ABSTRACT
Lignin is 1 of the 3 major components of lignocellulose. Its polymeric structure includes aromatic subunits that can be converted into high-value-added products, but this potential cannot yet been fully exploited because lignin is highly recalcitrant to degradation. Different approaches for the depolymerization of lignin have been tested, including pyrolysis, chemical oxidation, and hydrolysis under supercritical conditions. An additional strategy is the use of lignin-degrading enzymes, which imitates the natural degradation process. A versatile set of enzymes for lignin degradation has been identified, and research has focused on the production of recombinant enzymes in sufficient amounts to characterize their structure and reaction mechanisms. Enzymes have been analyzed individually and in combinations using artificial substrates, lignin model compounds, lignin and lignocellulose. Here we consider progress in the production of recombinant lignin-degrading peroxidases, the advantages and disadvantages of different expression hosts, and obstacles that must be overcome before such enzymes can be characterized and used for the industrial processing of lignin.
Introduction
Lignocellulose is the most abundant form of sustainable biomass on earth and is therefore highly promising for the production of renewable fuels and chemicals.Citation1 Two of the components of lignocellulose (cellulose and hemicellulose) are readily accessible for industrial processes because they are easily degraded by known enzymes and by chemical hydrolysis.Citation2 However, the third component (lignin) is much more resistant to degradation. Unlike the other components it is a non-carbohydrate polymer with aromatic subunits. Depending on the lignin source, up to 3 phenylpropanoid monomeric precursors are coupled to form the lignin polymer.Citation3 It is not currently possible to fully exploit the potential of this polymer due to the lack of knowledge and technologies for lignin degradation.Citation4 The lignin obtained from lignocellulosic biomass during industrial processes such as papermaking and biomass fractionation is therefore either incinerated to provide electricity for plants that produce cellulosic ethanolCitation5 or is chemically modified and used to manufacture products such as phenolic resins, polyurethane foams, epoxy resins and biodispersants.Citation6 However, lignin has the potential for conversion into high-value-added products such as vanillin, adipic acid, ferulic acid, vinyl guaiacol, optically-active lignans, the dimers of monolignols and p-coumaric acid.Citation7,8 Lignin could therefore replace the fossil-fuel-based feedstock used in many current industrial processes, increasing their sustainability and benefiting the environment.
Lignin-degrading enzymes
Studies focusing on the biological degradation of lignin date back to the 1980s. Enzymes from white-rot and brown-rot fungi have been studied more comprehensively than those from bacteria because they were initially considered to be more efficient, but recent reports have shown that bacteria also degrade lignin and produce potentially important peroxidases and laccases.Citation9 The first lignin-degrading enzymes to be identified were peroxidases from the white-rot fungus Phanerochaete chrysosporium.Citation10,11 Many further enzymes were subsequently identified and characterized, and these can be assigned to 2 major classes: heme peroxidases and laccases ().
Figure 1. Lignin subunits attacked by enzymes and their most common reaction mechanisms. The bulky lignin polymer structure represents part of an organosolv lignin substrate.Citation65 Both superfamilies of lignin-degrading enzymes (heme peroxidases and laccases) can oxidize phenolic lignin subunits. In order to oxidize non-phenolic lignin subunits, laccase requires the presence of a mediator. Lignin peroxidase, versatile peroxidase and DyP-type peroxidase do not require a mediator to attack non-phenolic structures. Manganese peroxidase and versatile peroxidase oxidize phenolic lignin subunits via the oxidation of manganese (Mn2+ → Mn3+). The inset box shows heme peroxidases reducing H2O2 to water to catalyze the oxidation reactions, whereas laccases reduce molecular oxygen to water, which is accompanied by the oxidation of the substrates or mediators. The atoms and bonds of the phenolic and non-phenolic lignin subunits are highlighted in bold within the bulky structure. Abbreviations: med = mediator, sub = substrate.
