ABSTRACT
Xanthine dehydrogenase (EC 1.17.1.4, XDH) is a typical and complex molybdenum-containing flavoprotein which has been extensively studied for over 110 years. This enzyme catalyzes the oxidation of purines, pterin and aldehydes with NAD+ or NADP+ as electron acceptor, and sometimes can be transformed to xanthine oxidase (EC 1.17.3.2, XOD) capable of utilizing oxygen as the electron acceptor. XDHs are widely distributed in all eukarya, bacteria and archaea domains, and are proposed to play significant roles in various cellular processes, including purine catabolism and production of reactive oxygen species (ROS) and nitric oxide (NO) in both physiological and pathological contexts. The recent applications of XDHs include clinical detections of xanthine and hypoxanthine content in body fluidics, and other diagnostic biomarkers like inorganic phosphorus, 5′-nucleotidase and adenosine deaminase. XDHs can also find applications in environmental degradation of pollutants like aldehydes and industrial application in nucleoside drugs like ribavirin. In this commentary, we would outline the latest knowledge on occurrence, structure, biosynthesis, and recent advances of production and applications of XDH, and highlighted the need to develop XDHs with improved performances by gene prospecting and protein engineering, and protocols for efficient production of active XDHs in response to the increasing demands.
Introduction
Xanthine dehydrogenase (EC 1.17.1.4, XDH) is an old typical and complex molybdenum-containing flavoenzyme, which has been extensively studied for its biochemical and structural properties ever since 1902.Citation1 It is well known that this oxidoreductase catalyzes the successive oxidation of hypoxanthine to xanthine and xanthine to uric acid with the concomitant reduction of NAD+ to NADH. In addition, XDH catalyzes the hydroxylation of sp2-hybridized carbon atoms of heterocyclic compounds including purines, pterins and aldehydes.Citation2 Some mammalian XDHs, like bovine milk XDH, can be converted from the dehydrogenase form into the xanthine oxidase (EC 1.17.3.2, XOD) form, a more convenient form capable of utilizing molecular oxygen in the air as the electron receptor.Citation3 Thus, XDHs have attracted wide attentions in physiological, clinical, environmental and industrial fields, such as clinical disease diagnosis, synthesis of nucleoside drugs, and detection and bioremediation of organic pollutants.Citation4,5
Since the first demonstration of the oxidation of formaldehyde by fresh milk XDH by Schardinger in 1902, dozens of XDHs have been characterized from eukaryotes and bacteria. Eukaryotic XDHs are originated from animals, including cow, chicken and rat, and from plants like Arabidopsis thaliana, and usually exist as α2 form.Citation6-9 Bacterial XDHs, including Rhodobacter capsulatus, Pseudomonas putida and Streptomyces cyanogenus, are found in the α2, α4, (αβ)2, (αβ)4 and αβγ forms.Citation10-14 XDH consists of 3 redox center domains, one of which contains 2 distinct iron-sulfur clusters ([2Fe-2S]), another that includes a flavin adenine dinucleotide (FAD), and a third that incorporates a sulfurated molybdenum cofactor (Moco). XDHs arrange in a highly similar way despite the differences in subunit composition, as disclosed by the structural analysis.Citation15,16 A few XDH genes have also been cloned and expressed in different hosts.Citation6,17,18 The bacterial XDHs usually show better catalytic activity and thermal stability than that of eukaryotic XDHs, however, both of them display optimum catalytic activity at nearly neutral pH and relatively low temperatures, and very limit pH-activity range, which limit their biotechnological applications (see the reviewCitation19).
In this addendum, we would describe the latest developments of occurrence, biosynthesis, structure, production, and the potential applications of the XDHs. We also have highlighted the need to develop XDHs with improved properties by gene prospecting and protein engineering, and protocols for efficient production of active XDH in response to the increasing demands.
