ABSTRACT
Cellulose is an abundant natural polysaccharide that is universally distributed. It can be extracted from corncobs, which are inexpensive, easily accessible, renewable, and environmentally friendly. A common strategy for effectively utilizing cellulose is efficient heterogeneous expression of cellulase genes in Saccharomyces cerevisiae. However, the improvement of cellulose utilization is a relevant issue. Based on our previous findings, we constructed an integrated secretion expression vector, pHBM368-pgk, containing a constitutive promoter sequence. Three genetically modified S. cerevisiae strains containing heterologous β-glucosidase, exoglucanase, and endoglucanase genes were constructed. The results of a 1-L bioreactor fermentation process revealed that the mixed recombinant S. cerevisiae could efficiently carry out simultaneous saccharification and fermentation (SSF) by using corncobs as the sole carbon source. The ethanol concentration reached 6.37 g/L after 96 hours of fermentation, which was about 3 times higher than that produced by genetically modified S. cerevisiae with the inducible promoter sequence. To investigate the microstructure characteristics of hydrolyzed corncobs during the fermentation process, corncob residues were detected by using a scanning electron microscope. This study provides a feasible method to improve the effect of SSF using corncobs as the sole carbon source.
Introduction
Cellulose is a major component of the plant cell wall, accounting for 50% of the structure, and as such carries out several important physiological functions.Citation1 Cellulose has significant potential to produce various economic products in industrial applications, such as ethanol and glycerol.Citation2 The main constituent of corncobs is cellulose and approximately 40 million tons of corncobs produced in China every year.Citation3 Moreover, it is a potent feedstock for the production of ethanol and glycerol. Bioethanol is considered a new energy alternative to traditional fossil fuels,Citation4 and glycerol has assumed tremendous importance in the food and drug industry. As it is an enormous natural biomass, improvement of cellulose utilization is a relevant issue to address.
Based on the effectiveness of different sites in cellulose hydrolysis, cellulolytic enzymes can be divided into 3 categories: β-glucosidase (BG), exoglucanase (CBH), and endoglucanase (EG). Generally, the accepted mechanism for cellulose hydrolysis is synergy of the cellulolytic enzymes.Citation5 Several different synergies were observed in different kinds of cellulolytic enzymes as well as in the same cellulolytic enzyme, such as synergy between 2 exoglucanases,Citation6 synergy between 2 endoglucanases, and synergy between β-glucosidase, exoglucanase, and endoglucanase.Citation7 Regardless of the type of synergy, the crystalline structure of cellulose can be disrupted by dispersal and defibrillation. Hydrolysis intermediates such as cellobiose and cellotriose easily decompose into sugar monomers, which are efficient carbon sources for bacteria and fungi.
A promoter is an important cis-acting element that induces the regulation of gene expression. A high-efficiency system chooses the appropriate promoter to initiate the transcription of downstream genes.Citation8-10 An inducible promoter can be controlled by inducers, but this increases the complexity of the process in practice. Therefore, many researchers have chosen constitutive promoters for exogenous gene expression.Citation11,12 Heterologous enzymes can be expressed in the preliminary growth stage by using a constitutive promoter. For the fermentation process, the use of genetically modified strains with constitutive promoters simplifies the process.
Separating hydrolysis fermentation (SHF) and simultaneous saccharification and fermentation (SSF) were applied, by using Saccharomyces cerevisiae, to produce bioethanol from cellulosic feedstock. In SHF, cellulose needs to initially be saccharified by cellulase or another method before producing bioethanol by yeast fermentation.Citation13 SSF is considered an ideal process for bioethanol production in the industry.Citation14,15 Glucose is utilized by yeast as a carbon source in fermentation during cellulose saccharification. The continuous consumption of glucose, which accumulates during saccharification, activates cellulase activity.
In our previous study, 3 recombinant S. cerevisiae cells containing different cellulase genes were constructed by using the inducible promoter gal1.Citation16 We developed a recombinant S. cerevisiae capable of efficient direct ethanol production from cellulose by genomic integration of the cellulase gene. This strategy enhanced cellulase activity and the growth rate of the strains compared to wild type strains. However, the activity of these cellulases was not optimized for SSF, and the addition of an inducer was required during SSF of genetically modified strains with inducible promoters. In this study, genetically modified strains with the constitutive promoter pgk1 were constructed for SSF. At the same time, corncob residues were detected by using a scanning electron microscope (SEM) to determine the microstructure characteristics of hydrolyzed corncob during SSF using corncobs are the sole carbon source.
