ABSTRACT
1,3-propanediol (1,3-PD) is an important compound from which many others can be synthesized. 2,3-butanediol (BDO) is the key by-product in the biosynthesis of 1,3-PD from glycerol, but it impedes its downstream purification. In Klebsiella, the budA, budB and budC genes encode enzymes that are responsible for the synthesis of BDO. In this study, 3 individual antisense RNAs were designed to repress the expression and hence activity of BudA-C. Compared with the parent strains, the activities of BudB and BudC were reduced by 60.5% and 70.5%, respectively, and the mRNA level of budA was reduced by 70%. Decreased BudC activity had no effect on cell growth or carbon distribution. However, reduced BudA and BudB activity decreased the BDO concentration by 35% and led to a 10% increase in the yield of 1,3-PD. This result suggests the activities of BudA and BudB could be key factors in the production of BDO from glycerol in Klebsiella. This study provides a deeper understanding of the role of budABC in glycerol metabolism in Klebsiella.
Introduction
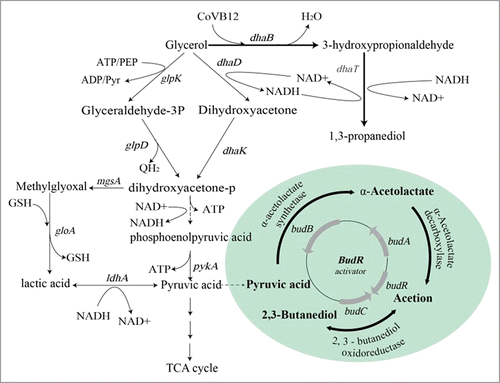
The rapid expansion of biodiesel production has led to the abundance of glycerol waste, and the conversion of glycerol to high value-added chemicals is receiving increasing attention. Among the glycerol derivatives, 1,3-propanediol (1,3-PD) is a particularly high value product,Citation1 with considerable commercial value in the synthesis of polytrimethylene terephthalate.Citation2 Klebsiella pneumoniae is one of the most efficient organisms identified to date for the biosynthesis of 1,3-PD. In Klebsiella, glycerol is assimilated via both reductive and oxidative pathways. In the reductive pathway, glycerol is converted to 1,3-PD by the coenzyme B12-dependent glycerol dehydratase (DhaB) and the 1,3-propanediol oxidoreductase (DhaT) in successive steps. In the oxidative pathway, glycerol is oxidized to dihydroxyacetone then pyruvate via the Embden-Meyerhof-Parnas (EMP) pathway before conversion to byproducts such as acetic acid, lactic acid, ethanol and 2,3-butanediol (BDO). This pathway competes for NADH and a carbon source with the 1,3-PD pathway.
Of all byproducts, BDO is the most abundant and shares a similar boiling point with 1,3-PD, which impedes the downstream purification of 1,3-PD.Citation3 In Klebsiella, the bud operon includes the transcriptional activator-like protein (budR)Citation4 and 3 enzymes; α-acetolactate synthase (bubB), α-acetolactate decarboxylase (bubA), and 2,3-butanediol dehydrogenase (bubC). Together, these proteins are responsible for the synthesis of BDOCitation5,6 (). Based on this knowledge, genetic manipulation has been performed to reduce the accumulation of BDO.Citation7,8 However, detailed knowledge of the role of these genes in BDO synthesis remains elusive.
Previous studies indicated that budR positively regulates budABC and BDO biosynthesis.Citation4,8,9 Generally, gene knockout is the method of choice for studying gene function, but manipulation of essential genes is not possible. The bud operon could also be responsible for the synthesis of branched chain amino acids such as valine and leucine, which are important to cell growth.Citation10 Unlike gene knockout, RNA-based approaches can be applied to repress the expression of essential genes in a more controlled manner.Citation11 In this study, expression of budA-C in K. pneumoniae CICIM B0057 was repressed using antisense RNAs to investigate the effect on 1,3-PD and BDO biosynthesis.
Material and methods
Strains, plasmid and reagents
K. pneumoniae CICIM B0057 was stored in our lab and used as the parent strain. Escherichia coli JM109 (Invitrogen) and pEtacCitation12 were utilized for plasmid construction. 1,3-propandiol and 2,3-butanediol were purchased from Sigma-Aldrich (Steinheim, Germany). DNA polymerase, restriction endonuclease, ligase (solution I), and Genomic DNA and Gel DNA Purification Kits were purchased from Takara (Dalian, China). Tryptone and yeast extract were bought from Oxoid (Basingstoke, UK). Isopropyl β-D-1-thiogalactopyranoside (IPTG) and kanamycin were bought from Sangon (Shanghai, China). All other compounds were of reagent grade or higher quality.
