?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Lipases can catalyze the hydrolysis of glycerol, esters and long chain fatty acids. A lipase producing isolate M35-15 was screened and identified as Thalassospira permensis using 16S rRNA gene sequence analysis. To our knowledge this is the first report on Thalassospira permensis producing lipases. In this paper the optimization of medium composition for the increase in bacterial lipase was achieved using statistical methods. Firstly the key ingredients were selected by Plackett-Burman experimental design, then the levels of the ingredients were optimized using central composite design of Response Surface Methodology. The predicted optimal lipase activity was 11.49 U under the conditions that medium composition were 5.15 g/l glucose, 11.74 g/l peptone, 6.74 g/l yeast powder and 22.90 g/l olive oil emulsifier.
Introduction
The deep sea covers 63.5% of the Earth's surface and supports the highest species diversity on Earth.Citation1,2 Because of exploration difficulties, there remain some environments the least studied and understood. The deep-sea bacteria exist in a cold environment with a constant temperature of 4-5 °Celsius below the depth of 1000 m. Therefore deep-sea bacteria considered to be unique and their metabolic enzymes provide potential for commercial development due to the high catalytic activity at low temperatures.
Lipases can catalyze the hydrolysis process of glycerol, esters and long chain fatty acids, and produced by many microorganisms, plants and animals.Citation3-5 Thalassospira are a genus created in 2002 and currently have 8 validly named species belonging to Rhodospirillaceae.Citation6-8 However, there was no report on Thalassospira permensis producing lipases to our knowledge.
Plackett-Burman experimental design and response surface methodology were widely used statistical methods to save time and number of tests for optimization of medium constituents. Central Composite Design was applied for determining the optimized levels of various ingredients with the interrelation between them.Citation9 The bacterial medium composition greatly influenced the lipase synthesis and its activity.Citation10-13 The objective of the study was to determine the medium composition of Thalassospira permensis M35-15 for maximum lipase production, using statistical designs of Plackett-Burman experimental design and central composite design of Response Surface Methodology.
Results and discussion
Isolation and screening of lipase producing microorganisms
Bacterial populations were successfully isolated from sea water and sediments samples using 2216E and R2A media, and the 3 study sites were in the South Atlantic Ocean. Three hundred ninety-one deep-sea bacteria were evaluated for their ability to produce lipases. Among the 391 strains 29.2% were isolated from sediments and 70.8% were isolated from the water column.
For the evaluation of lipase production on solid medium rhodamine B was used and the bacteria were cultivated at 28°C for 2 d. The orange fluorescent halo sizes surrounding the bacterial colonies correlated well with their lipase activities. Among the 391 bacteria, 88 strains (22.5%) showed lipase activity. Some deep-sea bacteria producing lipases have been reported in several papers.Citation14-16 According to a report of Odisi et al., among the 161 strains from sediment and water column in South Atlantic 14.3% strains exhibited lipolytic activities. The amount of lipolytic bacteria in sediment was larger than that in the water column.Citation17 The presence of lipolytic bacteria in the sediment could be explained by some kinds of fatty acids deposited on the sea floor.Citation18
The strain exhibited significant lipolytic activity and used in the optimization study was separated from sediment sample (−1523m), which collected from South Atlantic (W14.35°, S14.04°). It was identified as Thalassospira permensis M35-15 based on 16S rRNA gene sequence analysis (data not shown). Thalassospira are a genus created in 2002 and currently have 8 validly named species belonging to Rhodospirillaceae. To our knowledge there was no report on Thalassospira permensis producing lipases.
Optimization of bacterial medium composition
Since bacterial ability to produce lipases is strain dependent and the bacterial medium composition greatly influenced the lipase synthesis and its activity, optimization of medium composition for lipase production was performed in this study. Eight variables (glucose, fructose, yeast powder, peptone, ammonium chloride, ammonium sulfate, olive oil emulsifier and peanut oil emulsifier) were selected on the basis of its effect on the increase secretion of lipase using a 2-level Plackett-Burman experimental design.
The selected design matrix with 12 different medium composition and concentration was used to screen the significant variables, as shown in . The lipase activity results of the 12 experiments exhibited that run 6 was the best, and the variables showed significant influence were screened by analysis of test in .
Table 1. Plackett–Burman design matrix and lipase activity of Thalassospira permensis M35-15.
Table 2. Results of significant influence by analysis of t test using Plackett–Burman design.