Results
Construction of genetically modified yeast
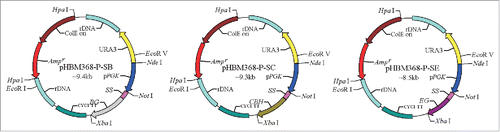
The α-factor signal sequence was amplified by using the SS-F-NotI and SS-R-SnaBI primers. The β-glucosidase gene was amplified by using the BG-F-SnaBI and BG-R-SpeI primers. Both products were digested with SnaBI and linked by using T4 DNA ligase. The exoglucanase and endoglucanase genes were synthetized and cloned into pPIC9K. The exoglucanase gene was amplified by using the SS-F-NotI and CBH-R-SpeI primers. The endoglucanase gene was amplified by using the SS-F-NotI and EG-R-SpeI primers. Three cellulase genes with an α-factor signal sequence were ligated into the pHBM368-pgk expression vector, which was digested with NotI and XbaI. The cellulase genes bg, cbh, and eg were cloned into the expression vector pHBM368-pgk with the ORF of the mature gene cloned in frame and downstream of the α-factor signal sequence for secreting recombinant protein into the medium. The α-factor signal sequence was identified by sequencing. The recombinant plasmids harboring bg, cbh, and eg were named pHBM368-P-SB, pHBM368-P-SC, and pHBM368-P-SE, respectively ().
Figure 1. Recombinant plasmid structures. The Pgk1 promoter, SS (α-factor signal sequence), bg, cbh, and eg cellulase genes were cloned in frame and downstream of the α-factor signal sequence. CYC1 TT, transcriptional terminator from CYC1gene; rDNA, rDNA fragment from S. cerevisiae for heterogeneous gene integration; Ampr, Ampicillin resistance gene; ColE: E. coli origin of replication; URA3, selection marker.
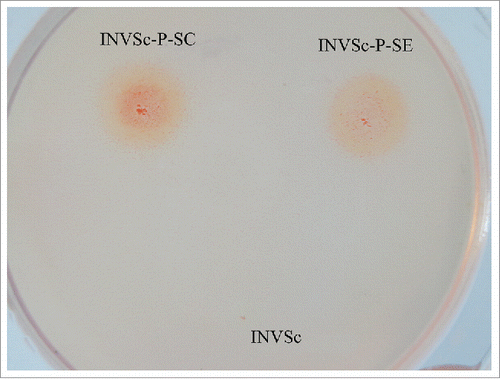
The recombinant plasmids were linearized with Hpa I and transformed into S. cerevisiae INVSc1 by electro transformation. The recombinants were screened by using YPD plates supplemented with carboxymethyl cellulose (CMC) as the substrate. The Congo red dye was used to screen for strains with cellulase activity. The expression patterns were determined by a halo around the colony (). A larger halo around the colony indicated higher cellulase activity. Based on this classification, transformants with maximum cellulase activity were selected. Since carboxymethyl cellulose is not the optimal β-glucosidase substrate, there was no obvious halo around recombinant S. cerevisiae INVSc-P-SB. By using total chromosome DNA of the recombinants as the template, DNA fragments of the same size as the cellulase genes, bg, cbh, and eg, were obtained by PCR amplification, and the fragments were verified by sequencing. These results confirmed that each gene was integrated into the chromosome of S. cerevisiae INVSc1. The recombinants were named S. cerevisiae INVSc-P-SB, S. cerevisiae INVSc-P-SC, and S. cerevisiae INVSc-P-SE, respectively.
Determination of cellulolytic enzyme activity
The recombinants were cultured in 50 ml YPD medium for about 20 h at 28°C. The supernatants of cultures were used for determining cellulase activity by the DNS method. The concentration of the reducing sugar was calculated based on OD540, and was used to calculate cellulolytic enzyme activity. One enzyme activity unit (U) is the amount of enzyme that releases 1 µmol of reducing sugar. The enzyme activity of β-glucosidase, exoglucanase, and endoglucanase in 1 ml of crude enzyme reached 45.22 U/ml, 72.11 U/ml, and 75.45 U/ml at 50°C, respectively.