Occurrence of XDHs and their homologous genes
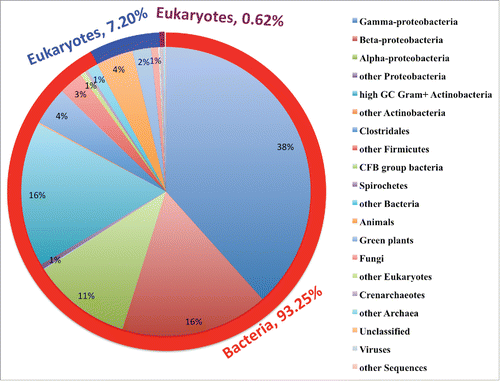
Bovine milk XDH is undoubtedly the first and best-characterized XDH so far, which has been reported as mostly pure form as early as 1939.Citation20 Since then, several eukaryotic XDHs have been purified and characterized from animals like human, rat, sheep, chicken and fruit fly.Citation21,22 In 1960, Bradshaw and Barker have reported the purification and characterization of the first bacterial XDHs from Clostridium cylindrosporum. Other bacterial XDHs, such as Micrococcus, Rhodobacter, Streptomyces genus, have also been purified and characterized.Citation23,24 With the progress of genome sequencing techniques, tens of thousands of new XDH genes have been disclosed. As of April 9, 2016, a total of 104, 303 XDH gene sequences have been deposited in the GenBank database. They are 96,077 bacterial XDHs (93.25%), 7513 eukaryotes XDHs (7.2%), 643 archaea XDHs (0.62%), 61 metagenomic ones (0.058%), 6 virus XDHs and 3 synthetic ones (). The top 3 subclasses (phyla) are the gamma-Proteobacteria (40045), β-Proteobacteria (17183) and high GC Gram+ Actinobacteria (16928). Up to now, only the bacteria and eukaryote XDHs are characterized experimentally, none of archaea and metagenomic XDHs have been identified experimentally. The only well-annotated virus XDH comes from Acinetobacter phage Ab105-3phi, and a homology search in GenBank database reveals 100% identity to the genomic XDH genes of Acinetobacter baumannii. It seems plausible that the similarity is a result of horizontal gene transfer. Further studies need to clarify the inheritance relationship between the XDHs of Acinetobacter genus and its virus in the future.
Figure 1. Distribution of 104, 303 XDH gene sequences retrieved from GenBank database as of April 9th, 2016. There are 96,077 bacterial XDHs (93.25%), 7513 eukaryotic XDHs (7.2%), 643 archaea XDHs (0.62%), 61 metagenomic XDHs (0.058%), 6 virus XDHs and 3 synthetic XDHs. In the bacteria domain, the dominant class is Proteobacteria, and the biggest subclass is the gamma Proteobacteria. This subclass consists mainly of the Pseudomonadales (18347) and Enterobacteria (18329) clusters, which are dominated by Pseudomonas genus (15570) and Escherichia genus (13258), respectively. In the eukaryotes domain, animals are the predominant contributor, which takes up 57.81% (4343). In the archaea domain, Sulfolobales genus with 335 species takes up the biggest sector (52.1%), of which Sulfolobus (278) is the majority genus. The number of the records in each class and its percentage that was rounded up are shown in each sector of the composite bar chart.
As shown in , pairwise alignments of primary sequences of XDH homologues reveal that: 1) eukaryotic XDHs share higher similarity and identity than those of bacteria, eubacteria and metagenomes; and 2) the [2Fe-2S] domainis are more conserved than the Moco domain, and FAD domain is the least conservative one. For example, the similarity and identity of [2Fe-2S] domain between the 9 XDH homologues representing the animal, plant, bacteria, archea and uncultured microorganisms distribute in a range of 70.1%–32.2% and 56.9%–22.4%, the Moco domain are 47.6%–34.7% and 47.6%–20.6%, and the FAD domain are only 41.3%–21.7% and 41.3%–9.9%, respectively (). Recently, we have firstly demonstrated a unique industrially applicable A.baumannii XDH, which shows only modest similarity to all the previous already-characterized XDHs.Citation19 This work may persuade the further exploitation of other low homologous XDHs, especially the never reported thermophiles, archaea and metagenomic ones.
Table 1. Amino acid sequence similarity (identity) of [2Fe-2S], FAD and Moco domains of 9 representative XDHs.
