolerance in Listeria.Citation18
Cronobacter sakazakii
My interest in osmotolerance did not end however with the conclusion of the Listeria story in 2001. Motivated by a 2008 joint United Nations-World Health Organization call, I returned to the subject of osmotolerance, almost a decade later, this time targeting the Gram-negative pathogen Cronobacter sakazakii. C. sakazakii is an extremely osmotolerant gastrointestinal pathogen.Citation19 The most common route of infection is via powdered infant formula (PIF), resulting in an infant mortality rate of ∼80%. Unravelling C. sakazakii's osmotolerance mechanisms is thus a key first step in controlling the pathogen. Unlike the situation with L. monocytogenes a decade earlier, the genome sequence of C. sakazakii was available from the outset. An exhaustive homology transfer based screening strategy, using the Escherichia coli K12 genome as the template, identified 53 putative osmotoleance loci.Citation20 Interestingly, while C. sakazakii contains homologs of all the principal E. coli osmotolerance loci; a key distinguishing feature is that C. sakazakii possesses multiple copies of certain osmotolerance genes; including seven copies of the E. coli proP homolog, each of which potentially encodes a separate osmolyte uptake system. This level of degeneracy is apparently unique to C. sakazakii (offering a plausible explanation for its distinctive osmotolerance profile). Indeed, gene expression analysis of each of the proP homologs demonstrated an increase in expression levels in response to osmotic upshift.Citation21 Furthermore, heterologous expression against the osmotically sensitive E. coli MKH13 host revealed that 6 of the 7 homologs conferred an osmotolerance phenotype, albeit to varying degrees. Further structural analysis of the C. sakazakii ProP homologs revealed that all but one (ProP1, which offers the greatest osmoprotective effect) are 60–70 amino acids shorter than the E. coli ProP; lacking the extended carboxyl tail. This immediately suggested a role for the C-terminal domain in modulating the protein's osmoprotective function. This hypothesis was proved by spicing the extended C-terminal domain of ProP1 (encoded by ESA_02131) onto the truncated C-terminal end of ProP2 (encoded by ESA_01706); creating a chimeric protein (ProPc) which exhibits increased osmotolerance relative to the wild type.Citation22 In tandem with the sequence based approach, functional screening of a C. sakazakii genomic bank, heterologously expressed against the E. coli MKH13 background, revealed a novel role for ProP1 (ESA_02131) as a carnitine uptake system.Citation23 Indeed, carnitine uptake via ProP1 allows growth at salt concentrations far in excess of that afforded by proline; a finding which has significant food safety implications, given that carnitine is often added to infant formula to boost its nutritional value. Furthermore, we showed that carnitine also promoted the growth of L. monocytoenes in infant formula stored at refrigeration temperatures.Citation9 Work in my lab has thus not only elucidated the C. sakazakii osmotoleance response, it has also helped to inform food safety policy – calling for a halt to the practice of adding carnitine to infant formula. Furthermore, the osmotolerance mechanisms identified now provide ideal targets for the control of the pathogen. One such approach, involves smugglin technology;Citation17,18 the use of antimicrobial compounds which structurally mimic carnitine, killing the cell rather than protecting it. This represents a viable alternative to antibiotic therapy and may lead to effective control of the pathogen in high risk foods such as PIF.
Metagenomics
In addition to focusing on individual gastrointestinal pathogens, my lab also employs a much broader metagenomics approach to identifying novel osmotolerance loci in the human gut microbiome (reviewed inCitation24,25). The human distal colon is one of the most complex and densely populated microbial ecosystems on Earth, with cell densities of up to 1012 per gram of faeces.Citation26-28 A functional screen of over 20,000 clones from a human gut microbiota library revealed 53 osmotolerant clones. A combined transposon mutagenesis and bioinformatic strategy revealed three genes (encoding GalE, MurB and MazG) from a single clone, exhibiting high levels of identity to a species from the genus Collinsella.Citation29 Additionally, in silico analysis of two other clones revealed the presence of an additional galE and mazG gene, from Akkermansia muciniphila and Eggerthella sp., respectively. Each of the three genes identified was found to be over-represented in the human gut metagenome and abundant among healthy subjects from the MetaHit data set,Citation24 suggesting an important role in gut colonisation. Indeed, as expected, cloning and heterologous expression of the genes in E. coli MKH13 resulted in increased salt tolerance of the transformed cells. A phenotype based microarray analysis identified one of the 53 clones as being positive for utilization/transport of carnitine,Citation30 while another of the loci was shown to be homologous to a brp/blh-family β-carotene 15,15′-monooxygenase.Citation31 In addition to the obvious osmotolerance phenotype, E. coli cell pellets expressing brpA adopt a red/orange pigmentation, when cultured in the presence of exogenous β-carotene, indicating the incorporation of carotenoids in the cell membrane. Finally, a significant advantage of functional screens over sequence based approaches is that they provide a powerful means to identify and assign function to novel genes, and their encoded proteins, without any prior sequence knowledge. In support of this, we describe the identification and subsequent analysis of an unknown gene (stlA, for “salt tolerance locus A”) with no currently known homologs in the databases.Citation32 The stlA gene is rare when searched against the human metagenome datasets, MetaHit and the Human Microbiome Project, and represents a novel and unique salt tolerance determinant which is apparently exclusive to the human gut environment.