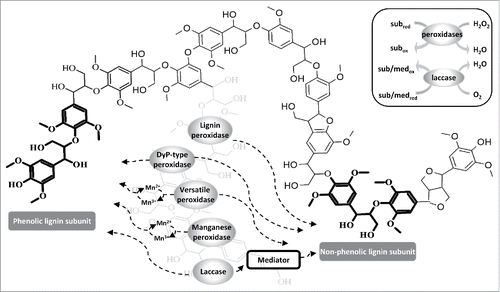
The first heme peroxidase superfamily comprises lignin peroxidase (LiP), manganese peroxidase (MnP) and versatile peroxidase (VP), which are typically produced by fungi.Citation12 Non-phenolic structures are thought to be degraded by LiP due to its high redox potential. MnP can oxidize phenolic model compounds via the oxidation of Mn2+ to Mn3+. VP is so called because it combines the catalytic properties of LiP and MnP.Citation3
The second heme peroxidase superfamily comprises the dye-decolorizing (DyP-type) peroxidases, which also oxidize β-O-4 non-phenolic and phenolic lignin model compounds. Although the first DyP-type peroxidase was isolated from fungi, subsequent genome sequence analysis has revealed that this superfamily of enzymes is also prominent in bacteria.Citation13 DyP-type peroxidases typically require hydrogen peroxide to form the oxo-ferryl intermediates of the enzymes, which subsequently oxidize the mediator or the substrate.
Unlike peroxidases, laccases reduce molecular oxygen to water using the copper atoms located within the active center in order to oxidize the mediator or substrate. Phenolic lignin units are oxidized directly whereas non-phenolic subunits are oxidized via a redox mediator system to overcome the low redox potential of laccase. The potential applications of laccases in lignocellulose degradation have been comprehensively reviewed.Citation14,15 The remaining enzymes that support lignin degradation are known as accessory enzymes.Citation3 These include enzymes that produce hydrogen peroxide and enzymes that reduce methoxy radicals generated by the peroxidases and laccases.
Recombinant lignin-degrading peroxidases
Although several groups have attempted to optimize the cultivation of P. chrysosporium to increase the yield of native LiP and MnP, a successful large-scale production system has yet to be reported. This could potentially be addressed by using heterologous expression systems to produce large amounts of unmodified or modified recombinant enzymes for targeted industrial applications.
The mechanism of cellulose degradation is well understood and recombinant cellulases have been produced successfully in various heterologous systems.Citation16,17 In contrast, the mechanisms and interactions of lignin-degrading enzymes (especially peroxidases) are poorly characterized. Even so, many recombinant lignin-degrading enzymes from diverse fungal () and bacterial () sources have been expressed in different host organisms, although it is challenging to achieve high yields and satisfactory catalytic activity due to the complex secondary and tertiary structures of such enzymes. Specific posttranslational modifications may be necessary during maturation, such as heme assembly, glycosylation and the formation of disulfide bridges, which may differ between the native source and heterologous production host.
Table 1. Recombinant fungal lignin-degrading peroxidases. Examples of successfully produced fungal lignin-degrading peroxidases and the substrates that have been used. The activity of the enzymes (if stated) is shown in parentheses. ABTS: 2,2-azinobis (3-ethylbenzothiazoline-6-sulfonate), DCP: 2,4-dichlorophenol, DFAD: 4-[(3,5-difluoro-4-hydroxyphenyl)azo]benzenesulfonic acid, DMP: 2,6-dimethoxyphenol, icb: inclusion body; MHG: methoxyhydroquinone, VA: veratryl alcohol.
Table 2. Recombinant bacterial lignin-degrading peroxidases. Examples of successfully produced recombinant bacterial lignin-degrading peroxidases and the substrates that have been used. The activity of the enzymes (if stated) is shown in parentheses. ABTS: 2,2-azinobis (3-ethylbenzothiazoline-6-sulfonate), DCP: 2,4-dichlorophenol, DMP: 2,6-dimethoxyphenol, DOPA: L-3,4-dihydroxyphenylalanine, VA: veratryl alcohol.