Structure of XDHs
As of April 9, 2016, only the crystal structure data of the bovine, rat, human and R. capsulatus XDHs are available from the PDB database.Citation25 The most-intensively studied structures are the bovine milk XDH and R. capsulatus XDH, which are the first and best resolved mammalian XDH and the only prokaryotic one, respectively. Compared to the bovine α2XDH, which consists of single polypeptide incorporating all the 3 redox center domains, the R. capsulatus α2β2XDH arranges the FAD and [2Fe-2S] domains and the Moco domain into 2 separate subunits. However, both the XDHs fold in a highly similar way, which lays the structural foundation for the same catalytic mechanism.Citation15,16
As shown in , multiple sequence alignment (see Supplementary Figs. S1, S2, and S3) of 9 representative XDH homologues reveals the conserved functional and structural features of primary sequences. First, [2Fe-2S] domain is the most conserved domain, and then comes the Moco domain, and the last the FAD domain. The two [2Fe-2S] cluster binding motifs: C-X4-C-X2-C-Xn-C and C-X2-C-Xn-C-X1-C, where X indicates any amino acid, and n means any number of amino acids, are completely identical (). This absolute conservation may provide the good explanation to the requirement of [2Fe-2S] cluster to transfer the electrons from the Moco domain to the FAD domain.
Figure 2. Graphical representation (sequence logo) of amino acid sequence conservation of Xanthine dehydrogenase [2Fe-2S] (A), FAD (B) and Moco domains (C) derived from a sequence alignment of 9 representative species. The species are 2 eukaryotes, Bos taurus and Arabidopsis thaliana, 3 eubacteria, which are Rhodobacter capsulatus B10, Acinetobacter baumannii CICC10254 and Escherichia coli K-12, one archaebacteria, Pyrobactulum neutrophilum V24Sta, and 3 metagenomes, which are uncultured Archaea, Leaf litter and marine bacteria. The multiple sequence alignment of the [2Fe-2S] domains correspond to the 1–166 fragment of B. taurus XDH (GI: 27806775), 1–237 fragment of A. thaliana XDH1 (GI: 332661034), 1–152 fragment of R. capsulatus B10XDHA (GI: 2956674), 1–176 fragment ofA. baumannii CICC10254 (GI: 966039133), 1–159 fragment of E. coli K-12 XDHC (GI: 16130770), 1–148 fragment of P. neutrophilum V24Sta (2Fe-2S) binding domain protein (GI: 170934475), 1–228 fragment of Leaf litter metagenomic XDH [2Fe-2S] binding domain (GI: 534502715), 1–169 fragment of Uncultured marine microorganism HF4000 XDH putative FAD binding domain (GI: 167041895), and 1–157 fragment of Uncultured Archaea metagenomic XDH [2Fe-2S] binding domain (GI: 452077216). The FAD domains correspond to the 228–541 fragment of B. taurus XDH (GI: 27806775), 256–566 fragment of A. thaliana XDH1 (GI: 332661034), 174–463 fragment of R. capsulatus B10XDHA (GI: 2956674), 198–499 fragment of A. baumannii CICC10254 XDHA (GI: 966039133), E. coli K-12 XDHB with a length of 292 AA (GI: 16130769), P. neutrophilum V24Sta FAD domain protein with a length of 272 AA (GI: 170934475), Leaf litter metagenomic XDH FAD domain with a length of 323 AA (GI: 534502714), 192–479 fragment of uncultured marine microorganism HF4000 XDH putative FAD binding domain (GI: 167041895), and 1–282 fragment of Uncultured Archaea metagenomic XDH FAD domain (GI: 452077215). The Moco domains are the 556–1332 fragment of B. taurus XDH (GI: 27806775), 581–1361 fragment of A. thaliana XDH1 (GI: 332661034), R. capsulatus B10XDHB with a length of 777 AA (GI: 13397863), A. baumannii CICC10254 XDHB with a length of 792 AA (GI: 966039134), E. coli K-12 XDHA with a length of 752 AA (GI: 16130768), P. neutrophilum V24Sta FAD domain protein with a length of 724 AA (GI: 170934473), Leaf litter metagenomic XDH FAD domain with a length of 734 AA (GI: 534502713), uncultured marine microorganism HF4000 XDH putative molybdopterin binding domain with a length of 764 AA (GI: 167041894), and uncultured Archaea metagenomic XDH molybdopterin