Bile tolerance
In silico analysis of the L. monocytogenes genome revealed a two-gene operon (formerly opuB), preceded by binding sites for PrfA (positive regulatory factor A) and the alternative stress sigma factor, σB.Citation18 Despite exhibiting significant sequence similarity to the betaine carnitine choline transporter (BCCT) family, further analysis suggested that the protein most likely functions as a bile efflux pump (actively extruding toxic bile out of the cell), a conclusion substantiated by radiolabelled bile exclusion studies. In addition, functionally inactivating BilE resulted in a significantly increased sensitivity to physiological concentrations of human bile, and a reduction in virulence potential when administered orally to a murine model of infection.Citation33 Renamed BilE (for Bile Exclusion), this protein not only represented a new and important virulence factor in L. monocytogenes, but also stimulated in me a new interest in bile tolerance as an important gastrointestinal associated virulence mechanism. This was followed by the functional analysis of bsh, pva, and btlB— three genes previously annotated as bile-associated loci in the public databases.Citation34 While deletion mutants revealed a role for all three genes in resisting the acute toxicity of bile, hydrolysis assays indicated that BSH is likely the only listerial bile salt hydrolase (leading to bile detoxification inside the cell). Transcriptional analyses and activity assays further revealed that, as with bilE, bsh is regulated by both PrfA and σB. The fact that both bilE and bsh are controlled by σB, along with betL, gbu and opuC (which also boasts a PrfA box), suggested a common function for all 4 proteins.Citation35 In support of this hypothesis, a systematic analysis of strains with mutations in BetL, OpuC and Gbu revealed roles for OpuC, and to a lesser extent BetL, in resisting the acute toxicity of bile. Furthermore, real-time gene expression profiling in the presence of bile, using a lux gene reporter system, revealed that both betL and opuC are induced by bile.Citation36
Following ingestion, the first physical stress encountered by the bacterium is the low pH of the stomach (∼pH 2), followed by elevated osmolarity (equivalent to 0.3M NaCl) and activity of the biological detergent bile in the upper small intestine. Significantly, pre-exposure to elevated osmolarity (0.3 M NaCl for 1 h) resulted in a dramatic increase in the ability of L. monocytogenes to deal with lethal concentrations of bile (adapted cells surviving 1,000 times better than naïve cellsCitation37). However, a low-pH challenge (as experienced during gastric transit) fails to similarly protect against subsequent osmotic or bile stress. Equally, pre-exposure to bile fails to protect against acid or salt. Thus, osmotic stress appears to be at the top of the hierarchy of stress responses during gastrointestinal transit.Citation38,39 Given that sigB is transcriptionally upregulated at elevated osmolarity, it is likely that the increased osmolarity of the gastrointestinal lumen may be interpreted by L. monocytogenes as an environmental cue, signaling gut entry (as is the case for Salmonella).Citation40 Furthermore, given that σB is known to modulate expression of prfA (the master regulator of the virulence gene cluster which coordinates the intracellular phase of L. monocytogenes infection) it is possible that osmotically induced stimulation of the σB regulon in the upper small intestine may not only facilitate successful gastrointestinal transit, but also prime the pathogen for the next phase of infection, which, in susceptible individuals, is the systemic invasive disease listeriosis.Citation41
Barotolerance
Prior to ingestion, gastrointestinal pathogens are often subject to a variety of hostile conditions; particularly stresses associated with food processing. One such stress is high-pressure processing (HPP); a non-thermal food processing method that subjects foods (liquid or solid) to pressures between 50 and 1,000 MPa. Indeed, in Considine et al.,Citation42 we outline how HPP can inactivate microorganisms and enzymes and modify macromolecular structures, with little or no impact on the nutritional and sensory quality of foods. As before, our goal was to identify the genetic loci which allow bacteria to survive HPP; thereby permitting a more in-depth assessment of the implications of HPP in the food industry, both in terms of food quality and safety. A L. monocytogenes genome bank was used to screen for barotolerance loci (i.e., resistance to 400 or 450 MPa for 5 minutes). The study, which represents the first use of agar plates as a novel methodology for screening genome libraries using HPP, identified systems involved in quorum sensing (Agr), flagellar machinery (Mot) and cytokinesis (EzrA) as conferring enhanced pressure resistance when expressed against an E. coli background.Citation43 In a follow-on study,Citation44 failure to disrupt EzrA (encoded by lmo1594) revealed that the protein is essential for the growth of L. monocytoenes. Thus, while gene deletion proved impossible; over-expression of lmo1594 confirmed a role for the protein in listerial barotolerance, resulting in significantly improved survival rates at 300 MPa.
In an interesting link with previous sections, and substantiating a role for osmolytes as a listerial passe-partout,Citation45 both osmolyte uptake and synthesis systems have been shown to play a role in listerial barotolerance. Indeed, listerial survival, following exposure to 400 MPa for 5 minutes, increased from 0.008 to 0.02% with added carnitine and to 0.05% with added betaine.Citation46 While proline synthesis is required for optimal survival when betaine and carnitine are absent or limiting.Citation47
Probiotics
In 2005 I was appointed as a principal investigator (PI) at the Alimentary Pharmabiotic Center (APC), UCC. With a mission statement “linking Irish science with industry and society through excellence in research, education and outreach in gastrointestinal health,” the APC is arguably one of the world's leading centers of gut health research. Indeed, Thomson Reuters Science Watch ranks the APC at number 2 in the world for probiotic research. As an APC PI, I developed an interest in probiotics,Citation48-50 specifically in applying the tools and techniques developed during my research on gastrointestinal pathogen stress responses, to develop improved probioticsCitation51 specifically with applications in the developing world.Citation52,53 Given the obvious commercial and clinical relevance of probiotic cultures; improving their stress tolerance profile and ability to overcome the physiochemical defenses of the host is, in my opinion, an important biological goal.Citation54,55
Patho-biotechnology
In 2006 Colin Hill and I coined the term ‘patho-biotechnology’ to describe the exploitation of pathogen derived stress survival strategies to engineer improved probiotic strains.Citation56-60 This approach showed promise not only for the design of more technologically robust probiotic cultures, with improved biotechnological applications, but also in the development of novel vaccine and drug delivery platforms. In support of this, Sheehan et al.,Citation61 describes how heterologous expression of the listerial betL gene in Lactobacillus salivarius significantly increased resistance of the probiotic to several ex vivo stresses, including elevated osmo-, cryo-, baro-, and chill tolerance, as well as increased resistance to spray- and freeze-drying (stresses associated with food processing and preservation). Furthermore, betL also improved in vivo stress survival in Bifidobacterium breve resulting in significantly improved tolerance to gastric juice and osmolarity conditions mimicking the gut environment. Indeed, B. breve strains expressing BetL were recovered at considerably higher levels in the faeces, intestines and cecum of inoculated animals and exhibited significantly lower levels of systemic infection compared to the control strain following oral challenge with L. monocytogenes.Citation57 Similar results were obtained for both B. breve and Lactococcus lactis when betL was replaced with bilE as the transgene of interest.Citation62 These physiologically enhanced probiotics function as ideal drug and vaccine delivery platforms;Citation63 targeting difficult to treat pathogens such as Clostridium difficile,Citation64 Acinetobacter baumanniiCitation65 and Mycobacteria.Citation66-68
In addition to the obvious clinical applications, described above, genetic manipulation of probiotics has provided fundamental insights into their natural life cycle within the host. In Cronin et al.,Citation69 for example, we describe the development of a non-invasive luciferase-based reporter system (pLuxMC1) for real-time tracking of Bifidobacterium species in vivo. This construct allowed us, for the first time, to track the colonisation potential and persistence of the probiotic in real time in live animal models. Indeed, a significant outcome of the study was the identification of the cecum as a niche environment for B. breve; a finding which may explain why appendectomies may lead to an increased risk of functional gastrointestinal disorders.Citation70
Molecular Diagnostics
Molecular diagnostics is arguably one of the fastest growing areas in gastrointestinal research. Growth of the so called ‘omics’ technologies has, over the last decade, led to a gradual migration away from the culture based ‘one test, one pathogen’ paradigm, toward DNA based multiplex approaches to infectious disease diagnosis, which have in turn led to significant improvements in clinical diagnostics and improved patient care.