Plasmid construction
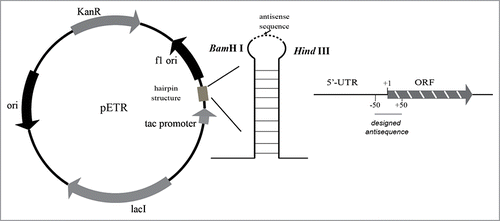
Primers P1 and P2 () were used for the construction of pETR by PCR using pEtac as a template. A hairpin structure was incorporated to improve RNA stability. Antisense DNA fragments of budA, budB, and budC were cloned using PCR and genomic DNA from K. pneumoniae CICIM B0057 based on genomic information from K. pneumoniae 342 (GenBank Accession Number: CP000964.1). The resultant DNA fragments were inserted into the BamHI and HindIII sites of pETR to generate pETR/anti-budA, pETR/anti-budB, and pETR/anti-budC. PCR amplification conditions were as follows: initial denaturation at 94°C for 10 min followed by 30 cycles of 98°C for 15 s, 60°C for 15 s, and 72°C for 2 min. Plasmids were transformed into K. pneumoniae CICIM B0057 to generate K. pneumoniae CICIM B0057 (pETR/anti-budA), K. pneumoniae CICIM B0057 (pETR/anti-budB), and K. pneumoniae CICIM B0057 (pETR/anti-budC).
Table 1. Primers used in this study.
Media and cultivation
Luria-Bertani medium (1% tryptone, 0.5% yeast extract, 1% sodium chloride) was used for the cultivation of K. pneumoniae CICIM B0057 and E. coli. The fermentation medium for K. pneumoniae CICIM B0057 contained the following (g/l): glycerol, 40; glucose, 5; K2HPO4, 7.5; (NH4)2SO4, 2; MgSO4·7H2O, 2; FeSO4·7H2O, 0.005; yeast extract, 7; vitamin B12, 0.015, and 1 ml trace element solution. The composition of the trace element solution was as follows (g/l): ZnCl2, 0.7; MnCl2·4H2O, 1; H3BO3, 0.6; CoCl2·6H2O, 2; CuCl2, 0.2; NiCl2·6H2O, 0.25; Na2MoO4·2H2O, 0.35. Kanamycin was added at a final concentration of 100 μg/ml where needed. Fermentation of K. pneumoniae CICIM B0057 was performed in 250 ml gauze and paper-covered conical flasks containing 50 ml medium at 37oC in a rotary shaker incubator at 100 rpm.
Enzyme activity assay
Recombinant K. pneumoniae CICIM B0057 cells were collected by centrifugation at 10,000×g for 10 min, washed twice, and suspended in 100 mM potassium phosphate buffer (pH 7.0). Cells were disrupted by sonication and centrifuged at 10,000×g for 10 min. The supernatant was collected as the crude protein extract. All procedures were performed at 4°C. Protein concentration was measured by the Bradford methodCitation13 using bovine serum albumin as the standard.
BudA activity was analyzed in 50 mM MES [2-(N-morpholino)ethanesulfonic acid buffer (pH 6.0)], 0.5 M NaCl, 0.2% Triton X-100, and 5.3 mM α-acetolactate at 30°C for 20 min as previously reported, and the substrate α-acetolactate was freshly prepared by saponifying ethyl-2-acetoxy-2-methylacetate at pH 11.5.Citation14 The acetoin content was analyzed as previously reported.Citation15 One unit of activity was defined as the formation of 1 μmol of acetoin in 1 min.
BudB activity was analyzed in 70 mM sodium acetate buffer (pH 5.3), 0.17 mM thiamine pyrophosphate and 80 mM pyruvate as previously reported.Citation16 The reaction was started by addition of cell-free extract at 37°C. The resultant α-acetolactate was converted to acetoin at 45°C for 30 min in the presence of 65 mM HCl, and acetoin was measured as previously reported.Citation15 One unit of activity was defined as the formation of 1 μmol of acetoin in 1 min.
BudC activity was analyzed in 120 mM BDO and 4 mM NAD+ in 33 mM sodium pyrophosphate (pH 8) as previously described.Citation17 The reaction was started by addition of cell-free extract at 37°C. One unit of activity was defined as the formation of 1 μmol of NADH in 1 min. The change in the concentration of NADH (ɛ340 = 6220 M-1·cm-1) was measured at 340 nm using a spectrophotometer (UV-2450; Shimadzu Co., Kyoto, Japan).