binding subunit with a length of 780 AA (GI: 452077214). Each logo consists of stacks of symbols, one stack for each position in the sequence. The overall height of the stack indicates the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each amino or nucleic acid at that position. Amino acids are colored according to their chemical properties: polar amino acids (G,S,T,Y,Q,N) are black except the C which is red, basic (K,R,H) green, acidic (D,E) blue and hydrophobic (A,V,L,I,P,W,F,M) amino acids are purple. The most conserved amino acids in the [2Fe-2S], FAD and Moco domains are indicated by hexagram, triangles and diamonds, and pentagram, respectively. The eight identical cysteines consist of the 2 [2Fe-2S] cluster binding motifs of [2Fe-2S] domain: C-X4-C-X2-C-Xn-C and C-X2-C-Xn-C-X1-C, where X indicates any amino acid, and n means any number of amino acids. The triangles indicate the 4 identical amino acids residues in the 4 conserved FAD binding motifs, and the diamonds show the conservative residue sites that affect the FAD electrostatic environment. The red box shows the loop423–433 residues (numbering in bovine XDH) that dominate the electrostatic potential of FAD binding, resulting in different electron transfer efficiency from Fe/S cluster to NAD. The pentagram stars indicate the conserved active site catalytic residues. Using the bovine XDH numbering, Glu802 binds the substrate and stabilizes the transition state, Glu1261 is the catalytic base, Arg880 and Thr1010 bind the substrate and decrease the reaction activation energy, Phe914 and Phe1009 orientate the substrate via π–π stacking, Val1011 is the key residue channeling the substrate, and Gln758 is responsible for releasing the product.
![Figure 2. Graphical representation (sequence logo) of amino acid sequence conservation of Xanthine dehydrogenase [2Fe-2S] (A), FAD (B) and Moco domains (C) derived from a sequence alignment of 9 representative species. The species are 2 eukaryotes, Bos taurus and Arabidopsis thaliana, 3 eubacteria, which are Rhodobacter capsulatus B10, Acinetobacter baumannii CICC10254 and Escherichia coli K-12, one archaebacteria, Pyrobactulum neutrophilum V24Sta, and 3 metagenomes, which are uncultured Archaea, Leaf litter and marine bacteria. The multiple sequence alignment of the [2Fe-2S] domains correspond to the 1–166 fragment of B. taurus XDH (GI: 27806775), 1–237 fragment of A. thaliana XDH1 (GI: 332661034), 1–152 fragment of R. capsulatus B10XDHA (GI: 2956674), 1–176 fragment ofA. baumannii CICC10254 (GI: 966039133), 1–159 fragment of E. coli K-12 XDHC (GI: 16130770), 1–148 fragment of P. neutrophilum V24Sta (2Fe-2S) binding domain protein (GI: 170934475), 1–228 fragment of Leaf litter metagenomic XDH [2Fe-2S] binding domain (GI: 534502715), 1–169 fragment of Uncultured marine microorganism HF4000 XDH putative FAD binding domain (GI: 167041895), and 1–157 fragment of Uncultured Archaea metagenomic XDH [2Fe-2S] binding domain (GI: 452077216). The FAD domains correspond to the 228–541 fragment of B. taurus XDH (GI: 27806775), 256–566 fragment of A. thaliana XDH1 (GI: 332661034), 174–463 fragment of R. capsulatus B10XDHA (GI: 2956674), 198–499 fragment of A. baumannii CICC10254 XDHA (GI: 966039133), E. coli K-12 XDHB with a length of 292 AA (GI: 16130769), P. neutrophilum V24Sta FAD domain protein with a length of 272 AA (GI: 170934475), Leaf litter metagenomic XDH FAD domain with a length of 323 AA (GI: 534502714), 192–479 fragment of uncultured marine microorganism HF4000 XDH putative FAD binding domain (GI: 167041895), and 1–282 fragment of Uncultured Archaea metagenomic XDH FAD domain (GI: 452077215). The Moco domains are the 556–1332 fragment of B. taurus XDH (GI: 27806775), 581–1361 fragment of A. thaliana XDH1 (GI: 332661034), R. capsulatus B10XDHB with a length of 777 AA (GI: 13397863), A. baumannii CICC10254 XDHB with a length of 792 AA (GI: 966039134), E. coli K-12 XDHA with a length