Bacterial diagnostics
In 2009 our group established a partnership with Serosep (an Irish molecular diagnostics company) and the Clinical Microbiology Department of Cork University Hospital (CUH), focused on the changing culture of medical microbiology in Ireland.Citation71 A retrospective analysis of faecal samples submitted to CUH in 2009 was performed to identify all Campylobacter species detected using Serosep's EntericBio® multiplex PCR system, in a single calendar year. The results confirmed that routine culture fails to detect over a third of Campylobacter positive samples. From a total of 7,194 diarrheal specimens, 349 Campylobacter-positive samples (23.8%) were shown to be Campylobacter ureolyticus. This represents the first report of C. ureolyticus in the faeces of patients presenting with gastroenteritis, suggesting a heretofore unreported role for this organism as an enteric pathogen.Citation72-74 Indeed, we showed that C. ureolyticus is second only to C. jejuni as the most common cause of Campylobacter associated gastroenteritis in Southern Ireland.Citation75 Furthermore, we reported a prominent seasonal distribution for campylobacteriosis (Spring), with C. ureolyticus (March) preceding C. jejuni/C. coli (April/May) and exhibiting a bimodal age and gender profile; being more commonly detected in elderly female patients.
Given that cultural isolation of C. ureolyticus was not possible using the established selective methods for the isolation of Campylobacter spp., we were forced to develop a new selective medium, nalidixic acid amphotericin B vancomycin (NAV), capable of isolating C. ureolyticus from faecal samples.Citation76 This medium made it possible, for the first time, to isolate pure cultures of C. ureolyticus and to determine the potential source of infections in humans. Building on this, Koziel et al.,Citation77 describes the collection and processing of 164 samples from various domestic animals over a period of 6 months. Significantly, Random Amplified Polymorphic DNA (RAPD) analysis identified a feline derived C. ureolyticus isolate as being genetically similar to a strain (CIT 007) isolated from an elderly female patient presenting with gastroenteritis. Together with the findings of Koziel et al.,,Citation75 which reported the detection of C. ureolyticus in bovine samples, it appeared likely that this emerging pathogen has a zoonotic potential. Whole genome sequence analysis of CIT 007 appeared to confirm this hypothesis; identifying a number of genetic loci necessary to cause gastrointestinal infection in humans.Citation78 Indeed, whole genome analysis of two further C. ureolyticus isolates, including the type strain, revealing 106 putative virulence associated factors, 52 of which are predicted to be secreted. Furthermore, this analysis also suggested that C. ureolyticus is likely to exist as a taxonomic continuum comprised of several species (genomospecies) that are likely to have varying impacts on human health and disease.Citation79
Interestingly, a C. ureolyticus screen in exotic animals led to the identification of strain CIT 045(T) in the faeces of captive lion-tailed macaques (Macaca silenus). Originally believed to be C. ureolyticus on the basis of colony morphology; 16S rRNA analysis showed it to be a completely new Campylobacter species;Citation80 a finding which we confirmed using matrix-assisted laser desorption/ionization time of flight (MALDI-TOF), together with whole genome sequence analysis.Citation81 We named this new species Campylobacter corcagiensis sp nov. (The Corkonian!), in honor of its home town of Cork.
In parallel with the identification and characterization of C. ureolyticus and C. corcagiensis, our lab has continued to work with Serosep; validating the next two iterations of the EntericBio® platform. The EntericBio Panel II® system, developed to detect Shiga toxin-producing Escherichia coli (STEC), differentiating Shiga-like toxins 1 and 2 (SLT-1 and SLT-2)Citation82 and the EntericBio real-time Gastro Panel I, which allows real-time detection of C. jejuni, coli, and lari, STEC, Shigella spp, and Salmonella spp. directly from faeces, without pre-enrichment.Citation83
Viral diagnostics
In addition to gastrointestinal specific bacterial diagnostics, we have also targeted gastrointestinal viruses; namely rotavirus, parvovirus and coronaviruses. Collins et al.,Citation84 investigated the occurrence of asymptomatic infections by procine group A rotaviruses in Irish swine between 2005–2007, while Cashman et al.,Citation85 examined the different bovine group A rotavirus genotypes circulating in the Irish bovine population from 2002–2009. Additionally, McElligott et al.,Citation86 described a nationwide screen, between 2008 and 2009, of canine parvovirus (CPV) and coronavirus (CCoV); the most common cause of acute gastroenteritis in dogs. A total of 250 faecal samples were collected from both symptomatic and asymptomatic dogs, from a diverse array of sources, representing the major canine groups in Ireland. Seven CPV strains and three CCoV strains were identified in symptomatic dogs, with one animal presenting with mixed CPV/CCoV infection. Significantly, from a public health perspective, each of these animal viruses represents a vast reservoir of genetic material for the diversification of their human equivalents. Thus, a key outcome of these studies is the need for constant genotype surveillance and vigilance. This is particularly relevant given that shifting genotype prevalence, the emergence of novel combinations and mixed infections, are all likely to impact negatively on existing vaccination programmes. In extreme cases this can lead to breakthrough infections.
In silico diagnostics
The development of ever faster and cheaper next generation sequencing strategies has resulted in a paradigm shift in molecular diagnostics; changing the focus from single target genes to whole genomes. Since 2008, genomic data has outpaced Moore's Law by a factor of 4, with a global annual sequencing capacity estimated to be 13 quadrillion bases; enough data to fill a stack of DVDs 2 miles high! Recognizing the potential of biology's big data sets in developing improved diagnostic techniques, our lab began to focus on phylogenetics,Citation87,88 genomics,Citation89-92 protein structure/function predictionCitation93-98 and the emerging field of synthetic biology.Citation99-107 Indeed, this changing focus allowed me to establish links with the Department of Computing at Cork Institute of Technology (CIT).Citation108,109 This union produced CIT's first commercial spinout company, nSilico, which I co-founded in 2012. Combining all of the above elements, I wrote and coordinated ClouDx-i, a €1.3 million EU FP7 funded project, with CIT, nSilico and the University of Edinburgh as partners. The principal scientific and technological objective of ClouDx-i was to develop computational approaches to support rapid molecular diagnosis of infection, embedding these techniques in an efficient end-to-end diagnostic process.Citation110,111 The project has sequenced and analyzed the genomes of 12 bacterial pathogens, identifying biomarkers for the rapid diagnosis of neonatal infections.Citation112-118
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
Sleator is Coordinator of the EU FP7 project ClouDx-i.