This challenge is clearly illustrated by recent progress toward the production of high enzyme titers with activity against complex lignin model compounds, as well as lignin and wood (). The first report describing the production of recombinant fungal MnP and LiP was published in 1991, but although the authors used insect cells and the reliable baculovirus expression system, only low yields were achieved and the cost of the process was high ().Citation18-20 P. chrysosporium was used for the homologous production of MnP and LiP with yields of up to 2 mg of purified LiP per liter of culture.Citation20,21 Later, Pleurotus ostreatus was used to express the versatile peroxidase MnP2 homologously, with a yield of up to 21 mg L−1.Citation22 However, the extensive screening required to identify mutant production strains, as well as contamination with the native enzyme, led to disappointing results. The model bacterium Escherichia coli has been extensively developed as an expression platform for fungal enzymes, and many recombinant enzymes have been isolated successfully from inclusion bodies and refolded with 1–28% efficiencyCitation23-30 and maximum yields of 1.5–14 mg L−1 after in vitro activation and purification.Citation29 A soluble form of a VP was produced using a thioredoxin tagCitation31,32 or following intensive screening.Citation31 Few publications have described the activity of such recombinant enzymes against complex lignin model compounds or synthetic lignin.Citation29,30,33 Fungal and yeast expression systems have been used to ensure that posttranslational modifications such as glycosylation are carried out, because these are thought to increase the stability of many fungal enzymes. MnP, LiP and VP have been expressed in various Pichia and Saccharomyces strains, the recombinant enzymes were secreted into the medium (e.g. 100 mg L−1 and 21.6 mg L−1 for Pichia methanolica and Saccharomyces cerevisiae, respectively)Citation34,35 and catalytic activities were determined using common substrates such as 2,2'azinobis(3-ethylbenzothiazoline-6-sulfonate) (ABTS) and veratryl alcohol (VA).Citation34-38 MnP produced in this manner was suitable for the treatment of Kraft lignin as determined by measuring the kappa number and the increase in Klason lignin residue.Citation39 MnP from P. chrysosporium has also been expressed in the filamentous fungi Aspergillus oryzae and A. niger,Citation40,41 yielding up to 100 mg L−1 in A. niger. A VP from Pleurotus eryngii was expressed successfully in another Aspergillus strain (A. nidulans) with a yield of 0.4 mg L-1 after protein purification.Citation42 Recently, native LiP and MnP were expressed in P. chrysosporium in concert with an exogenous VP, generating a recombinant strain with higher LiP and MnP activity and detectable VP activity.Citation43 Further strain engineering included the introduction of an additional laccase.Citation44 In addition to LiP, MnP and VP, fungal DyP-type peroxidases have also been expressed in various heterologous production platforms,Citation45-48 with yields of 0.1 mg L−1 purified enzyme achieved using E. coliCitation47 and up to 62 mg L−1 recovered from the supernatant of A. oryzae expression cultures.Citation45
The production of several recombinant bacterial lignin-degrading enzymes has also been achieved over the last few years (). The first recombinant LiP was produced successfully in 1998 by cloning a 4.1-kb fragment of Streptomyces viridosporus genomic DNA and expressing it in the yeast Pichia pastoris.Citation49 Several DyP-type peroxidases from bacteria have been expressed in E. coli, including enzymes from Thermobifida fusca,Citation50,51 Amycolatopsis sp.,Citation52 Pseudomonas aeruginosa,Citation53 Anabaena sp.,Citation54 Pseudomonas putida,Citation55 Bacillus subtilis,Citation55,56 Saccharomonospora viridis,Citation57 Pseudomonas fluorescens,Citation58 and 2 enzymes (DypA and DypB) from Rhodococcus jostii RHA1.Citation59,60 The yields were generally in the range 3–25 mg L−1, but exceptional yields of 106.5 mg L−1 and 2.1 g L−1 have been achieved in E. coli for S. viridisCitation57 and B. subtilisCitation56 peroxidases, respectively. The recombinant enzymes have been extensively characterized and optimized using a range of dyes as well as simple and also more complex lignin model compounds (). These studies revealed that DyPs are active against both non-phenolic and phenolic substrates and can oxidize polymeric lignin. Another DyP from R. jostii, which was expressed in its native host, was shown to be active against wheat straw lignocellulose and produced potentially valuable products, namely vanillin and smaller amounts of ferulic acid and 4-hydroxybenzaldehyde.Citation61