of 752 AA (GI: 16130768), P. neutrophilum V24Sta FAD domain protein with a length of 724 AA (GI: 170934473), Leaf litter metagenomic XDH FAD domain with a length of 734 AA (GI: 534502713), uncultured marine microorganism HF4000 XDH putative molybdopterin binding domain with a length of 764 AA (GI: 167041894), and uncultured Archaea metagenomic XDH molybdopterin binding subunit with a length of 780 AA (GI: 452077214). Each logo consists of stacks of symbols, one stack for each position in the sequence. The overall height of the stack indicates the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each amino or nucleic acid at that position. Amino acids are colored according to their chemical properties: polar amino acids (G,S,T,Y,Q,N) are black except the C which is red, basic (K,R,H) green, acidic (D,E) blue and hydrophobic (A,V,L,I,P,W,F,M) amino acids are purple. The most conserved amino acids in the [2Fe-2S], FAD and Moco domains are indicated by hexagram, triangles and diamonds, and pentagram, respectively. The eight identical cysteines consist of the 2 [2Fe-2S] cluster binding motifs of [2Fe-2S] domain: C-X4-C-X2-C-Xn-C and C-X2-C-Xn-C-X1-C, where X indicates any amino acid, and n means any number of amino acids. The triangles indicate the 4 identical amino acids residues in the 4 conserved FAD binding motifs, and the diamonds show the conservative residue sites that affect the FAD electrostatic environment. The red box shows the loop423–433 residues (numbering in bovine XDH) that dominate the electrostatic potential of FAD binding, resulting in different electron transfer efficiency from Fe/S cluster to NAD. The pentagram stars indicate the conserved active site catalytic residues. Using the bovine XDH numbering, Glu802 binds the substrate and stabilizes the transition state, Glu1261 is the catalytic base, Arg880 and Thr1010 bind the substrate and decrease the reaction activation energy, Phe914 and Phe1009 orientate the substrate via π–π stacking, Val1011 is the key residue channeling the substrate, and Gln758 is responsible for releasing the product.](/cms/asset/00f9daa6-cacb-4ea0-a147-922fb8683b29/kbie_a_1206168_f0002_oc.gif)
Second, as for the Moco domain, where the catalytic activity takes place, catalytic amino acid residues and the substrate-binding pocket region are very conserved (). Using bovine XDH numbering, Glu802 binds the substrate and stabilizes the transition state, Glu1261 is the catalytic base, Arg880 and Thr1010 bind the substrate and decrease the reaction activation energy, Phe914 and Phe1009 orientate the substrate via π–π stacking, Val1011 is the key residue channeling the substrate, and Gln758 is responsible for releasing the product. The results are in strong consistent with structural superimposition results revealed by a previous research.Citation19 There is an obvious variation of key residues channeling the substrate and binding pocket, which affect the substrate entry and product release, resulting in different catalytic activity and enzymatic properties. Surprisingly, the 2 pairs of cysteines, C535 and C992, and C1316 and C1324 numbering in bovine XDH, which are proposed to control the reversible post-translational conversion from XDH to XOD by forming 2 cysteine disulfide bonds, are totally absent in all other 8 XDHs, even the eukaryotic A. thaliana XDH1. This absence may be the reasons for the true XDH, like R. capsulatus and chicken XDH, that can't be converted to oxidase form by neither proteolytic cleavage nor oxidation of specific cysteine residues.Citation17
Third, as for the FAD domain, the FAD cofactor binding motifs are conserved in 4 regions each with an identical amino acid residue (). There is also a good conservation in the loop423–433 residues (numbering in bovine XDH), which dominates the electrostatic potential difference between the XDH and XO.Citation26 The conservations probably lay the foundation for the reduction of NAD by transferring the electron received from the Fe/S cluster to NAD.Citation15 The conservative substitutions in the above motifs except the identical sites, together with the variations in the loop423–433 determine the electron transfer efficiency from Fe/S cluster to NAD, bringing up the different enzymatic properties.Citation15,19