References
- Stack HM, Sleator RD, Bowers M, Hill C, Gahan CG. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl Environ Microbiol 2005; 71:4241-7; PMID:16085809; http://dx.doi.org/10.1128/AEM.71.8.4241-4247.2005
- Sleator RD, Hill C. A novel role for the LisRK two-component regulatory system in listerial osmotolerance. Clin Microbiol Infect 2005; 11:599-601; PMID:16008610; http://dx.doi.org/10.1111/j.1469-0691.2005.01176.x
- Van de Velde S, Carryn S, Van Bambeke F, Hill C, Tulkens PM, Sleator RD. Penicillin-binding proteins (PBP) and Lmo0441 (a PBP-like protein) play a role in Beta-lactam sensitivity of Listeria monocytogenes. Gut Pathog 2009; 1:23; PMID:20003484; http://dx.doi.org/10.1186/1757-4749-1-23
- Sleator RD, Gahan CG, Abee T, Hill C. Identification and disruption of BetL, a secondary glycine betaine transport system linked to the salt tolerance of Listeria monocytogenes LO28. Appl Environ Microbiol 1999; 65:2078-83; PMID:10224004
- Sleator RD, Gahan CGM, O'Driscoll B, Hill C. Analysis of the role of betL in contributing to the growth and survival of Listeria monocytogenes LO28. Int J Food Microbiol 2000; 60:261-8; PMID:11016615; http://dx.doi.org/10.1016/S0168-1605(00)00316-0
- Sleator RD, Wood JM, Hill C. Transcriptional regulation and posttranslational activity of the betaine transporter BetL in Listeria monocytogenes are controlled by environmental salinity. J Bacteriol 2003; 185:7140-4; PMID:14645273; http://dx.doi.org/10.1128/JB.185.24.7140-7144.2003
- Sleator RD. A role for translational control in listerial osmoregulation and strain variation? Foodborne Pathog Dis 2007; 4:395-6; PMID:18041949; http://dx.doi.org/10.1089/fpd.2007.0043
- Sleator RD, Wouters J, Gahan CG, Abee T, Hill C. Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl Environ Microbiol 2001; 67:2692-8; PMID:11375182; http://dx.doi.org/10.1128/AEM.67.6.2692-2698.2001
- Sleator RD, Banville N, Hill C. Carnitine enhances the growth of Listeria monocytogenes in infant formula at 7 degrees C. J Food Prot 2009; 72:1293-5; PMID:19610343
- Wemekamp-Kamphuis HH, Wouters JA, Sleator RD, Gahan CG, Hill C, Abee T. Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl Environ Microbiol 2002; 68:4710-6; PMID:12324311; http://dx.doi.org/10.1128/AEM.68.10.4710-4716.2002
- Wemekamp-Kamphuis HH, Sleator RD, Wouters JA, Hill C, Abee T. Molecular and physiological analysis of the role of osmolyte transporters BetL, Gbu, and OpuC in growth of Listeria monocytogenes at low temperatures. Appl Environ Microbiol 2004; 70:2912-8; PMID:15128551; http://dx.doi.org/10.1128/AEM.70.5.2912-2918.2004
- Dreux N, Albagnac C, Sleator RD, Hill C, Carlin F, Morris CE, Nguyen-the C. Glycine betaine improves Listeria monocytogenes tolerance to desiccation on parsley leaves independent of the osmolyte transporters BetL, Gbu and OpuC. J Appl Microbiol 2008; 104:1221-7; PMID:17976173; http://dx.doi.org/10.1111/j.1365-2672.2007.03623.x
- Sleator RD, Francis GA, O'Beirne D, Gahan CG, Hill C. Betaine and carnitine uptake systems in Listeria monocytogenes affect growth and survival in foods and during infection. J Appl Microbiol 2003; 95:839-46; PMID:12969299; http://dx.doi.org/10.1046/j.1365-2672.2003.02056.x
- Sleator RD, Gahan CG, Hill C. Identification and disruption of the proBA locus in Listeria monocytogenes: role of proline biosynthesis in salt tolerance and murine infection. Appl Environ Microbiol 2001; 67:2571-7; PMID:11375165; http://dx.doi.org/10.1128/AEM.67.6.2571-2577.2001
- Sleator RD, Gahan CG, Hill C. Mutations in the listerial proB gene leading to proline overproduction: effects on salt tolerance and murine infection. Appl Environ Microbiol 2001; 67:4560-5; PMID:11571156; http://dx.doi.org/10.1128/AEM.67.10.4560-4565.2001
- Hoffmann RF, McLernon S, Feeney A, Hill C, Sleator RD. A single point mutation in the listerial betL sigma(A)-dependent promoter leads to improved osmo- and chill-tolerance and a morphological shift at elevated osmolarity. Bioengineered 2013; 4:401-7; PMID:23478432; http://dx.doi.org/10.4161/bioe.24094
- Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 2002; 26:49-71; PMID:12007642; http://dx.doi.org/10.1111/j.1574-6976.2002.tb00598.x
- Sleator RD, Gahan CG, Hill C. A postgenomic appraisal of osmotolerance in Listeria monocytogenes. Appl Environ Microbiol 2003; 69:1-9; PMID:12513970; http://dx.doi.org/10.1128/AEM.69.1.1-9.2003
- Feeney A, Kropp KA, O'Connor R, Sleator RD. Cronobacter sakazakii: stress survival and virulence potential in an opportunistic foodborne pathogen. Gut Microbes 2014; 5:711-8; PMID:25562731; http://dx.doi.org/10.4161/19490976.2014.983774
- Feeney A, Sleator RD. An in silico analysis of osmotolerance in the emerging gastrointestinal pathogen Cronobacter sakazakii. Bioeng Bugs 2011; 2:260-70; PMID:21918371; http://dx.doi.org/10.4161/bbug.2.5.17238
- Feeney A, Johnston CD, Govender R, O'Mahony J, Coffey A, Sleator RD. Analysis of the role of the Cronobacter sakazakii ProP homologues in osmotolerance. Gut Pathog 2014; 6:15; PMID:24910715; http://dx.doi.org/10.1186/1757-4749-6-15
- Feeney A, Johnston CD, Lucid A, O'Mahony J, Coffey A, Lucey B, Sleator RD. The role of the Cronobacter sakazakii ProP C-terminal coiled coil domain in osmotolerance. Gut Pathog 2014; 6:46; PMID:25530808; http://dx.doi.org/10.1186/s13099-014-0046-9