The regression equation was calculated using Minitab16.0 software and exhibited as below:(1)
(1) The results showed that the most effective 3 factors in the production of lipase were olive oil emulsifier, yeast powder and glucose.
In this study, Central Composite Design was applied for determining the optimized levels of various ingredients (olive oil emulsifier, yeast powder, glucose and peptone) with the interrelation between them. The design matrix of 31 experiments with the 4 various ingredients at different concentration was shown in . The regression equation of the CCD design for T. permensis M35-15 lipase production was calculated using Minitab16.0 software and exhibited as below:(2)
(2)
Table 3. The design matrix of CCD with 4 various ingredients at different concentration.
The regression equation of the CCD design for T. permensis M35-15 lipase production was checked by F test. The ANOVA results calculated by Minitab16.0 were given in . P values and t values exhibited the significance of the CCD design for T. permensis M35-15 lipase production were shown in . The R2 result of 0.9769 showed the fit goodness of the equation.
Table 4. ANOVA results of the CCD design for T. permensis M35-15 lipase production calculated by Minitab16.0.
Table 5. Significance of the CCD design for T. permensis M35-15 lipase production.
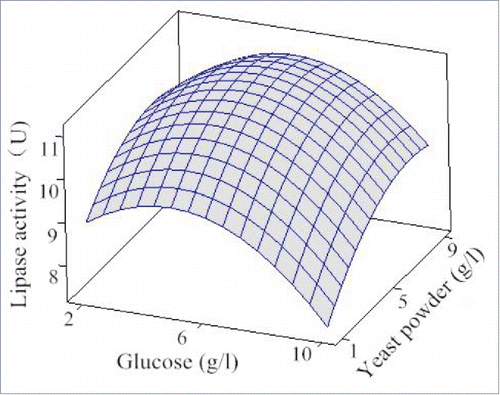
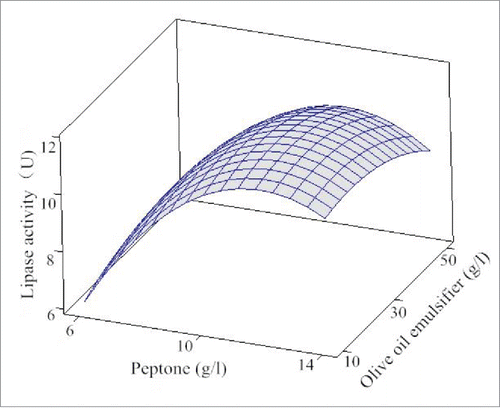
Three-dimensional response surfaces using Minitab 16.0 software were graphical results of the regression equation. The response surfaces were exhibited in to help visualize the effects of olive oil emulsifier, yeast powder, glucose and peptone on lipase activity. From , it is evident that the increase of glucose, yeast powder, peptone and olive oil emulsifier concentrations enhanced the lipase production, followed by a decrease in lipase production. The optimized results of the 4 various ingredients for obtaining the maximum lipase production calculated by Minitab16.0 software were 5.15 g/l glucose, 11.74 g/l peptone, 6.74 g/l yeast powder and 22.90 g/l olive oil emulsifier. In these conditions, the lipase activity was predicted to be 11.49 U for an increase of 1.85 times.
Materials and methods
Media composition
2216E medium (% w/v): yeast extract, 0.5; ferric citrate, 0.01; peptone, 0.5; NaCl, 1.945; MgCl2, 0.59; MgSO4 · 7H2O, 0.324; CaCl2 · 2H2O, 0.18; KCl, 0.055; NaHCO3, 0.016; KBr, 0.008; SrCl2, 3.4 mg; H3BO3, 2.2 mg; Na2SiO3·9H2O, 0.4 mg; NaF, 0.24 mg; NH4NO3, 0.16 mg; NaH2PO4, 0.8 mg; agar, 1.5; pH in nature.
R2A medium (% w/v Seawater): peptone, 0.05; yeast extract, 0.05; lactium, 0.05; dextrose, 0.05; amylogen, 0.05; Sodium pyruvate, 0.03; K2HPO4, 0.03; MgSO4 · 7H2O, 0.005; agar, 1.5; pH in nature.
Isolation and screening of lipase producing microorganisms
Deep-sea water and sediment samples were taken from different depths of the South Atlantic Ocean during No. Twenty-six ocean Cruise by R/V Dayang 1 in 2012. Samples were taken from the sediment and water column of about 3000m using SBE-911 plus CTD. The microorganisms from sediment and sea water samples were isolated using 2216E and R2A media. All plates were incubated at 20°C-22°C and bacterial strains were obtained for 3-7 d. Pure culture isolates were preserved for further research on lipase production screening.