Enzymatic properties of XDHs
The majority of the characterized XDHs display optimum catalytic activity at neutral pH values (from 6.5 to 9.0) and relatively low temperatures (25°C–55°C), and narrow pH-activity and temperature-activity profiles, which can not meet the diversified requirements, such as very harsh operational conditions in biotechnological applications (see a review in the Supplementary Table 1 in referenceCitation19). Comparatively speaking, eukaryotic XDHs have preferable substrate affinity (lower Kmvalues), but the bacterial ones usually perform better activity, stability and broader ranges of pH-activity and temperature-activity relationships. The newly characterized A. baumannii XDH extends the pH tolerance to no less than pH 11.0, and the catalytic activity to the turnover number of 25 s−1 and catalytic efficiency of 2.74 μM−1.s−119. A. baumannii XDH exhibits very promising applicable properties, suggesting that bacteria could be used as a source for obtaining novel and useful XDHs. To satisfy the various specific biotechnological applications, like extreme pH values, it is urgent to develop more excellent XDHs with better catalytic activity, pH tolerance, thermo-tolerant performances. Further investigation of the newly annotated XDH genes in GenBank, especially those derived from extremophiles utilizing substrates including purines, pterines, and aldehydes, may potentially hit the right nail on the head. Meanwhile, it is also very promising to tailor the already existing efficient bacterial XDHs for specific properties, such as better substrate affinity, by using the protein engineering and directed evolution approaches, which have not been applied in XDHs except some mutational research for identifying the functional roles of specific active site residues.
Biosynthesis of XDHs and heterologous production
The biosynthesis of active XDH is a multistep process, which requires the synthesis and formation of multimeric apoprotein, and ordered incorporation of redox centers into the apoprotein.Citation17,27 Leimkuhler group has proposed a pioneering mechanism for the assembly and cofactor insertion of R. capsulatus (αβ)2XDH.Citation28 According to the mechanism, the biosynthesis of active XDH is proposed to involve 5 processes: 1) synthesis of apoproteins consisting of the 3 domains, depicted as S, M and L; 2) formation of heteromultimer apoprotein in the case of heteromultimer multi-subunit XDH; 3) insertion of 2 [2Fe-2S] clusters and a FAD into the S and M domains respectively; 4) dimerization of apoproteins assembled with [2Fe-2S] and FAD cofactors via the L domains; 5) insertion of sulfurated Moco into the L domains with the help of chaperone protein XdhC (). The Moco insertion is the last step to form active XDH and occurs after the formation of dimerization of apoproteins assembled with [2Fe-2S] and FAD cofactors. The sulfurated Moco is synthesized from molybdenum by a series of helper proteins, including molybdenum insertase, chaperone XdhC and Moco sulfurase (NifS4).Citation29 XdhC binds stoichiometric amount of Moco, interacts with NifS4 for the sulfuration of Moco, protects sulfurated Moco from oxidation, and further transfers to XDHCitation29 (). The efficient synthesis and insertion of sulfurated Moco is the bottleneck to produce the active XDHs. Some mammalian XDHs including bovine milk and rat liver can be converted reversibly into active XOD form by forming disulfide bond or irreversibly by limited proteolysisCitation3,30 (). These post-translational conversion steps become another hint for efficient production of active XOD.