- Feeney A, Sleator RD. Functional Screening of the Cronobacter sakazakii BAA-894 Genome reveals a role for ProP (ESA_02131) in carnitine uptake. Bioengineered 2015:1-5; PMID:25915804
- Culligan EP, Marchesi JR, Hill C, Sleator RD. Mining the human gut microbiome for novel stress resistance genes. Gut Microbes 2012; 3:394-7; PMID:22688726; http://dx.doi.org/10.4161/gmic.20984
- Culligan EP, Sleator RD, Marchesi JR, Hill C. Metagenomics and novel gene discovery: promise and potential for novel therapeutics. Virulence 2014; 5:399-412; PMID:24317337; http://dx.doi.org/10.4161/viru.27208
- Sleator RD, Shortall C, Hill C. Metagenomics. Lett Appl Microbiol 2008; 47:361-6; PMID:19146522; http://dx.doi.org/10.1111/j.1472-765X.2008.02444.x
- Sleator RD. The human superorganism - of microbes and men. Med Hypotheses 2010; 74:214-5; PMID:19836146; http://dx.doi.org/10.1016/j.mehy.2009.08.047
- Feeney A, Sleator RD. The human gut microbiome: the ghost in the machine. Fut Microbiol 2012; 7:1235-7; PMID:23075440; http://dx.doi.org/10.2217/fmb.12.105
- Culligan EP, Sleator RD, Marchesi JR, Hill C. Functional metagenomics reveals novel salt tolerance loci from the human gut microbiome. ISME J 2012; 6:1916-25; PMID:22534607; http://dx.doi.org/10.1038/ismej.2012.38
- Culligan EP, Marchesi JR, Hill C, Sleator RD. Combined metagenomic and phenomic approaches identify a novel salt tolerance gene from the human gut microbiome. Front Microbiol 2014; 5:189; PMID:24808895; http://dx.doi.org/10.3389/fmicb.2014.00189
- Culligan EP, Sleator RD, Marchesi JR, Hill C. Metagenomic identification of a novel salt tolerance gene from the human gut microbiome which encodes a membrane protein with homology to a brp/blh-family beta-carotene 15,15′-monooxygenase. PloS One 2014; 9:e103318; PMID:25058308; http://dx.doi.org/10.1371/journal.pone.0103318
- Culligan EP, Sleator RD, Marchesi JR, Hill C. Functional environmental screening of a metagenomic library identifies stlA; a unique salt tolerance locus from the human gut microbiome. PloS One 2013; 8:e82985; PMID:24349412; http://dx.doi.org/10.1371/journal.pone.0082985
- Sleator RD, Wemekamp-Kamphuis HH, Gahan CG, Abee T, Hill C. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol Microbiol 2005; 55:1183-95; PMID:15686563; http://dx.doi.org/10.1111/j.1365-2958.2004.04454.x
- Begley M, Sleator RD, Gahan CG, Hill C. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun 2005; 73:894-904; PMID:15664931; http://dx.doi.org/10.1128/IAI.73.2.894-904.2005
- Sleator RD, Hill C. Compatible solutes: the key to Listeria's success as a versatile gastrointestinal pathogen? Gut Pathog 2010; 2:20; PMID:21143981; http://dx.doi.org/10.1186/1757-4749-2-20
- Watson D, Sleator RD, Casey PG, Hill C, Gahan CG. Specific osmolyte transporters mediate bile tolerance in Listeria monocytogenes. Infect Immun 2009; 77:4895-904; PMID:19737907; http://dx.doi.org/10.1128/IAI.00153-09
- Sleator RD, Clifford T, Hill C. Gut osmolarity: a key environmental cue initiating the gastrointestinal phase of Listeria monocytogenes infection? Med Hypotheses 2007; 69:1090-2; PMID:17433559; http://dx.doi.org/10.1016/j.mehy.2007.02.028
- Sleator RD, Hill C. Food reformulations for improved health: A potential risk for microbial food safety? Med Hypotheses 2007; 69:1323-4; PMID:17452083; http://dx.doi.org/10.1016/j.mehy.2007.03.007
- Sleator RD, Hill C. Molecular analysis of the microbial food safety implications of food reformulations for improved health. Foodborne Pathog Dis 2008; 5:499-504; PMID:18666862; http://dx.doi.org/10.1089/fpd.2008.0089
- Sleator RD, Watson D, Hill C, Gahan CG. The interaction between Listeria monocytogenes and the host gastrointestinal tract. Microbiology 2009; 155:2463-75; PMID:19542009; http://dx.doi.org/10.1099/mic.0.030205-0
- McLaughlin HP, Xiao Q, Rea RB, Pi H, Casey PG, Darby T, Charbit A, Sleator RD, Joyce SA, Cowart RE, et al. A putative P-type ATPase required for virulence and resistance to haem toxicity in Listeria monocytogenes. PloS One 2012; 7:e30928; PMID:22363518; http://dx.doi.org/10.1371/journal.pone.0030928
- Considine KM, Kelly AL, Fitzgerald GF, Hill C, Sleator RD. High-pressure processing–effects on microbial food safety and food quality. FEMS Microbiol Lett 2008; 281:1-9; PMID:18279335; http://dx.doi.org/10.1111/j.1574-6968.2008.01084.x
- Considine KM, Sleator RD, Kelly AL, Fitzgerald GF, Hill C. Novel listerial genetic loci conferring enhanced barotolerance in Escherichia coli. J Appl Microbiol 2011; 110:618-30; PMID:21223465; http://dx.doi.org/10.1111/j.1365-2672.2010.04924.x
- Considine KM, Sleator RD, Kelly AL, Fitzgerald GF, Hill C. Identification and characterization of an essential gene in Listeria monocytogenes using an inducible gene expression system. Bioeng Bugs 2011; 2:150-9; PMID:21637009; http://dx.doi.org/10.4161/bbug.2.3.15476
- Sleator RD, Hill C. Compatible solutes: A listerial passe-partout? Gut Microbes 2010; 1:77-9; PMID:21326913; http://dx.doi.org/10.4161/gmic.1.2.10968
- Smiddy M, Sleator RD, Patterson MF, Hill C, Kelly AL. Role for compatible solutes glycine betaine and L-carnitine in listerial barotolerance. Appl Environ Microbiol 2004; 70:7555-7; PMID:15574960; http://dx.doi.org/10.1128/AEM.70.12.7555-7557.2004