Future challenges
Detailed studies describing the activity of recombinant enzymes on complex lignin model compounds, lignin and lignocellulose substrates have been published only in the last few years. Activity assays using lignin as a substrate are time-consuming because both the measurements and data analysis take a long time. The oligomeric nature of lignin model compounds affects the catalytic activity of lignin-degrading enzymes, e.g., the activity of LiP on the model compound β-O-4 lignin was 25-fold lower than on the simple artificial substrate VA.Citation62 To increase the convenience of protein characterization and engineering approaches, artificial substrates such as ABTS, VA and DMP are used for initial testing and simple phenolic or non-phenolic model compounds are used in subsequent assays. However, the degradation of these simple aromatic compounds may require different catalytic mechanisms, which makes it difficult to extrapolate the results to typical lignin and lignocellulose samples. For example, LiP has two substrate interaction sites: a heme-edge site which is typical for peroxidases and a second site for the oxidation of VA.Citation24 The optimization of enzyme activity by screening will only select for increases in activity against the test substrate under the specific conditions of the test and will not necessarily have a positive impact on the degradation of typical lignin substrates found in plant biomass or organosolv lignin. It is also clear that the successful degradation of lignin and lignocellulose requires multiple peroxidases and other lignin-degrading enzymes and accessory proteins.Citation63 Each enzyme individually may not show promising results in vitro because optimal activity requires the presence of other enzymes, or conversely an individual enzyme may show promising in vitro activity alone but this may not be fulfilled under natural conditions when other enzymes are present. Lignin degradation is not an isolated process in nature, but is instead embedded in the more complex process of lignocellulose degradation. For example, the oxidative activity of lytic polysaccharide monooxygenases against cellulose was recently linked to lignin via long-range electron transfer.Citation64
In conclusion, the production and characterization of recombinant single enzymes is necessary to understand the different reaction mechanisms required for successful and tailored lignin degradation. Optimized expression, intensive characterization and the analysis of enzyme structures and reaction mechanisms will help to determine the optimal reaction conditions of these enzymes. Nevertheless, successful lignin degradation currently appears to require a combination of enzymes. In the future, the combination of certain lignin-degrading enzymes will enable the effective and targeted degradation of lignin feedstock sources to obtain specific value-added products.
Abbreviations
| ABTS | = | 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonate) |
| DMP | = | 2,6-dimethoxyphenol |
| DyP-type peroxidase | = | dye-decolorizing peroxidase |
| LiP | = | lignin peroxidase |
| MnP | = | manganese peroxidase |
| VA | = | veratryl alcohol |
| VP | = | versatile peroxidase |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank R. M. Twyman for manuscript editing.
Funding
This work was supported by the Cluster of Excellence “Tailor-made Fuels from Biomass” (EXC 236), which is funded through the Excellence Initiative by the German federal and state governments to promote science and research at German universities, and by the European Commission (SuBiCat Initial Training Network, Call FP7-PEOPLE-2013-ITN, grant no. 607044).
References