Simultaneous saccharification and fermentation by using the recombinants
Three recombinant yeast strains (S. cerevisiae INVSc-P-SB, S. cerevisiae INVSc-P-SC, and S. cerevisiae INVSc-P-SE) and the wild type yeast strain (S. cerevisiae INVSc) were activated as fermentation strains. When the OD600 reached 2.0, the recombinants were inoculated into 1 L of bioreactor, which included YPC liquid medium with microcrystalline cellulose powder as the sole carbon source. A mixed inoculation of equal parts of the 3 recombinant yeasts was performed. The biomass was determined every 12 h and repeated 3 times.
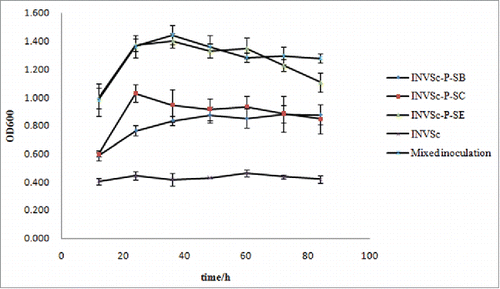
The biomass of recombinant yeasts was evaluated by OD600 determination, and was substantially higher than that of the wild type yeast, INVSc1 (). The standard deviation was calculated to reflect the degree of dispersion within the data set. The wild type control plasmid could only maintain an OD600 of approximately 0.4, indicating no growth. The OD600 of the 3 recombinant plasmids, INVSc-P-SB, INVSc-P-SC, and INVSc-P-SE, reached above 0.80, 1.20, and 1.40, respectively, indicating better growth than the wild type strain. However, the OD600 of the mixed inoculation group reached 1.44, demonstrating the best growth. The genetically modified yeasts were able to use the microcrystalline cellulose as a carbon source for growth, whereas the wild type yeast could not. This indicates that biomass production positively correlated with cellulase activity. Based on the biomass trend, increase in cellulase hydrolysis efficiency by mixed inoculation provides evidence of cellulase synergy during the hydrolysis process.
Figure 3. Biomass curve generated from fermentation with cellulose. The biomass of each fermentation system was determined by the OD600. The fermentation process lasted for 84 h, and biomass was determined every 12 h. The OD600 of the mixed inoculation group reached 1.44, which suggested that this group generated the maximum biomass among the yeast groups tested, thus indicating the best growth. The stationary phase of the mixed inoculation group was also longer than that of the other groups.
SSF was then conducted in 1 L of bioreactor, and corncob powders served as the sole carbon source. Ethanol concentrations fermented with S. cerevisiae INVSc-P-SC and S. cerevisiae INVSc-P-SE reached 2.35 g/L and 3.26 g/L, respectively. However, almost no ethanol could be detected from fermentation of S. cerevisiae INVSc-P-SB. The concentration of ethanol produced from fermentation with the mixed genetically modified S. cerevisiae containing the constitutive promoter was 6.37 g/L after 96 h. This production was higher than that of the other groups, and was about 3 times higher than the genetically modified S. cerevisiae containing the inducible promoter sequence, as previously reported.Citation16 The results showed that the synergistic effect of different cellulases was necessary and sufficient for 1-step bioethanol fermentation by using corncob as the sole carbon source.
Microstructure of corncobs after fermentation
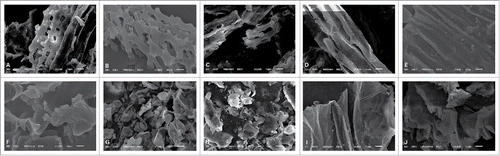
The microstructure of corncobs was observed after fermentation by using SEM at 1000X magnification (). The microstructures of the controls are shown in , and the microstructures of the mixed inoculation are shown in .