Figure 3. Biosynthesis of XDH and its conversion to XOD. (I) Biosynthesis of sulfurated molybdenum cofactor. Mo (molybdenum) is inserted in the MPT (molybdopterin or metal-containing pterin) to form the molybdenumcofactor (Moco) by molybdenum insertase, like Cnx1 in plants, Geph in Humans, or MogA and MoeA in E. coli. Then, Moco is sulfurated by the Moco sulfurase, that is ABA3 in plants or NifS in E. coli. The chaperone XdhC binds stoichiometric amount of Moco, interacts with NifS4 for the sulfuration of Moco, protects sulfurated Moco from oxidation, and further transfers to XDH. (II) Biosynthesis of active XDH. It involves 5 processes: 1) synthesis of apoproteins consisting of the 3 domains, depicted as S, M and L; 2) formation of heteromultimer apoprotein in the case of multi-subunit heteromultimer XDH; 3) insertion of 2 [2Fe-2S] and a FAD cofactor into the S and M domains respectively; 4) dimerization of apoprotein assembled with [2Fe-2S] and a FAD cofactors via the L domain; 5) insertion of sulfurated Moco into the L subunits with the help of chaperone protein XdhC. The Moco insertion is the last step to form active XDH and occurs after the formation of dimerization of multi-domain apoprotein or heteromultimer assembled with [2Fe-2S] and FAD cofactors. (III) Conversion from active XDH to XOD. The active XDH is reversibly converted into XOD by forming 2 cysteine disulfide bonds that causes the conformational rearrangement around the FAD domain to prefer the oxygen as electron acceptor. The irreversible conversion is carried out by limited proteolysis via trypsin or chymotrypsin. The five arrows marked with numbers indicate the possible keys for efficient production of active XDH/XODs.
![Figure 3. Biosynthesis of XDH and its conversion to XOD. (I) Biosynthesis of sulfurated molybdenum cofactor. Mo (molybdenum) is inserted in the MPT (molybdopterin or metal-containing pterin) to form the molybdenumcofactor (Moco) by molybdenum insertase, like Cnx1 in plants, Geph in Humans, or MogA and MoeA in E. coli. Then, Moco is sulfurated by the Moco sulfurase, that is ABA3 in plants or NifS in E. coli. The chaperone XdhC binds stoichiometric amount of Moco, interacts with NifS4 for the sulfuration of Moco, protects sulfurated Moco from oxidation, and further transfers to XDH. (II) Biosynthesis of active XDH. It involves 5 processes: 1) synthesis of apoproteins consisting of the 3 domains, depicted as S, M and L; 2) formation of heteromultimer apoprotein in the case of multi-subunit heteromultimer XDH; 3) insertion of 2 [2Fe-2S] and a FAD cofactor into the S and M domains respectively; 4) dimerization of apoprotein assembled with [2Fe-2S] and a FAD cofactors via the L domain; 5) insertion of sulfurated Moco into the L subunits with the help of chaperone protein XdhC. The Moco insertion is the last step to form active XDH and occurs after the formation of dimerization of multi-domain apoprotein or heteromultimer assembled with [2Fe-2S] and FAD cofactors. (III) Conversion from active XDH to XOD. The active XDH is reversibly converted into XOD by forming 2 cysteine disulfide bonds that causes the conformational rearrangement around the FAD domain to prefer the oxygen as electron acceptor. The irreversible conversion is carried out by limited proteolysis via trypsin or chymotrypsin. The five arrows marked with numbers indicate the possible keys for efficient production of active XDH/XODs.](/cms/asset/b28f0e67-a4b0-4771-b1d7-1680a9247922/kbie_a_1206168_f0003_oc.gif)
The first commercial XDH product is the bovine milk XDH, which can be tracked to the 1970s and was extracted from bovine milk, and it is still the most important market product. Other available commercial XDHs include the E. cloacae XOD and A. luteus XOD, produced by Kikkoman Company and Toyobo Company, respectively (). However, all these commercial products are extracted from native host and usually mixed with a large quantity of inactive form XDHs, including desulfo-, and demolybdo-XORs,Citation21 because of the complex biosynthesis and multiple-step production processes. Several XDH genes have been cloned and expressed in exogenous hosts, which includes the rat liver XDH expression in insect cell system, A. thalianas XDH in Pichia pastoris, and R. capsulates and A. baumannii XDHs in E. coli.Citation6,17,18 A recombinant E. coli XDH can even be obtained from Sigma Company in the market. However, all the commercial XDHs are suffered in low catalytic activity and efficiency, in comparison to other industrial enzymes with hundreds of thousands of turnover numbers. There is an urgent need to improve the productivity of active XDH and XOD, it is worth trying to overexpress the helper proteins to assist the synthesis and assembly processes of active XDH and then its posttranslational modification to XOD.