- Moreno AD, Ibarra D, Alvira P, Tomas-Pejo E, Ballesteros M. A review of biological delignification and detoxification methods for lignocellulosic bioethanol production. Crit Rev Biotechnol 2015; 35:342-54; PMID:24506661; http://dx.doi.org/10.3109/07388551.2013.878896
- Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol 2009; 60:165-82; PMID:19014348; http://dx.doi.org/10.1146/annurev.arplant.043008.092125
- Abdel-Hamid AM, Solbiati JO, Cann IK. Insights into lignin degradation and its potential industrial applications. Adv Appl Microbiol 2013; 82:1-28; PMID:23415151; http://dx.doi.org/10.1016/B978-0-12-407679-2.00001-6
- Calvo-Flores FG, Dobado JA. Lignin as renewable raw material. Chem Sus Chem 2010; 3:1227-35; PMID:20839280; http://dx.doi.org/10.1002/cssc.201000157
- Schubert C. Can biofuels finally take center stage? Nat Bio Technol 2006; 24:777-84; PMID:16841058; http://dx.doi.org/10.1038/nbt0706-777
- Lora JH, Glasser WG. Recent industrial applications of lignin: A sustainable alternative to nonrenewable materials. J Polym Environ 2002; 10:39-48; http://dx.doi.org/10.1023/A:1021070006895
- Ghaffar SH, Fan MZ. Lignin in straw and its applications as an adhesive. Int J Adhes Adhes 2014; 48:92-101; http://dx.doi.org/10.1016/j.ijadhadh.2013.09.001
- Bugg TDH, Rahmanpour R. Enzymatic conversion of lignin into renewable chemicals. Curr Opin Chem Biol 2015; 29:10-7; PMID:26121945; http://dx.doi.org/10.1016/j.cbpa.2015.06.009
- Bugg TDH, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Bio Tech 2011; 22:394-400; PMID:21071202; http://dx.doi.org/10.1016/j.copbio.2010.10.009
- Glenn JK, Morgan MA, Mayfield MB, Kuwahara M, Gold MH. An extracellular H2O2-requiring enzyme preparation involved in lignin biodegradation by the white rot basidiomycete Phanerochaete chrysosporium. Biochem Biophys Res Commun 1983; 114:1077-83; PMID:6615503; http://dx.doi.org/10.1016/0006-291X(83)90672-1
- Tien M, Kirk TK. Lignin-Degrading Enzyme from the Hymenomycete Phanerochaete chrysosporium Burds. Science 1983; 221:661-3; PMID:17787736; http://dx.doi.org/10.1126/science.221.4611.661
- Pollegioni L, Tonin F, Rosini E. Lignin-degrading enzymes. FEBS J 2015; 282:1190-213; PMID:25649492; http://dx.doi.org/10.1111/febs.13224
- Colpa DI, Fraaije MW, van Bloois E. DyP-type peroxidases: a promising and versatile class of enzymes. J Ind Microbiol Biot 2014; 41:1-7; http://dx.doi.org/10.1007/s10295-013-1371-6
- Munk L, Sitarz AK, Kalyani DC, Mikkelsen JD, Meyer AS. Can laccases catalyze bond cleavage in lignin? Bio Technol Adv 2015; 33:13-24; PMID:25560931; http://dx.doi.org/10.1016/j.biotechadv.2014.12.008
- Roth S, Spiess AC. Laccases for biorefinery applications: a critical review on challenges and perspectives. Bio Process Bio Syst Eng 2015; 38:2285-313; PMID:26437966; http://dx.doi.org/10.1007/s00449-015-1475-7
- Garvey M, Klose H, Fischer R, Lambertz C, Commandeur U. Cellulases for biomass degradation: comparing recombinant cellulase expression platforms. Trends Bio Technol 2013; 31:581-93; PMID:23910542; http://dx.doi.org/10.1016/j.tibtech.2013.06.006
- Lambertz C, Garvey M, Klinger J, Heesel D, Klose H, Fischer R, Commandeur U. Challenges and advances in the heterologous expression of cellulolytic enzymes: a review. Bio Technol Bio Fuels 2014; 7:135; PMID:25356086; http://dx.doi.org/10.1186/s13068-014-0135-5
- Johnson TM, Li JK. Heterologous expression and characterization of an active lignin peroxidase from Phanerochaete chrysosporium using recombinant baculovirus. Arch Bio Chem Bio Phys 1991; 291:371-8; PMID:1952950; http://dx.doi.org/10.1016/0003-9861(91)90148-C
- Pease EA, Aust SD, Tien M. Heterologous expression of active manganese peroxidase from Phanerochaete chrysosporium using the baculovirus expression system. Bio Chem Bio Phys Res Commun 1991; 179:897-903; PMID:1898410; http://dx.doi.org/10.1016/0006-291X(91)91903-P
- Mayfield MB, Kishi K, Alic M, Gold MH. Homologous expression of recombinant manganese peroxidase in Phanerochaete chrysosporium. Appl Environ Microbiol 1994; 60:4303-9; PMID:7811070
- Gelpke MDS, Mayfield-Gambill M, Cereghino GPL, Gold MH. Homologous expression of recombinant lignin peroxidase in Phanerochaete chrysosporium. Appl Environ Microb 1999; 65:1670-4.