- Considine KM, Sleator RD, Kelly AL, Fitzgerald GF, Hill C. A role for proline synthesis and transport in Listeria monocytogenes barotolerance. J Appl Microbiol 2011; 110:1187-94; PMID:21338448; http://dx.doi.org/10.1111/j.1365-2672.2011.04982.x
- Sleator RD, Hill C. New frontiers in probiotic research. Lett Appl Microbiol 2008; 46:143-7; PMID:18028323; http://dx.doi.org/10.1111/j.1472-765X.2007.02293.x
- Culligan EP, Hill C, Sleator RD. Probiotics and gastrointestinal disease: successes, problems and future prospects. Gut Pathog 2009; 1:19; PMID:19930635; http://dx.doi.org/10.1186/1757-4749-1-19
- Sleator RD. Probiotics – a viable therapeutic alternative for enteric infections especially in the developing world. Discov Med 2010; 10:119-24; PMID:20807472
- Sleator RD. Probiotic therapy - recruiting old friends to fight new foes. Gut Pathog 2010; 2:5; PMID:20579345; http://dx.doi.org/10.1186/1757-4749-2-5
- Sleator RD. Designer probiotics: Development and applications in gastrointestinal health. World J Gastrointest Pathophysiol 2015; 6:73-8; PMID:26301121
- Sleator RD. Not so neglected diseases. J Infect Dev Ctries 2007; 1:350-1; PMID:19734620
- Sleator RD, Hill C. Engineered pharmabiotics with improved therapeutic potential. Hum Vaccin 2008; 4:271-4; PMID:18682694; http://dx.doi.org/10.4161/hv.4.4.6315
- Sleator RD, Hill C. Rational design of improved pharmabiotics. J Biomed Biotechnol 2009; 2009:275287; PMID:19753318; http://dx.doi.org/10.1155/2009/275287
- Sleator RD, Hill C. Patho-biotechnology: using bad bugs to do good things. Curr Opin Biotechnol 2006; 17:211-6; PMID:16459072; http://dx.doi.org/10.1016/j.copbio.2006.01.006
- Sheehan VM, Sleator RD, Hill C, Fitzgerald GF. Improving gastric transit, gastrointestinal persistence and therapeutic efficacy of the probiotic strain Bifidobacterium breve UCC2003. Microbiology 2007; 153:3563-71; PMID:17906153; http://dx.doi.org/10.1099/mic.0.2007/006510-0
- Sleator RD, Hill C. Patho-biotechnology; using bad bugs to make good bugs better. Sci Prog 2007; 90:1-14; PMID:17455762; http://dx.doi.org/10.3184/003685007780440530
- Sleator RD, Hill C. 'Bioengineered Bugs' - a patho-biotechnology approach to probiotic research and applications. Med Hypotheses 2008; 70:167-9; PMID:17452084; http://dx.doi.org/10.1016/j.mehy.2007.03.008
- Sleator RD, Hill C. Battle of the bugs. Science 2008; 321:1294-5; PMID:18772416; http://dx.doi.org/10.1126/science.321.5894.1294b
- Sheehan VM, Sleator RD, Fitzgerald GF, Hill C. Heterologous expression of BetL, a betaine uptake system, enhances the stress tolerance of Lactobacillus salivarius UCC118. Appl Environ Microbiol 2006; 72:2170-7; PMID:16517668; http://dx.doi.org/10.1128/AEM.72.3.2170-2177.2006
- Watson D, Sleator RD, Hill C, Gahan CG. Enhancing bile tolerance improves survival and persistence of Bifidobacterium and Lactococcus in the murine gastrointestinal tract. BMC Microbiol 2008; 8:176; PMID:18844989; http://dx.doi.org/10.1186/1471-2180-8-176
- Johnston C, Coffey A, J OM, Sleator RD. Development of a novel oral vaccine against Mycobacterium avium paratuberculosis and Johne disease: a patho-biotechnological approach. Bioeng Bugs 2010; 1:155-63; PMID:21326921; http://dx.doi.org/10.4161/bbug.1.3.10408
- Sleator RD, Hill C. Designer probiotics: a potential therapeutic for Clostridium difficile? J Med Microbiol 2008; 57:793-4; PMID:18480340; http://dx.doi.org/10.1099/jmm.0.47697-0
- Howard A, O'Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 2012; 3:243-50; PMID:22546906; http://dx.doi.org/10.4161/viru.19700
- Talip BA, Sleator RD, Lowery CJ, Dooley JS, Snelling WJ. An Update on Global Tuberculosis (TB). Infect Dis 2013; 6:39-50; PMID:24847176
- Johnston CD, Bannantine JP, Govender R, Endersen L, Pletzer D, Weingart H, Coffey A, O'Mahony J, Sleator RD. Enhanced expression of codon optimized Mycobacterium avium subsp. paratuberculosis antigens in Lactobacillus salivarius. Front Cell Infect Microbiol 2014; 4:120; PMID:25237653; http://dx.doi.org/10.3389/fcimb.2014.00120
- Johnston C, Douarre PE, Soulimane T, Pletzer D, Weingart H, MacSharry J, Coffey A, Sleator RD, O'Mahony J. Codon optimisation to improve expression of a Mycobacterium avium ssp. paratuberculosis-specific membrane-associated antigen by Lactobacillus salivarius. Pathog Dis 2013; 68:27-38; PMID:23620276; http://dx.doi.org/10.1111/2049-632X.12040
- Cronin M, Sleator RD, Hill C, Fitzgerald GF, van Sinderen D. Development of a luciferase-based reporter system to monitor Bifidobacterium breve UCC2003 persistence in mice. BMC Microbiol 2008; 8:161; PMID:18816375; http://dx.doi.org/10.1186/1471-2180-8-161
- Sleator RD, Cronin M, Hill C. Why appendectomies may lead to an increased risk of functional gastrointestinal disorders. Med Hypotheses 2008; 71:814-6; PMID:18691832; http://dx.doi.org/10.1016/j.mehy.2008.06.008
- Bullman S, Lucey B, Sleator RD. Molecular diagnostics: the changing culture of medical microbiology. Bioeng Bugs 2012; 3:1-7; PMID:22179143; http://dx.doi.org/10.4161/bbug.3.1.19011