Discussion
In this study, 3 genetically modified S. cerevisiae containing the pgk1 constitutive promoter were constructed. The enzyme activity of β-glucosidase, exoglucanase, and endoglucanase were 45.22 U/ml, 72.11 U/ml, and 75.45 U/ml, respectively. As previous reported,Citation16 the enzyme activity of the recombinants containing the β-glucosidase, exoglucanase, and endoglucanase genes with the inducible promoter gal1 were 3.89 U/ml, 3.96 U/ml, and 3.01 U/ml respectively. This suggests that a strong constitutive promoter is more suitable for the expression of cellulase genes than an inducible promoter.
By using microcrystalline cellulose as the sole carbon source, the maximum biomass of the mixed inoculation was higher than that of all the single inoculations tested, and its stationary phase lasted longer. In the SSF using corncob powders as the sole carbon source, the mixed genetically modified S. cerevisiae demonstrated the highest ethanol yield (6.37 g/L). This demonstrated that the synergism of β-glucosidase, exoglucanase, and endoglucanase is very important for cellulose hydrolysis and SSF. The genetically modified S. cerevisiae containing the constitutive promoter are more convenient and efficient for converting cellulose to bioethanol in industrial application than containing the inducible promoter.
The microstructures of corncobs during SSF were evaluated. The original corncob structure resembles long bundle nets (). However, after preliminary hydrolysis, the structure was severed lengthwise and changed to resemble short bundle nets (). Moreover, structures with cracks in the profile () and structures with both lengthwise and profile cracks were observed (). After further hydrolysis, structures demonstrating lengthwise cracks were transformed into irregular granules (), whereas those with cracks in the profile unfolded like a cloth (). Understanding the changes in the structure of corncobs when they are used as the sole carbon source during SSF may prove to be very useful.
In this study, 3 genetically modified S. cerevisiae containing the pgk1 constitutive promoter were constructed, and SSF was performed by using corncobs as the sole carbon source. This method was convenient and efficient, and is overall a feasible method to improve cellulose hydrolysis and bioethanol fermentation using natural cellulose as the carbon source.
Materials and methods
Strains, plasmids and media
and summarize the genetic properties of the strains and plasmids used in this study. Briefly, Escherichia coli strain XL10-Gold served as the host for recombinant DNA manipulations, and cellulolytic enzymes were expressed in the auxotroph S. cerevisiae INVSc1. Recombinant S. cerevisiae INVSc-P-SB, INVSc-P-SC, INVSc-P-SE were constructed as described below. The primers for the α-factor signal sequence and the 3 cellulolytic enzyme genes are shown in .
Table 1. Characteristics of the bacterial and yeast strains used in this study.
Table 2. Characteristics of plasmids used in this study.
Table 3. List of primers used for amplification in this study.
E. coli was grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.0). S. cerevisiae INVSc1 was used as the host for expression of exogenous cellulolytic enzyme genes. The yeast cells were grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) at 28°C. The transformants were screened in SC medium (0.67% YNB, 1% (NH4)2SO4, 2% Glucose) with 0.01% His, Leu, and Trp at 28°C. Recombinant proteins with cellulase activity were screened in CMC-YPD medium (1% yeast extract, 2% peptone, 1% glucose, 1% carboxymethyl cellulose) at 28°C. They were fermented in YPM medium (1% yeast extract, 2% peptone, 2% microcrystalline cellulose) and YPC medium (1% yeast extract, 2% peptone, 2% corncobs). The modified integrated secretion expression vector pHBM368 was conserved in our laboratory.
Vector construction
DNA was purified and manipulated as previously described.Citation17 We searched for the following gene sequences in the NCBI database (www.ncbi.nlm.nih.gov), and synthesized the genes accordingly (Generay Biotech Co., Ltd, Shanghai): constitutive promoter pgk1 sequence (GeneBank: FJ415226), β-glucosidase gene (GeneBank: EU169241), exoglucanase gene from Chaetomium thermophilum (GeneBank: AY861348), and endoglucanase gene (GeneBank: EU169241). Furthermore, the restriction enzyme sites NdeI and NotI were synthesized in the 5′- and 3′-terminals of the pgk1 promoter sequence. The expression vector pHBM368-pgk was constructed by linking the pgk1 promoter to pHBM368 which was digested by NdeI and Not I. The recombinant plasmid was named pHBM368-pgk. The α-factor signal sequence was cloned from pPIC9K, and linked with 3 cellulase genes in vitro. The restriction enzyme sites NotI and SpeI in the 5′- and 3′-terminals of the sequences were used for linking. Three expression vectors were constructed by linking with pHBM368-pgk digested by using NotI and XbaI.