- Tsukihara T, Honda Y, Sakai R, Watanabe T, Watanabe T. Exclusive overproduction of recombinant versatile peroxidase MnP2 by genetically modified white rot fungus, Pleurotus ostreatus. J Bio Technol 2006; 126:431-9; PMID:16820241; http://dx.doi.org/10.1016/j.jbiotec.2006.05.013
- Whitwam R, Tien M. Heterologous expression and reconstitution of fungal Mn peroxidase. Arch Bio Chem Biophys 1996; 333:439-46; PMID:8809085; http://dx.doi.org/10.1006/abbi.1996.0413
- Doyle WA, Blodig W, Veitch NC, Piontek K, Smith AT. Two substrate interaction sites in lignin peroxidase revealed by site-directed mutagenesis. Bio Chemistry-Us 1998; 37:15097-105; http://dx.doi.org/10.1021/bi981633h
- Nie GJ, Reading NS, Aust SD. Expression of the lignin peroxidase H2 gene from Phanerochaete chrysosporium in Escherichia coli. Bio Chem Bioph Res Co 1998; 249:146-50; http://dx.doi.org/10.1006/bbrc.1998.9106
- Timofeevski SL, Nie G, Reading NS, Aust SD. Addition of veratryl alcohol oxidase activity to manganese peroxidase by site-directed mutagenesis. Bio Chem Bioph Res Co 1999; 256:500-4; http://dx.doi.org/10.1006/bbrc.1999.0360
- Perez-Boada M, Doyle WA, Ruiz-Duenas FJ, Martinez MJ, Martinez AT, Smith AT. Expression of Pleurotus eryngii versatile peroxidase in Escherichia coli and optimisation of in vitro folding. Enzyme Microb Tech 2002; 30:518-24; http://dx.doi.org/10.1016/S0141-0229(02)00008-X
- Miki Y, Morales M, Ruiz-Duenas FJ, Martinez MJ, Wariishi H, Martinez AT. Escherichia coli expression and in vitro activation of a unique ligninolytic peroxidase that has a catalytic tyrosine residue. Protein Expr Purif 2009; 68:208-14; PMID:19505579; http://dx.doi.org/10.1016/j.pep.2009.06.003
- Fernandez-Fueyo E, Ruiz-Duenas FJ, Martinez AT. Engineering a fungal peroxidase that degrades lignin at very acidic pH. Bio Technol Bio Fuels 2014; 7:114; PMID: 25788979; http://dx.doi.org/10.1186/1754-6834-7-114
- Fernandez-Fueyo E, Ruiz-Duenas FJ, Martinez MJ, Romero A, Hammel KE, Medrano FJ, Martinez AT. Ligninolytic peroxidase genes in the oyster mushroom genome: heterologous expression, molecular structure, catalytic and stability properties, and lignin-degrading ability. Bio Technol Bio Fuels 2014; 7:2; PMID:24387130; http://dx.doi.org/10.1186/1754-6834-7-2
- Mohorcic M, Bencina M, Friedrich J, Jerala R. Expression of soluble versatile peroxidase of Bjerkandera adusta in Escherichia coli. Bio Resour Technol 2009; 100:851-8; PMID:18707878; http://dx.doi.org/10.1016/j.biortech.2008.07.005
- Bao X, Liu A, Lu X, Li JJ. Direct over-expression, characterization and H2O2 stability study of active Pleurotus eryngii versatile peroxidase in Escherichia coli. Bio Technol Lett 2012; 34:1537-43; PMID:22566208; http://dx.doi.org/10.1007/s10529-012-0940-5
- Mester T, Ambert-Balay K, Ciofi-Baffoni S, Banci L, Jones AD, Tien M. Oxidation of a tetrameric nonphenolic lignin model compound by lignin peroxidase. J Biol Chem 2001; 276:22985-90; PMID:11304528; http://dx.doi.org/10.1074/jbc.M010739200
- Wang HK, Lu FP, Sun WF, Du LX. Heterologous expression of lignin peroxidase of Phanerochaete chrysosporium in Pichia methanolica. Bio Technol Lett 2004; 26:1569-73; PMID:15604798; http://dx.doi.org/10.1023/B:BILE.0000045654.66689.b4
- Garcia-Ruiz E, Gonzalez-Perez D, Ruiz-Duenas FJ, Martinez AT, Alcalde M. Directed evolution of a temperature-, peroxide- and alkaline pH-tolerant versatile peroxidase. Bio Chem J 2012; 441:487-98; PMID:21980920; http://dx.doi.org/10.1042/BJ20111199
- Gu L, Lajoie C, Kelly C. Expression of a Phanerochaete chrysosporium manganese peroxidase gene in the yeast Pichia pastoris. Bio Technol Prog 2003; 19:1403-9; PMID:14524699; http://dx.doi.org/10.1021/bp025781h