- Bullman S, Corcoran D, O'Leary J, Lucey B, Byrne D, Sleator RD. Campylobacter ureolyticus: an emerging gastrointestinal pathogen? FEMS Immunol Med Microbiol 2011; 61:228-30; PMID:21320172; http://dx.doi.org/10.1111/j.1574-695X.2010.00760.x
- Bullman S, Corcoran D, O'Leary J, Byrne D, Lucey B, Sleator RD. Epsilonproteobacteria in humans, New Zealand. Emerg Infect Dis 2012; 18:1709-10; author reply 10-1; PMID:23017215; http://dx.doi.org/10.3201/eid1810.120369
- O'Donovan D, Corcoran GD, Lucey B, Sleator RD. Campylobacter ureolyticus: a portrait of the pathogen. Virulence 2014; 5:498-506; PMID:24717836; http://dx.doi.org/10.4161/viru.28776
- Koziel M, Lucey B, Bullman S, Corcoran GD, Sleator RD. Molecular-based detection of the gastrointestinal pathogen Campylobacter ureolyticus in unpasteurized milk samples from two cattle farms in Ireland. Gut Pathog 2012; 4:14; PMID:23151337; http://dx.doi.org/10.1186/1757-4749-4-14
- O'Doherty A, Koziel M, De Barra L, Corcoran D, Bullman S, Lucey B, Sleator RD. Development of nalidixic acid amphotericin B vancomycin (NAV) medium for the isolation of Campylobacter ureolyticus from the stools of patients presenting with acute gastroenteritis. Br J Biomed Sci 2014; 71:6-12; PMID:24693569
- Koziel M, Corcoran GD, Sleator RD, Lucey B. Detection and molecular analysis of Campylobacter ureolyticus in domestic animals. Gut Pathog 2014; 6:9; PMID:24739468; http://dx.doi.org/10.1186/1757-4749-6-9
- Lucid A, Bullman S, Koziel M, Corcoran GD, Cotter PD, Sleator RD, Lucey B. Draft genome sequence of campylobacter ureolyticus strain CIT007, the first whole-genome sequence of a clinical Isolate. Gen Announc 2014; 2; PMID:24723712; http://dx.doi.org/10.1128/genomeA.00262-14
- Bullman S, Lucid A, Corcoran D, Sleator RD, Lucey B. Genomic investigation into strain heterogeneity and pathogenic potential of the emerging gastrointestinal pathogen Campylobacter ureolyticus. PloS One 2013; 8:e71515; PMID:24023611; http://dx.doi.org/10.1371/journal.pone.0071515
- Koziel M, O'Doherty P, Vandamme P, Corcoran GD, Sleator RD, Lucey B. Campylobacter corcagiensis sp. nov., isolated from faeces of captive lion-tailed macaques (Macaca silenus). Int J Syst Evol Microbiol 2014; 64:2878-83; PMID:24876239; http://dx.doi.org/10.1099/ijs.0.063867-0
- Koziel M, Lucid A, Bullman S, Corcoran GD, Lucey B, Sleator RD. Draft genome sequence of campylobacter corcagiensis strain CIT045T, a representative of a novel campylobacter species isolated from lion-tailed macaques (Macaca silenus). Gen Announc 2014; 2; PMID:24744327; http://dx.doi.org/10.1128/genomeA.00248-14
- Koziel M, Corcoran D, O'Callaghan I, Sleator RD, Lucey B. Validation of the EntericBio Panel II(R) multiplex polymerase chain reaction system for detection of Campylobacter spp., Salmonella spp., Shigella spp., and verotoxigenic E. coli for use in a clinical diagnostic setting. Diagn Microbiol Infect Dis 2013; 75:46-9; PMID:23182078; http://dx.doi.org/10.1016/j.diagmicrobio.2012.09.007
- Koziel M, Kiely R, Blake L, O'Callaghan I, Corcoran GD, Lucey B, Sleator RD. Improved detection of bacterial pathogens in patients presenting with gastroenteritis by use of the EntericBio real-time Gastro Panel I assay. J Clin Microbiol 2013; 51:2679-85; PMID:23761157; http://dx.doi.org/10.1128/JCM.00809-13
- Collins PJ, Martella V, Sleator RD, Fanning S, O'Shea H. Detection and characterisation of group A rotavirus in asymptomatic piglets in southern Ireland. Arch Virol 2010; 155:1247-59; PMID:20526785; http://dx.doi.org/10.1007/s00705-010-0713-1
- Cashman O, Lennon G, Sleator RD, Power E, Fanning S, O'Shea H. Changing profile of the bovine rotavirus G6 population in the south of Ireland from 2002 to 2009. Vet Microbiol 2010; 146:238-44; PMID:20541335; http://dx.doi.org/10.1016/j.vetmic.2010.05.012
- McElligott S, Collins PJ, Sleator RD, Martella V, Decaro N, Buonavoglia C, O'Shea H. Detection and genetic characterization of canine parvoviruses and coronaviruses in southern Ireland. Arch Virol 2011; 156:495-503; PMID:21107617; http://dx.doi.org/10.1007/s00705-010-0861-3
- Sleator RD. Phylogenetics. Arch Microbiol 2011; 193:235-9; PMID:21249334; http://dx.doi.org/10.1007/s00203-011-0677-x
- Sleator RD. A beginner's guide to phylogenetics. Microb Ecol 2013; 66:1-4; PMID:23624570; http://dx.doi.org/10.1007/s00248-013-0236-x
- Henry M, O'Sullivan O, Sleator RD, Coffey A, Ross RP, McAuliffe O, O'Mahony JM. In silico analysis of Ardmore, a novel mycobacteriophage isolated from soil. Gene 2010; 453:9-23; PMID:20064590; http://dx.doi.org/10.1016/j.gene.2009.12.007
- Sleator RD. An overview of the current status of eukaryote gene prediction strategies. Gene 2010; 461:1-4; PMID:20430068; http://dx.doi.org/10.1016/j.gene.2010.04.008
- O'Mahony J, Fenton M, Henry M, Sleator RD, Coffey A. Lysins to kill - a tale of viral weapons of mass destruction. Bioeng Bugs 2011; 2:306-8; PMID:22008941; http://dx.doi.org/10.4161/bbug.2.6.16804
- Lynch KM, Lucid A, Arendt EK, Sleator RD, Lucey B, Coffey A. Genomics of Weissella cibaria with an examination of its metabolic traits. Microbiology 2015; 161:914-30; PMID:25678547; http://dx.doi.org/10.1099/mic.0.000053
- Sleator RD. An overview of the processes shaping protein evolution. Sci Prog 2010; 93:1-6; PMID:20222353; http://dx.doi.org/10.3184/003685009X12605492662844