Analytical methods
Glycerol, 1,3-PD, 3-HP and other metabolites were analyzed by High Performance Liquid Chromatography fitted with a refractive index detector and a Bio-Rad Aminex organic acids HPX-87H column at 60°C with 5 mM H2SO4 as the mobile phase and an elution rate of 0.6 ml/min.Citation18
Results and discussion
Construction of an antisense RNA expression system
To improve the stability of antisense RNA (asRNA) against degradation by RNAase, a hairpin structureCitation19 was inserted into the multiple cloning site of the plasmid pEtac, resulting in plasmid pETR (). A previous study suggested that an antisense DNA sequence complementary to a nucleotide sequence close to the ribosome-binding site (RBS) is better able to inhibit the expression of target genes.Citation20 Thus, we designed asRNAs based on the DNA sequence between -50 and 50 nucleotides from the RBS for all 3 genes (). The obtained antisense fragments were cloned into pETR, resulting in recombinant plasmids pETR/anti-budA, pETR/anti-budB, and pETR/anti-budC. These plasmids were transformed into K. pneumoniae CICIM B0057 to generate K. pneumoniae CICIM B0057 (pETR/anti-budA), K. pneumoniae CICIM B0057 (pETR/anti-budB), and K. pneumoniae CICIM B0057 (pETR/anti-budC).
Effect of antisense RNAs on enzyme activity and cell growth
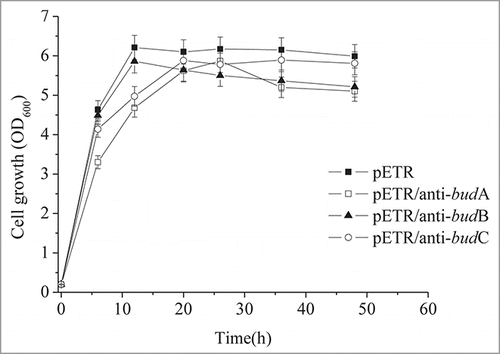
In K. pneumoniae, pyruvate is decarboxylated by BduB to form α-acetolactate, which is converted to acetoin and 2,3-butanediol by BudA and BudC, respectively. To repress the expression of these enzymes, antisense RNAs were transformed into the host to generate K. pneumoniae CICIM B0057 (pETR/anti-budA), K. pneumoniae CICIM B0057 (pETR/anti-budB), and K. pneumoniae CICIM B0057 (pETR/anti-budC). The BudB activity of the anti-budB strain was reduced by 60.5% (from 11.90 to 4.69 U/mg), the BudC activity of the anti-budC strain was decreased by 70.5%, and the BudA activity of the anti-budA strain was reduced by 68% (0.015 to 0.0048 U/mg). The antisense RNAs therefore successfully repressed the expression of their corresponding enzymes, leading to dramatically decreased enzyme activity.
Antisense RNAs for budA and budC also repressed cell growth at the log phase (). However, the final biomass of the anti-budC strain was comparable to the parent K. pneumoniae CICIM B0057 (pETR) strain. The final biomass of K. pneumoniae CICIM B0057 (pETR/anti-budA) and K. pneumoniae CICIM B0057 (pETR/anti-budB) was reduced by 14.8% and 13.2%, respectively, suggesting that anti-budA and anti-budB led to a decreased biomass.
Effect of antisense RNAs on 1,3-PD and BDO production
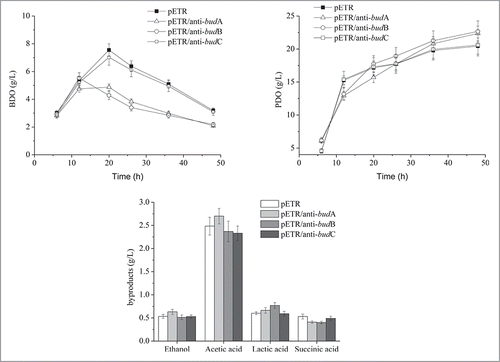
As shown in , the BDO concentration in all strains was similar before 12 h. However, after this time, BDO accumulation in the anti-budA and anti-budB strains was reduced from ˜5 g/l to 2 g/l. In contrast, BDO production in the parent and anti-budC strains peaked at ˜7 g/l after 20 h, then decreased to ˜3 g/l at the end of the fermentation. These results indicated that BDO synthesis was impeded by the introduction of antisense RNAs for budA and budB. The 1,3-PD yield of the anti-budA and anti-budB strains increased by less than 10% compared to the parent strain, indicating a positive effect of the 2 antisense RNAs on 1,3-PD biosynthesis. No significant differences in by-products were detected between the strains tested; ethanol, lactic acid and succinic acid all accumulated to ˜0.6 g/l, whereas acetic acid reached 2.5 g/l.