- Ryu K, Kang JH, Wang L, Lee EK. Expression in yeast of secreted lignin peroxidase with improved 2,4-dichlorophenol degradability by DNA shuffling. J Bio Technol 2008; 135:241-6; PMID:18514942; http://dx.doi.org/10.1016/j.jbiotec.2008.04.007
- Wang W, Wen X. Expression of lignin peroxidase H2 from Phanerochaete chrysosporium by multi-copy recombinant Pichia strain. J Environ Sci (China) 2009; 21:218-22; PMID:19402425; http://dx.doi.org/10.1016/S1001-0742(08)62254-8
- Jiang F, Kongsaeree P, Charron R, Lajoie C, Xu HW, Scott G, Kelly C. Production and separation of manganese peroxidase from heme amended yeast cultures. Bio Technol Bio Eng 2008; 99:540-9; PMID:17680655; http://dx.doi.org/10.1002/bit.21590
- Stewart P, Whitwam RE, Kersten PJ, Cullen D, Tien M. Efficient expression of a Phanerochaete chrysosporium manganese peroxidase gene in Aspergillus oryzae. Appl Environ Microbiol 1996; 62:860-4; PMID:8975615
- Conesa A, van den Hondel CA, Punt PJ. Studies on the production of fungal peroxidases in Aspergillus niger. Appl Environ Microbiol 2000; 66:3016-23; PMID:10877800; http://dx.doi.org/10.1128/AEM.66.7.3016-3023.2000
- Ruiz-Duenas FJ, Martinez MJ, Martinez AT. Heterologous expression of Pleurotus eryngii peroxidase confirms its ability to oxidize Mn(2+) and different aromatic substrates. Appl Environ Microbiol 1999; 65:4705-7; PMID:10508113
- Coconi-Linares N, Magana-Ortiz D, Guzman-Ortiz DA, Fernandez F, Loske AM, Gomez-Lim MA. High-yield production of manganese peroxidase, lignin peroxidase, and versatile peroxidase in Phanerochaete chrysosporium. Appl Microbiol Bio Technol 2014; 98:9283-94; PMID:25269601; http://dx.doi.org/10.1007/s00253-014-6105-9
- Coconi-Linares N, Ortiz-Vazquez E, Fernandez F, Loske AM, Gomez-Lim MA. Recombinant expression of four oxidoreductases in Phanerochaete chrysosporium improves degradation of phenolic and non-phenolic substrates. J Biotechnol 2015; 209:76-84; PMID:26113215; http://dx.doi.org/10.1016/j.jbiotec.2015.06.401
- Sugano Y, Muramatsu R, Ichiyanagi A, Sato T, Shoda M. DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family: ASP171 replaces the distal histidine of classical peroxidases. J Biol Chem 2007; 282:36652-8; PMID:17928290; http://dx.doi.org/10.1074/jbc.M706996200
- Sugano Y, Nakano R, Sasaki K, Shoda M. Efficient heterologous expression in Aspergillus oryzae of a unique dye-decolorizing peroxidase, DyP, of Geotrichum candidum Dec 1. Appl Environ Microbiol 2000; 66:1754-8; PMID:10742277; http://dx.doi.org/10.1128/AEM.66.4.1754-1758.2000
- Sugano Y, Ishii Y, Shoda M. Role of H164 in a unique dye-decolorizing heme peroxidase DyP. Biochem Biophys Res Commun 2004; 322:126-32; PMID:15313183; http://dx.doi.org/10.1016/j.bbrc.2004.07.090
- Liers C, Pecyna MJ, Kellner H, Worrich A, Zorn H, Steffen KT, Hofrichter M, Ullrich R. Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood- and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl Microbiol Bio Technol 2013; 97:5839-49; PMID:23111597; http://dx.doi.org/10.1007/s00253-012-4521-2
- Thomas L, Crawford DL. Cloning of clustered Streptomyces viridosporus T7A lignocellulose catabolism genes encoding peroxidase and endoglucanase and their extracellular expression in Pichia pastoris. Can J Microbiol 1998; 44:364-72; PMID:9674109; http://dx.doi.org/10.1139/w98-010
- van Bloois E, Torres Pazmino DE, Winter RT, Fraaije MW. A robust and extracellular heme-containing peroxidase from Thermobifida fusca as prototype of a bacterial peroxidase superfamily. Appl Microbiol Bio Technol 2010; 86:1419-30; PMID:19967355; http://dx.doi.org/10.1007/s00253-009-2369-x
