ABSTRACT
We propose a mechanism of action for the betL* mutation which is based on DNA topology. Removing a single thymine residue from the betL σA promoter's −10 and −35 spacer results in a ‘twist’-mediated activation of transcription which accounts for the osmotolerance phenotype observed for strains expressing betL*.
KEYWORDS:
Work in our labs,Citation1-5 and others,Citation6 has shown osmoregulatory control in the intracellular foodborne bacterial pathogen Listeria monocytogenes to be elaborately orchestrated. This is particularly true of BetL, a key component of the listerial osmotolerance responseCitation7-10 and a versatile stress tolerance locus.Citation11-13 One of the primary respondents in the secondary response to osmotic up-shock, betL (and its encoded membrane protein, BetLCitation7) is regulated at the transcriptional,Citation8 translationalCitation14 and post-translational levels.Citation9 Indeed, we have shown that betL is controlled by at least 2 putative promoter elementsCitation10: σA and σB. While the latter is a global stress regulator and a key component of the pathogen's gastrointestinal phase of infectionCitation15-17; the former is more usually associated with general housekeeping activities. However, we recently described a single point mutation (deletion of a thymine residue) in the putative betL σA promoter which dramatically improves the pathogen's osmotolerance profile; suggesting a previously unreported role for this promoter in the listerial osmoregulatory response.Citation10 Herein, we describe the use of pPL2lux,Citation18 a luciferase-based reporter system, to monitor the transcriptional profile of betL* in real-time, thereby enabling us to pinpoint when and to what extent the mutation influences transcription, both in the presence and absence of salt stress.
A derivative of the listerial integration vector pPL2,Citation19 which exhibits site-specific, single-copy integration into the L. monocytogenes chromosome, pPL2lux harbours a synthetic luxABCDE operon encoding both the substrate and enzyme required to produce measurable quantities of light.Citation20 Furthermore, given that the luxABCDE operon was derived from pSB2025 with the introduction of a blunt-end SwaI restriction site overlapping the ATG start codon of luxA; cloning between SalI-SwaI facilitates exact translational fusions to the lux reporter, mimicking transcription and translation initiation as it occurs at the native chromosomal location of the promoter under investigation.Citation18 Using this system, the level of light emitted in real-time is directly proportional to the level of transcription.
In the current study, the betL and betL* promoter elements (PbetL and PbetL*), were PCR amplified from previously constructed plasmids, pRS3 and pRS2 respectively,Citation10 using KOD polymerase (Merck) with primers betLPF (5′-CAATGTCGACCCCACGCTCACCGGCTCCAG-3′ SalI restriction site underlined) and betLPR (5′-CAATACATCACTTCCCTTTATTTTC-3′). The resulting ∼0.3 kb PCR products contained the regulatory regions immediately upstream of betL and betL*, respectively. The PbetL and PbetL* amplicons were digested with SalI and cloned into SwaI-SalI-digested pPL2lux, yielding pPL2lux-PbetL and pPL2lux-PbetL*, respectively. Subsequently, pPL2lux (negative control), pPL2lux-PbetL, and pPL2lux-P betL* were transformed into L. monocytogenes LO28, and candidate integrants were checked for site-specific integration by PCR, using primers PL95 (5′-ACATAATCAGTCCAAAGTAGATGC-3′) and PL102 (5′-TATCAGACCTAACCCAAACCTTCC-3′).Citation19 From each transformation, several colonies with the correct genotype were selected for phenotypic confirmation by bioluminescent imaging, using the Xenogen IVIS 100 system (Xenogen, Alameda, CA).
Overnight cultures of the confirmed strains (LO28::pPL2lux, LO28::pPL2lux-PbetL, and LO28::pPL2lux-PbetL*) were diluted 1:50 in Tryptone Soya Broth (TSB). Growth and bioluminescence were monitored simultaneously from lag to stationary phase, in the presence and absence of added salt ().
Figure 1. Expression profiles for betL and betL* during growth of LO28::pPL2lux-PbetL (•) and LO28::pPL2lux-PbetL* (○) at 37°C in TSB (A) in the absence of added NaCl and (B) in the presence of 4% added NaCl. Bars indicate the average luciferase expression profiles for LO28::pPL2lux-PbetL (gray) and LO28::pPL2lux-PbetL* (white). As expected, no light was observed for the negative control strain, LO28::pPL2lux, which for clarity was not shown on the graph. BLC: bioluminescence counts. OD595: optical density at 595 nm. The data presented are representative of 3 independent experiments.