The isolates were screened for the lipase production using rhodamine B agar plate method. Bacteria were spot inoculated onto rhodamine B medium (containing 3.0 g/l glucose, 10.0 g/l peptone, 5.0 g/l yeast powder, 0.5 g/l rhodamine B, 40.0 g/l olive oil emulsifier, 5.0 g/l NaCl and 15.0 g/l agar powder, pH 7.5-8.) and incubated at 28°C for 2 d. The lipolytic activities were determined by evaluating the size of the orange fluorescent halo surrounding the bacterial colonies. The orange fluorescent halo sizes correlated well with their lipase activities.
The bacterial isolates that exhibited obvious orange fluorescent halo were tested for lipase activity. Lipase activity was measured by titrimetric assay using an olive oil emulsifier as substrate.Citation19
Design of experiment
The optimization of medium composition to improve lipase activity of Thalassospira permensis M35-15 was carried out through Plackett-Burman experimental design and response surface methodology method. Eight variables including carbon sources, nitrogen sources, and lipase inducers were selected on the basis of its effect on the increase secretion of lipase using a 2-level Plackett-Burman experimental design, as shown in . The significance results of Plackett Burman experimental design were analyzed using Minitab16.0 software.
Table 6. Medium composition at different concentration for lipase secretion using Plackett-Burman experimental design.
The levels of the selected ingredients were optimized for maximum lipase production using central composite design of Response Surface Methodology. The four selected ingredients were glucose, peptone, yeast powder and olive oil emulsifier, and the 31 experiments were designed by Minitab16.0 software ().
Conclusions
Three hundred ninety-one deep-sea bacteria obtained from the South Atlantic Ocean were evaluated for their ability to produce lipases. Among them, 88 strains (22.5%) showed lipase activity. The strain M35-15 exhibited significant lipolytic activity was identified as Thalassospira permensis based on 16S rRNA gene sequence analysis.
The medium composition with increased production of lipase from T. permensis M35-15 was optimized using Plackett-Burman experimental design and central composite design. Maximum lipase activity was predicted to be 11.49 U for an increase of 1.85 times under the conditions of medium composition with 5.15 g/l glucose, 11.74 g/l peptone, 6.74 g/l yeast powder and 22.90 g/l olive oil emulsifier.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Supported by Foundation for Outstanding Young Scientist in Shandong Province (Grand No. BS2014NY012), COMRA project (No. DY125-15-R-01), China Postdoctoral Science Foundation (Grand No. 2015M581456) and Guidance Funds for Discipline Construction (Grand No. WH 20140202).
References
- Danovaro R, Company JB, Corinaldesi C, D'Onghia G, Galil B, Gambi C, Gooday AJ, Lampadariou N, Luna GM, Morigi C, et al. Deep-sea biodiversity in the Mediterranean Sea: the known, the unknown, and the unknowable. PLoS One 2010; 5:e11832; PMID:20689848; http://dx.doi.org/10.1371/journal.pone.0011832
- German CR, Ramirez-Llodra E, Baker MC, Tyler PA. The ChEss Scientific Steering Committee. Deep-water chemosynthetic ecosystem research during the census of marine life decade and beyond: a proposed deep-ocean road map. PLoS One 2011; 6(8):e23259; PMID:21829722; http://dx.doi.org/10.1371/journal.pone.0023259
- Gupta R, Gupta N, Rathi P. Bacterial lipases: an overview of production, purification and biochemical properties. Appl Microbiol Biotechnol 2004; 64:763-81; PMID:14966663; http://dx.doi.org/10.1007/s00253-004-1568-8
- Zhang J, Zeng R. Molecular cloning and expression of a cold-adapted lipase gene from an Antarctic deep sea psychrotrophic bacterium Pseudomonas sp. 7323. Mar Biotechnol 2008; 10(5):612-21; PMID:18461394; http://dx.doi.org/10.1007/s10126-008-9099-4