- Rahmanpour R, Rea D, Jamshidi S, Fulop V, Bugg TD. Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Arch Bio Chem Biophys 2016; 594:54-60; PMID:26901432; http://dx.doi.org/10.1016/j.abb.2016.02.019
- Brown ME, Barros T, Chang MC. Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium. ACS Chem Biol 2012; 7:2074-81; PMID:23054399; http://dx.doi.org/10.1021/cb300383y
- Li J, Liu C, Li B, Yuan H, Yang J, Zheng B. Identification and molecular characterization of a novel DyP-type peroxidase from Pseudomonas aeruginosa PKE117. Appl Biochem Biotechnol 2012; 166:774-85; PMID:22161141; http://dx.doi.org/10.1007/s12010-011-9466-x
- Ogola HJ, Kamiike T, Hashimoto N, Ashida H, Ishikawa T, Shibata H, Sawa Y. Molecular characterization of a novel peroxidase from the cyanobacterium Anabaena sp. strain PCC 7120. Appl Environ Microbiol 2009; 75:7509-18; PMID:19801472; http://dx.doi.org/10.1128/AEM.01121-09
- Santos A, Mendes S, Brissos V, Martins LO. New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications. Appl Microbiol Biot 2014; 98:2053-65; http://dx.doi.org/10.1007/s00253-013-5041-4
- Min K, Gong G, Woo HM, Kim Y, Um Y. A dye-decolorizing peroxidase from Bacillus subtilis exhibiting substrate-dependent optimum temperature for dyes and β-ether lignin dimer. Sci Rep 2015; 5:8245; PMID:25650125; http://dx.doi.org/10.1038/srep08245
- Yu W, Liu W, Huang H, Zheng F, Wang X, Wu Y, Li K, Xie X, Jin Y. Application of a novel alkali-tolerant thermostable DyP-type peroxidase from Saccharomonospora viridis DSM 43017 in biobleaching of eucalyptus kraft pulp. PLoS One 2014; 9:e110319; PMID:25333297; http://dx.doi.org/10.1371/journal.pone.0110319
- Rahmanpour R, Bugg TD. Characterisation of Dyp-type peroxidases from Pseudomonas fluorescens Pf-5: Oxidation of Mn(II) and polymeric lignin by Dyp1B. Arch Biochem Biophys 2015; 574:93-8; PMID:25558792; http://dx.doi.org/10.1016/j.abb.2014.12.022
- Ahmad M, Roberts JN, Hardiman EM, Singh R, Eltis LD, Bugg TD. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Bio Chemistry-Us 2011; 50:5096-107; http://dx.doi.org/10.1021/bi101892z
- Roberts JN, Singh R, Grigg JC, Murphy MEP, Bugg TDH, Eltis LD. Characterization of Dye-Decolorizing Peroxidases from Rhodococcus jostii RHA1. Bio Chemistry-Us 2011; 50:5108-19; http://dx.doi.org/10.1021/bi200427h
- Sainsbury PD, Hardiman EM, Ahmad M, Otani H, Seghezzi N, Eltis LD, Bugg TD. Breaking down lignin to high-value chemicals: the conversion of lignocellulose to vanillin in a gene deletion mutant of Rhodococcus jostii RHA1. ACS Chem Biol 2013; 8:2151-6; PMID:23898824; http://dx.doi.org/10.1021/cb400505a
- Baciocchi E, Fabbri C, Lanzalunga O. Lignin peroxidase-catalyzed oxidation of nonphenolic trimeric lignin model compounds: fragmentation reactions in the intermediate radical cations. J Org Chem 2003; 68:9061-9; PMID:14604381; http://dx.doi.org/10.1021/jo035052w
- Camarero S, Martinez MJ, Martinez AT. Understanding lignin biodegradation for the improved utilization of plant biomass in mod ern biorefineries. Biofuel Bioprod Bior 2014; 8:615-25; http://dx.doi.org/10.1002/bbb.1467
- Westereng B, Cannella D, Wittrup Agger J, Jorgensen H, Larsen Andersen M, Eijsink VG, Felby C. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci Rep 2015; 5:18561; PMID:26686263; http://dx.doi.org/10.1038/srep18561
- Barta K, Matson TD, Fettig ML, Scott SL, Iretskii AV, Ford PC. Catalytic disassembly of an organosolv lignin via hydrogen transfer from supercritical methanol. Green Chem 2010; 12:1640-7; http://dx.doi.org/10.1039/c0gc00181c