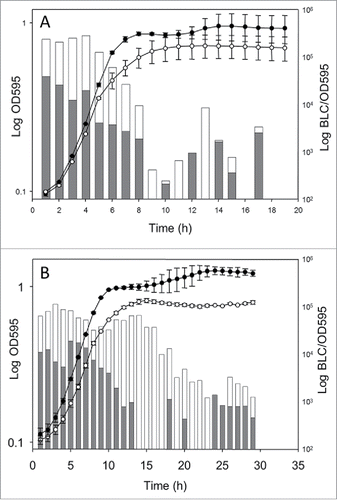
While previously no significant differences in transcript levels were observed between betL* and the wild-type gene in the absence of salt stress (using an RT-PCR based approachCitation10) this was not the case in the current study. Indeed, expression levels from LO28::pPL2lux-PbetL* appeared markedly higher than that of LO28::pPL2lux-PbetL, particularly during the lag and log phases of growth. Furthermore, while LO28::pPL2lux-PbetL expression levels began to decrease in mid log phase, LO28::pPL2lux-PbetL* expression remained steady until late log phase (). Interestingly, for both cultures, expression rebounded in stationary phase, suggesting the involvement of a promoter other than the putative σA; possibly σB, or an as yet unidentified stationary phase specific promoter.
In the presence of 4% added NaCl (the optimal salt concentration for BetL activityCitation9), similar, albeit more dramatic trends were observed. Expression levels for LO28::pPL2lux-PbetL* were significantly higher than those of LO28::pPL2lux-PbetL from the outset and remained so until early stationary phase. Indeed, while LO28::pPL2lux-PbetL expression levels were no longer detectable after 13 hours, LO28::pPL2lux-PbetL* expression remained steady up to hour 16, only gradually decreasing to hour 24. Resurgence in expression in stationary phase was again observed for both cultures in the presence of 4% NaCl, further suggesting the involvement of an alternative stationary phase promoter.
Finally, based on the above observations, we propose a mechanism of action which is based on DNA topology. If, as we suspect, the betL σA-like promoter belongs to a class of DNA twist-sensitive promotersCitation21; removing a single thymine residue from the promoter's −10 and −35 spacer region would, as we have observed, boost transcription of betL even in the absence of salt stress. Furthermore, given previously well documented links between osmolarity and DNA supercoilingCitation22-25; the addition of salt is likely to lead to further activation of betL expression. This exogenous ‘twist’-mediated activation, in the presence of NaCl, boosts already elevated transcript levels; resulting in the dramatic osmotolerance phenotype observed for strains expressing betL*.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
SMK is a CIT RISAM Scholar. EPC is funded by an Irish Research Council Government of Ireland Postdoctoral Fellowship (GOIPD/2015/53). RFH was funded by a Genetics Society student workplace grant. RDS is Coordinator of the EU FP7 project ClouDx-i.
References
- Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 2002; 26:49-71; PMID:12007642; http://dx.doi.org/10.1111/j.1574-6976.2002.tb00598.x
- Sleator RD, Gahan CG, Hill C. A postgenomic appraisal of osmotolerance in Listeria monocytogenes. Appl Environ Microbiol 2003; 69:1-9; PMID:12513970; http://dx.doi.org/10.1128/AEM.69.1.1-9.2003
- Sleator RD, Hill C. A novel role for the LisRK two-component regulatory system in listerial osmotolerance. Clin Microbiol Infect 2005; 11:599-601; PMID:16008610; http://dx.doi.org/10.1111/j.1469-0691.2005.01176.x
- Sleator RD, Hill C. Compatible solutes: A listerial passe-partout? Gut microbes 2010; 1:77-9; PMID:21326913; http://dx.doi.org/10.4161/gmic.1.2.10968
- Sleator RD, Hill C. Compatible solutes: the key to Listeria's success as a versatile gastrointestinal pathogen? Gut pathogens 2010; 2:20; PMID:21143981; http://dx.doi.org/10.1186/1757-4749-2-20
- O'Byrne CP, Karatzas KA. The role of sigma B (sigma B) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Advan Appl Microbiol 2008; 65:115-40; PMID:19026864; http://dx.doi.org/10.1016/S0065-2164(08)00605-9
- Sleator RD, Gahan CG, Abee T, Hill C. Identification and disruption of BetL, a secondary glycine betaine transport system linked to the salt tolerance of Listeria monocytogenes LO28. Appl Environ Microbiol 1999; 65:2078-83; PMID:10224004