Although the designed antisense RNAs exhibited similar target enzyme inhibition efficiencies, cell growth and BDO and 1,3-PD production differed significantly. Unlike BudC, a reduction in BudB and BudA activity decreased BDO production, suggesting BudA and BudB might be rate-limiting factors in BDO biosynthesis. Actually, BDO synthesis in the budC deficient K. pneumoniae CICIM B0057 was still observed in our previous report.Citation7 It has been reported that glycerol dehydrogenase might also contribute to the bioconversion of acetoin to BDO,Citation21 suggesting isoenzymes of BudC may be present in Klebsiella. This may explain why a reduction in BudC activity did not appreciably decrease the BDO concentration. Previous studies suggested that BDO protects against acidification.Citation10,22 The reduced BDO synthesis capacity of the anti-budB and anti-budA strains may also decrease their tolerance to acetic acid, which could explain the decreased biomass. Interestingly, BDO production decreased during the late stages of fermentation in all tested strains. This phenomenon can be explained by the reversibility of the BDO synthesis pathway.Citation23 The introduced budB and budA antisense RNAs lowered the production of BDO, which decreased NADH consumption by the BDO pathway. This resulted in increased NADH availability for the synthesis of 1,3-PD, explaining the increased abundance of this compound in the anti-budB and anti-budA strains.
In summary, the activities of BudA and BudB proved to be the limiting factors affecting BDO biosynthesis in K. pneumoniae CICIM B0057. This knowledge on the role of budABC in glycerol metabolism could prove useful for reducing BDO and other by-products, and improving the downstream purification of 1,3-PD from glycerol in future commercial bioproduction.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National High Technology Research and Development Program of China (863 Program, NO.2012AA021201), Natural Science Foundation of China (NO.31270080, 31570052), Natural Science Foundation of Jiangsu Province (No. BK20140138), the Fundamental Research Funds for the Central Universities, and the 111 Project (No. 111-2-06).
References
- Bozell JJ, Petersen GR. Technology development for the production of biobased products from biorefinery carbohydrates—the US Department of Energy's “top 10” revisited. Green Chem 2010; 12:539-54; http://dx.doi.org/10.1039/b922014c
- Kurian JV. A new polymer platform for the future - Sorona (R) from corn derived 1,3-propanediol. J Polym Environ 2005; 13:159-67; http://dx.doi.org/10.1007/s10924-005-2947-7
- Xiu ZL, Zeng AP. Present state and perspective of downstream processing of biologically produced 1,3-propanediol and 2,3-butanediol. Appl Microbiol Biot 2008; 78:917-26; http://dx.doi.org/10.1007/s00253-008-1387-4
- Nicholson WL. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl Environ Microbiol 2008; 74:6832-8; PMID:18820069; http://dx.doi.org/10.1128/AEM.00881-08
- Blomqvist K, Nikkola M, Lehtovaara P, Suihko ML, Airaksinen U, Straby KB, Knowles JK, Penttila ME. Characterization of the genes of the 2,3-butanediol operons from Klebsiella terrigena and Enterobacter aerogenes. J Bacteriol 1993; 175:1392-404; PMID:8444801
- Wood BE, Yomano LP, York SW, Ingram LO. Development of industrial-medium-required elimination of the 2,3-butanediol fermentation pathway to maintain ethanol yield in an ethanologenic strain of Klebsiella oxytoca. Biotechnol Prog 2005; 21:1366-72; PMID:16209539; http://dx.doi.org/10.1021/bp050100e
- Guo X, Fang H, Zhuge B, Zong H, Song J, Zhuge J, Du X. budC knockout in Klebsiella pneumoniae for bioconversion from glycerol to 1,3-propanediol. Biotechnol Appl Biochem 2013; 60:557-63; PMID:23586646; http://dx.doi.org/10.1002/bab.1114