- Sleator RD, Walsh P. An overview of in silico protein function prediction. Arch Microbiol 2010; 192:151-5; PMID:20127480; http://dx.doi.org/10.1007/s00203-010-0549-9
- Henry M, Coffey A, J OM, Sleator RD. Comparative modelling of LysB from the mycobacterial bacteriophage Ardmore. Bioeng Bugs 2011; 2:88-95; PMID:21636995; http://dx.doi.org/10.4161/bbug.2.2.14138
- Fenton M, Cooney JC, Ross RP, Sleator RD, McAuliffe O, O'Mahony J, Coffey A. In silico modeling of the staphylococcal bacteriophage-derived peptidase CHAP(K). Bacteriophage 2011; 1:198-206; PMID:23050213; http://dx.doi.org/10.4161/bact.1.4.18245
- Sleator RD. Proteins: form and function. Bioeng Bugs 2012; 3:80-5; PMID:22095055; http://dx.doi.org/10.4161/bbug.18303
- Sleator RD. Prediction of protein functions. Methods Mol Biol 2012; 815:15-24; PMID:22130980; http://dx.doi.org/10.1007/978-1-61779-424-7_2
- Sleator RD. The story of Mycoplasma mycoides JCVI-syn1.0: the forty million dollar microbe. Bioeng Bugs 2010; 1:229-30; PMID:21327053
- Sleator RD. Ferretting out the facts behind the H5N1 controversy. Bioeng Bugs 2012; 3:139-43; PMID:22402712; http://dx.doi.org/10.4161/bbug.20136
- Sleator RD. Digital biology: a new era has begun. Bioengineered 2012; 3:311-2; PMID:23099453; http://dx.doi.org/10.4161/bioe.22367
- Sleator RD. Synthetic ribosomes: making molecules that make molecules. Bioengineered 2013; 4:63-4; PMID:23324614; http://dx.doi.org/10.4161/bioe.23640
- Sleator RD, O'Driscoll A. Digitizing humanity. Artificial DNA, PNA & XNA 2013; 4:37-8; PMID:23912716; http://dx.doi.org/10.4161/adna.25489
- Sleator RD. The synthetic biology future. Bioengineered 2014; 5:69-72; PMID:24561910; http://dx.doi.org/10.4161/bioe.28317
- Sleator RD. Genetics just got SEXY: Sequences encoding XY. Bioengineered 2014; 5:214-5; PMID:24847983; http://dx.doi.org/10.4161/bioe.29306
- Sleator RD. The genetic code. Rewritten, revised, repurposed. Artificial DNA, PNA & XNA 2014; 5:e29408; PMID:25483933; http://dx.doi.org/10.4161/adna.29408
- O'Driscoll A, Daugelaite J, Sleator RD. 'Big data', Hadoop and cloud computing in genomics. J Biomed Inform 2013; 46:774-81; PMID:23872175; http://dx.doi.org/10.1016/j.jbi.2013.07.001
- Manning T, Sleator RD, Walsh P. Biologically inspired intelligent decision making: a commentary on the use of artificial neural networks in bioinformatics. Bioengineered 2014; 5:80-95; PMID:24335433; http://dx.doi.org/10.4161/bioe.26997
- Manning T, Sleator RD, Walsh P. Naturally selecting solutions: the use of genetic algorithms in bioinformatics. Bioengineered 2013; 4:266-78; PMID:23222169; http://dx.doi.org/10.4161/bioe.23041
- Walsh P, Carroll J, Sleator RD. Accelerating in silico research with workflows: a lesson in Simplicity. Comput Biol Med 2013; 43:2028-35; PMID:24290918; http://dx.doi.org/10.1016/j.compbiomed.2013.09.011
- O'Driscoll A, Belogrudov V, Carroll J, Kropp K, Walsh P, Ghazal P, Sleator RD. HBLAST: Parallelised sequence similarity - A Hadoop MapReducable basic local alignment search tool. J Biomed Inform 2015; 54:58-64; PMID:25625550; http://dx.doi.org/10.1016/j.jbi.2015.01.008
- Walsh P, Bekaert M, Carroll J, Manning T, Kelly B, O'Driscoll A, Lu X, Smith C, Dickinson P, Templeton K, et al. Draft genome sequences of six different staphylococcus epidermidis clones, isolated individually from preterm neonates presenting with sepsis at Edinburgh's royal infirmary. Gen Announc 2015; 3; PMID:25977439; http://dx.doi.org/10.1128/genomeA.00471-15
- Kropp KA, Lucid A, Carroll J, Belgrudov V, Walsh P, Kelly B, Templeton K, Smith C, Dickinson P, O'Driscoll A, et al. Draft Genome Sequence of a Pantoea sp. Isolated from a Preterm Neonatal Blood Sepsis Patient. Gen Announc 2014; 2; PMID:25212624
- Kropp KA, Lucid A, Carroll J, Belgrudov V, Walsh P, Kelly B, Templeton K, Smith C, Dickinson P, O'Driscoll A, et al. Draft genome sequence of a staphylococcus aureus isolate taken from the blood of a preterm neonatal blood sepsis patient. Gen Announc 2014; 2; PMID:25212625
- Kropp KA, Lucid A, Carroll J, Belgrudov V, Walsh P, Kelly B, Smith C, Dickinson P, O'Driscoll A, Templeton K, et al. Draft genome sequence of a streptococcus agalactiae strain isolated from a preterm neonate blood sepsis patient at the royal infirmary, Edinburgh, Scotland. Gen Announc 2014; 2; PMID:25189584
- Kropp KA, Lucid A, Carroll J, Belgrudov V, Walsh P, Kelly B, Smith C, Dickinson P, O'Driscoll A, Templeton K, et al. Draft genome sequence of a staphylococcus warneri strain isolated from a preterm neonate blood sepsis patient at the royal infirmary, Edinburgh, Scotland. Gen Announc 2014; 2; PMID:25189586
- Kropp KA, Lucid A, Carroll J, Belgrudov V, Walsh P, Kelly B, Smith C, Dickinson P, O'Driscoll A, Templeton K, et al. Draft genome sequence of an enterococcus faecalis strain isolated from a neonatal blood sepsis patient. Gen Announc 2014; 2; PMID:25212626
- Kropp KA, Lucid A, Carroll J, Belgrudov V, Walsh P, Kelly B, Smith C, Dickinson P, O'Driscoll A, Templeton K, et al. Draft genome sequence of a serratia marcescens strain isolated from a preterm neonatal blood sepsis patient at the royal infirmary, Edinburgh, Scotland, United Kingdom. Gen Announc 2014; 2; PMID:25212627