- Sleator RD, Gahan CGM, O'Driscoll B, Hill C. Analysis of the role of betL in contributing to the growth and survival of Listeria monocytogenes LO28. Int J Food Microbiol 2000; 60:261-8; PMID:11016615; http://dx.doi.org/10.1016/S0168-1605(00)00316-0
- Sleator RD, Wood JM, Hill C. Transcriptional regulation and posttranslational activity of the betaine transporter BetL in Listeria monocytogenes are controlled by environmental salinity. J Bacteriol 2003; 185:7140-4; PMID:14645273; http://dx.doi.org/10.1128/JB.185.24.7140-7144.2003
- Hoffmann RF, McLernon S, Feeney A, Hill C, Sleator RD. A single point mutation in the listerial betL sigma(A)-dependent promoter leads to improved osmo- and chill-tolerance and a morphological shift at elevated osmolarity. Bioengineered 2013; 4:401-7; PMID:23478432; http://dx.doi.org/10.4161/bioe.24094
- Watson D, Sleator RD, Casey PG, Hill C, Gahan CG. Specific osmolyte transporters mediate bile tolerance in Listeria monocytogenes. Infect Immun 2009; 77:4895-904; PMID:19737907; http://dx.doi.org/10.1128/IAI.00153-09
- Sheehan VM, Sleator RD, Fitzgerald GF, Hill C. Heterologous expression of BetL, a betaine uptake system, enhances the stress tolerance of Lactobacillus salivarius UCC118. Appl Environ Microbiol 2006; 72:2170-7; PMID:16517668; http://dx.doi.org/10.1128/AEM.72.3.2170-2177.2006
- Sheehan VM, Sleator RD, Hill C, Fitzgerald GF. Improving gastric transit, gastrointestinal persistence and therapeutic efficacy of the probiotic strain Bifidobacterium breve UCC2003. Microbiology 2007; 153:3563-71; PMID:17906153; http://dx.doi.org/10.1099/mic.0.2007/006510-0
- Sleator RD. A role for translational control in listerial osmoregulation and strain variation? Foodborne pathogens and disease 2007; 4:395-6; PMID:18041949; http://dx.doi.org/10.1089/fpd.2007.0043
- Sleator RD, Watson D, Hill C, Gahan CG. The interaction between Listeria monocytogenes and the host gastrointestinal tract. Microbiology 2009; 155:2463-75; PMID:19542009; http://dx.doi.org/10.1099/mic.0.030205-0
- Sleator RD, Wemekamp-Kamphuis HH, Gahan CG, Abee T, Hill C. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol Microbiol 2005; 55:1183-95; PMID:15686563; http://dx.doi.org/10.1111/j.1365-2958.2004.04454.x
- Begley M, Sleator RD, Gahan CG, Hill C. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun 2005; 73:894-904; PMID:15664931; http://dx.doi.org/10.1128/IAI.73.2.894-904.2005
- Bron PA, Monk IR, Corr SC, Hill C, Gahan CG. Novel luciferase reporter system for in vitro and organ-specific monitoring of differential gene expression in Listeria monocytogenes. Appl Environ Microbiol 2006; 72:2876-84; PMID:16597994; http://dx.doi.org/10.1128/AEM.72.4.2876-2884.2006
- Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol 2002; 184:4177-86; PMID:12107135; http://dx.doi.org/10.1128/JB.184.15.4177-4186.2002
- Qazi SN, Counil E, Morrissey J, Rees CE, Cockayne A, Winzer K, et al. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect Immun 2001; 69:7074-82; PMID:11598083; http://dx.doi.org/10.1128/IAI.69.11.7074-7082.2001
- Wang JY, Syvanen M. DNA twist as a transcriptional sensor for environmental changes. Mol Microbiol 1992; 6:1861-6; PMID:1508037; http://dx.doi.org/10.1111/j.1365-2958.1992.tb01358.x
- Higgins CF, Dorman CJ, Stirling DA, Waddell L, Booth IR, May G, et al. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 1988; 52:569-84; PMID:2830029; http://dx.doi.org/10.1016/0092-8674(88)90470-9
- Auble DT, deHaseth PL. Promoter recognition by Escherichia coli RNA polymerase. Influence of DNA structure in the spacer separating the −10 and −35 regions. J Mol Biol 1988; 202:471-82; PMID:3050126; http://dx.doi.org/10.1016/0022-2836(88)90279-3
- Jordi BJ, Owen-Hughes TA, Hulton CS, Higgins CF. DNA twist, flexibility and transcription of the osmoregulated proU promoter of Salmonella typhimurium. EMBO J 1995; 14:5690-700; PMID:8521826
- Alice AF, Sanchez-Rivas C. DNA supercoiling and osmoresistance in Bacillus subtilis 168. Curr Microbiol 1997; 35:309-15; PMID:9462962; http://dx.doi.org/10.1007/s002849900260