- Lee S, Kim B, Jeong D, Oh M, Um Y, Kim YR, Kim J, Lee J. Observation of 2,3-butanediol biosynthesis in Lys regulator mutated Klebsiella pneumoniae at gene transcription level. J Biotechnol 2013; 168:520-6; PMID:24076264; http://dx.doi.org/10.1016/j.jbiotec.2013.09.015
- Effantin G, Rivasseau C, Gromova M, Bligny R, Hugouvieux-Cotte-Pattat N. Massive production of butanediol during plant infection by phytopathogenic bacteria of the genera Dickeya and Pectobacterium. Mol Microbiol 2011; 82:988-97; PMID:22032684; http://dx.doi.org/10.1111/j.1365-2958.2011.07881.x
- Mayer D, Schlensog V, Bock A. Identification of the transcriptional activator controlling the butanediol fermentation pathway in Klebsiella terrigena. J Bacteriol 1995; 177:5261-9; PMID:7665514
- Na D, Yoo SM, Chung H, Park H, Park JH, Lee SY. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol 2013; 31:170-4; PMID:23334451; http://dx.doi.org/10.1038/nbt.2461
- Zhuge B, Zhang C, Fang H, Zhuge J, Permaul K. Expression of 1,3-propanediol oxidoreductase and its isoenzyme in Klebsiella pneumoniae for bioconversion of glycerol into 1,3-propanediol. Appl Microbiol Biotechnol 2010; 87:2177-84; PMID:20499228; http://dx.doi.org/10.1007/s00253-010-2678-0
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248-54; PMID:942051; http://dx.doi.org/10.1016/0003-2697(76)90527-3
- Diderichsen B, Wedsted U, Hedegaard L, Jensen B, Sjøholm C. Cloning of aldB, which encodes α-acetolactate decarboxylase, an exoenzyme from Bacillus brevis. J Bacteriol 1990; 172:4315-21; PMID:2198252
- Westerfield WW. A colorimetric determination of blood acetoin. J Biol Chem 1945; 161:495-502; PMID:21006932
- Cavin JF, Dartois V, Labarre C, Divies C. Cloning of branched chain amino acid biosynthesis genes and assays of α-acetolactate synthase activities in Leuconostoc mesenteroides subsp. cremoris. Res Microbiol 1999; 150:189-98; PMID:10229948; http://dx.doi.org/10.1016/S0923-2508(99)80035-7
- Gonzalez E, Fernandez MR, Larroy C, Sola L, Pericas MA, Pares X, Biosca JA. Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene product. Disruption and induction of the gene. J Biol Chem 2000; 275:35876-85; PMID:10938079; http://dx.doi.org/10.1074/jbc.M003035200
- Ji X-J, Huang H, Zhu J-G, Ren L-J, Nie Z-K, Du J, Li S. Engineering Klebsiella oxytoca for efficient 2, 3-butanediol production through insertional inactivation of acetaldehyde dehydrogenase gene. Appl Microbiol Biot 2010; 85:1751-8; http://dx.doi.org/10.1007/s00253-009-2222-2
- Nakashima N, Tamura T. Conditional gene silencing of multiple genes with antisense RNAs and generation of a mutator strain of Escherichia coli. Nucleic Acids Res 2009; 37:e103; PMID:19515932; http://dx.doi.org/10.1093/nar/gkp498
- Wu JJ, Yu O, Du GC, Zhou JW, Chen J. Fine-Tuning of the Fatty Acid Pathway by Synthetic Antisense RNA for Enhanced (2S)-Naringenin Production from L-Tyrosine in Escherichia coli. Appl Environ Microb 2014; 80:7283-92; http://dx.doi.org/10.1128/AEM.02411-14
- Wang Y, Tao F, Xu P. Glycerol dehydrogenase plays a dual role in glycerol metabolism and 2,3-butanediol formation in Klebsiella pneumoniae. J Biol Chem 2014; 289:6080-90; PMID:24429283; http://dx.doi.org/10.1074/jbc.M113.525535
- Moons P, Van Houdt R, Vivijs B, Michiels CW, Aertsen A. Integrated regulation of acetoin fermentation by quorum sensing and pH in Serratia plymuthica RVH1. Appl Environ Microbiol 2011; 77:3422-7; PMID:21441339; http://dx.doi.org/10.1128/AEM.02763-10
- Fu J, Wang Z, Chen T, Liu W, Shi T, Wang G, Tang YJ, Zhao X. NADH plays the vital role for chiral pure D-(-)-2,3-butanediol production in Bacillus subtilis under limited oxygen conditions. Biotechnol Bioeng 2014; 111:2126-31; PMID:24788512; http://dx.doi.org/10.1002/bit.25265