Yeast transformation
S. cerevisiae INVSc1 was transformed with linearized plasmids by electroporation.Citation18 The electroporator was set to 1800 V for a 2-mm cuvette in order to achieve maximum transformation efficiency. For each transformation, 10 μl linearized plasmid DNA and 130 μl competent cells were mixed. Sorbitol was added to aliquots of this mixture, which were then plated onto SC screen medium and incubated at 28°C.
Enzymatic analysis
The concentration of the reducing sugar was assayed by using the 3.5-dinitrosalicylic acid (DNS) method,Citation19 and cellulolytic enzyme activity was calculated based on the reducing sugar concentration.
Simultaneous saccharification and fermentation
Microcrystalline cellulose or corncobs were used as the sole carbon source for fermentation in order to confirm whether they could serve as the carbon source for culturing recombinant S. cerevisiae in medium. The recombinant and control proteins were cultured in 50 ml YPD medium for 16 hours at 28°C at 200 rpm. The yeast cells were used as the inoculation seed when the OD600 reached 2.0. Fermentation was performed at 28°C in a 1 L bioreactor. The inoculation amount was 10–15% (v/v). Ethanol concentration was analyzed by gas chromatography (Agilent 7890A GC system, USA) with a flame ionization detector. Isopropanol served as an internal standard.Citation20
Microstructure of corncobs after fermentation
The corncobs were collected using a 500-mesh filter cloth after YPC fermentation and washed with sterile water. The microstructure of corncobs was observed by using a SEM system (JSM6510LV, JEOL, Japan) at 1000X magnification.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Notes on contributors
HTS and SHL carried out construction of the recombinants, and drafted the manuscript. YG and YMY carried out simultaneous saccharification and fermentation. WJX and WCX carried out analysis. ZLL, RL and XDM participated in scanning electron microscopy. ZBJ conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the following grants: Nation Natural Science Foundation of China (2167061089), Science and Technology Support Program of Hubei Province (2015BBA177), National High Technology Research and Development Program (“863” Program: 2012AA020403, 2012AA02A701).
References
- Brás JL, Cartmell A, Carvalho AL, Verzé G, Bayer EA, Vazana Y, Correia MA, Prates JA, Ratnaparkhe S, Boraston Ab, et al. Structural insights into a unique cellulase fold and mechanism of cellulose hydrolysis. Proc Natl Acad Sci U S A 2011; 108(13):5237-42; http://www.pnas.org/cgi/pmidlookup?view=long&pmid=21393568; http://dx.doi.org/10.1073/pnas.1015006108
- Camargo D, Gomes SD, Sene L. Ethanol production from sunflower meal biomass by simultaneous saccharification and fermentation (SSF) with Kluyveromyces marxianus ATCC 36907. Bioprocess Biosyst Eng 2014; 37(11):2235-42; PMID:24794173; http://dx.doi.org/10.1007/s00449-014-1201-x
- Xu SF. Talking about the Comprehensive Utilizationof Corncob. Sci-Tech Infomation Dev Economy 2011; 21(17):174-5
- Dai Z, Liu Y, Huang L, Zhang X. Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnol Bioeng 2012; 109(11):2845-53; PMID:22566191; http://dx.doi.org/10.1002/bit.24547
- Engels B, Dahm P, Jennewein S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab Eng 2008; 10(3–4):201-6; http://linkinghub.elsevier.com/retrieve/pii/S1096-7176(08)00014-1; PMID:18485776; http://dx.doi.org/10.1016/j.ymben.2008.03.001
- Gusakov AV, Kondratyeva EG, Sinitsyn AP. Comparison of two methods for assaying reducing sugars in the determination of carbohydrase activities. Int J Anal Chem 2011; 2011:283658; PMID:21647284; http://dx.doi.org/10.1155/2011/283658