- Suzuki T, Nakayama T, Kurihara T, Nishino T, Esaki N. Cold-active lipolytic activity of psychrotrophic Acinetobacter sp. strain No. Six. J Bio Sci Bio Eng 2001; 92:144-8
- Lai Q, Liu Y, Yuan J, Du J, Wang L, Sun F, Shao Z. Multilocus sequence analysis for assessment of phylogenetic diversity and biogeography in Thalassospira bacteria from diverse marine environments. PLoS One 2014; 9(9):e106353; PMID:25198177; http://dx.doi.org/10.1371/journal.pone.0106353
- Lopez-Lopez A, Pujalte MJ, Benlloch S, Mata-Roig M, Rossello-Mora R, Garay E, Rodríguez-Valera F. Thalassospira lucentensis gen. nov., sp. nov., a new marine member of the α-Proteobacteria. Int J Syst Evol Microbiol 2002; 52:1277-83; PMID:12148640
- Kiseleva L, Garushyants SK, Briliute J, Simpson DJ, Cohen MF, Goryanin I. Genome sequence of the electrogenic petroleum-degrading Thalassospira sp. strain HJ. Genome Announc 2015; 3(3):e00483-15; PMID:25977412; http://dx.doi.org/10.1128/genomeA.00483-15
- Wang K, Yan P, Cao L. Chitinase from a Novel Strain of Serratia marcescens JPP1 for biocontrol of Aflatoxin: molecular characterization and production optimization using response surface methodology. BioMed Res Int 2014; 2014:482623; PMID:24812619
- Rajendran A, Thangavelu V. Optimization of medium composition for lipase production by Candida rugosa NCIM 3462 using response surface methodology. Can J Microbiol 2007; 53(5):643-55; PMID:17668023; http://dx.doi.org/10.1139/W07-017
- Yan J, Yan Y. Optimization for producing cell-bound lipase from Geotrichum sp. and synthesis of methyl oleate in microaqueous solvent. Appl Microbiol Biotechnol 2008; 78(3):431-9; PMID:18193214; http://dx.doi.org/10.1007/s00253-007-1331-z
- Sifour M, Zaghloul TI, Saeed HM, Berekaa MM, Abdel-Fattah YR. Enhanced production of lipase by the thermophilic Geobacillus stearothermophilus strain-5 using statistical experimental designs. N Biotechnol 2010; 27(4):330-6; PMID:20412872; http://dx.doi.org/10.1016/j.nbt.2010.04.004
- Salgado JM, Abrunhosa L, Venâncio A, Domínguez JM, Belo I. Integrated use of residues from olive mill and winery for lipase production by solid state fermentation with Aspergillus sp. Appl Biochem Biotechnol 2014; 172(4):1832-45; PMID:24276916; http://dx.doi.org/10.1007/s12010-013-0613-4
- Yu Y, Li HR, Zeng YX, Chen B. Bacterial Diversity and Bioprospecting for Cold-Active Hydrolytic Enzymes from Culturable Bacteria Associated with Sediment from Nella Fjord, Eastern Antarctica. Mar Drugs 2011; 9(2):184-95; PMID:21566794; http://dx.doi.org/10.3390/md9020184
- Sun QL, Wang MQ, Sun L. Characteristics of the cultivable bacteria from sediments associated with two deep-sea hydrothermal vents in Okinawa Trough. World J Microbiol Biotechnol 2015; 31(12):2025-37; PMID:26410427; http://dx.doi.org/10.1007/s11274-015-1953-8
- Alcaide M, Tchigvintsev A, Martínez-Martínez M, Popovicb A, Revac ON, Lafrayaa Á, Bargielaa R, Nechitaylod TY, Matesanze R, Cambon-Bonavitaf MA, et al. Identification and characterization of carboxyl esterases of gill chamber-associated microbiota in the deep-sea shrimp Rimicaris exoculata by using functional metagenomics. Appl Environ Microbiol 2015; 81(6):2125-36; PMID:25595762; http://dx.doi.org/10.1128/AEM.03387-14
- Odisi EJ, Silvestrin MB, Takahashi RYU, Castro da Silva MA, Lima AOS. Bioprospection of cellulolytic and lipolytic South Atlantic deep-sea bacteria. Electron J Biotechnol 2012; 15(5):18.
- Jørgensen BB, Boetius A. Feast and famine-microbial life in the deep-sea bed. Nat Rev Microbiol 2007; 5(10):770-81; PMID:17828281; http://dx.doi.org/10.1038/nrmicro1745
- Pignède G, Wang H, Fudalej F, Gaillardin C, Seman M, Nicaud JM. Characterization of an extracellular lipase encoded by LIP2 in Yarrowia lipolytica. J Bacteriol 2000; 182(10):2802-10; http://dx.doi.org/10.1128/JB.182.10.2802-2810.2000