- Knust B, von Wettstein D. Expression and secretion of pea-seed lipoxygenase isoenzymes in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 1992; 37(3):342-51; PMID:1368907; http://dx.doi.org/10.1007/BF00210990
- Koppram R, Olsson L. Combined substrate, enzyme and yeast feed in simultaneous saccharification and fermentation allow bioethanol production from pretreated spruce biomass at high solids loadings. Biotechnol Biofuels 2014; 7(1):54; PMID:24713027; http://dx.doi.org/10.1186/1754-6834-7-54
- Lamed R, Kenig R, Morag E, Calzada J. Efficient cellulose solubilization by a combined cellulosome glucosidase system. Appl Biochem Biotechnol 1991; 27:173-83; http://dx.doi.org/10.1007/BF02921525
- Mansfield SD, Mooney C, Saddler JN. Substrate and Enzyme Characteristics that Limit Cellulose Hydrolysis. Biotechnol Prog 1999; 15(5):804-16; PMID:10514250; http://dx.doi.org/10.1021/bp9900864
- Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, Ladisch M. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 2005; 96(6):673-86; http://linkinghub.elsevier.com/retrieve/pii/S0960-8524(04)00253-6; PMID:15588770; http://dx.doi.org/10.1016/j.biortech.2004.06.025
- Nacken V, Achstetter T, Degryse E. Probing the limits of expression levels by varying promoter strength and plasmid copy number in Saccharomyces cerevisiae. Gene 1996; 175(1–2):253-60; PMID:8917107; http://dx.doi.org/10.1016/0378-1119(96)00171-0
- Oberoi HS, Babbar N, Sandhu SK, Dhaliwal SS, Kaur U, Chadha BS, Bhargav VK. Ethanol production from alkali-treated rice straw via simultaneous saccharification and fermentation using newly isolated thermotolerant Pichia kudriavzevii HOP-1. J Ind Microbiol Biotechnol 2012; 39(4):557-66; PMID:22131104; http://dx.doi.org/10.1007/s10295-011-1060-2
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edition Cold Spring Harbor Laboratory Press, 2001
- Thomas L, Joseph A, Gottumukkala LD. Xylanase and cellulase systems of Clostridium sp: an insight on molecular approaches for strain improvement. Bioresour Technol 2014; 158:343-50; http://linkinghub.elsevier.com/retrieve/pii/S0960-8524(14)00165-5; PMID:24581864; http://dx.doi.org/10.1016/j.biortech.2014.01.140
- Song H, Liu J, Liu Y, Leng H, Li X, Zhao Y, Yang X, Jiang Z. Genetically modified Saccharomyces cerevisiae for one-step fermentation of bioalcohol using corncob as sole carbon source. Annals Microbiol 2014; 64:781-5; http://dx.doi.org/10.1007/s13213-013-0714-x
- Uppugundla N, da Costa Sousa L, Chundawat SP, Yu X, Simmons B, Singh S, Gao X, Kumar R, Wyman CE, Dale BE, et al. A comparative study of ethanol production using dilute acid, ionic liquid and AFEX™ pretreated corn stover. Biotechnol Biofuels 2014; 7:72; PMID:24917886; http://dx.doi.org/10.1186/1754-6834-7-72
- Zhou YJ, Gao W, Rong Q, Jin G, Chu H, Liu W, Yang W, Zhu Z, Li G, Zhu G, et al. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc 2012; 134(6):3234-41; PMID:22280121; http://dx.doi.org/10.1021/ja2114486
- Zuo Q, Zhao XQ, Xiong L, Liu HJ, Xu YH, Hu SY, Ma ZY, Zhu QW, Bai FW. Fine-tuning of xylose metabolism in genetically engineered Saccharomyces cerevisiae by scattered integration of xylose assimilation genes. Biochem Biophys Res Commun 2013; 440(2):241-4; PMID:24051089; http://dx.doi.org/10.1016/j.bbrc.2013.09.046
- Liu R, Shen F. Impacts of main factors on bioethanol fermentation from stalk juice of sweet sorghum by immobilized Saccharomyces cerevisiae (CICC 1308). Bioresour Technol 2008; 99(4):847-54; PMID:17360181; http://dx.doi.org/10.1016/j.biortech.2007.01.009