Table 2. Comparison of some commercial XODs and 2 bacterial XDHs.
Physiological roles and applications of bacterial XDHs
The most recognized roles of XDH/XOD are key enzymes in catabolizing purines and other heterocyclic compounds with sp2-hybridized carbon atoms like aldehydes. The former directly involves the formation of uric acid and its abnormity becomes the main cause of gout, and the latter takes part in detoxification of drugs and harmful intermediate metabolites. In addition, XOD has been implicated as a key oxidative enzyme to produce superoxide radicals and hydrogen peroxide molecules, of which the increase could lead to oxidative stress damage, metabolic syndrome, cardiovascular diseases such as hypertension and ischemia reperfusion.Citation31 In recent years, the generation of reactive oxygen species (ROS) and nitric oxide (NO) in both physiological and pathological contexts like in ischemia-reperfusion injury has also been proposed.Citation21 However, the structural complexity and specialized tissue distribution and highly regulation of XDH/XOD suggest other not fully identified functions, which require additional work to elucidate. The extensively distributed bacterial XDHs in different habitants and the largely different activity and substrate specificity may also relate to some physiological roles in comparison to mammal XDH/XODs.
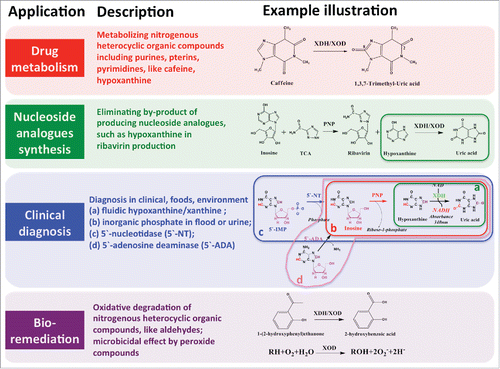
XDH finds use in diverse applications, which include: drug treatment of heterocyclic compounds such as caffeine and hypoxanthine, nucleoside analogs production by eliminating by-products, diagnosis of purines and purine-relating enzymes in clinical, foods and environment, and biological remediation (). Of these, an important commercialized industrial application of XDH/XOD is the production of ribavirin, which is an anti-viral drug retrieved on the World Health Organization's List of Essential Medicines, a list of the most important medication needed in a basic health system.Citation32 The timely removal of by-product of hypoxanthine by application of XDH/XOD shifts the reaction equilibrium to the direction of synthesising ribavirin, resulting in increased productivity.Citation33 Another inspiring applications are the clinical detections of xanthine and hypoxanthine content in body fluidics, and other disease biomarkers like inorganic phosphorus, 5′-nucleotidase and adenosine deaminase by coupling with purine-nucleoside phosphorylase and uricase. There must be promising prospects in the widely adopted XOD-based test kits markets for the true XDHs, like RcXDH and AbXDH, which selectively utilize NAD but not molecular oxygen as electron receptor and can eliminate the requirement of unmanageable oxygen in test samples in comparison to XOD.
Conclusions
Although the biosynthesis of active XDH and its conversion into XOD form are complex multistep processes and require several helper proteins, the XDHs exist ubiquitously in all organisms of 3 domains of life. The XDHs show great variability in gene sequence, protein sequence, and the subunit composition, but they fold in a highly similar way, which lays the structural foundation for the same catalytic mechanism. The wide range of enzymatic activities and properties of XDHs can be a great advantage of survival in many different niches including extreme habitats and an efficient adaptive mechanism, although their specific physiological and pathological roles require further investigation. During the past more than one hundred years of research history, the XDHs are proposed to play significant roles in various cellular processes, including purine catabolism and production of reactive oxygen species (ROS) and nitric oxide (NO) in both physiological and pathological contexts and show great applicable prospects in drug metabolism, industrial synthesis of nucleoside analogs, clinical diagnosis and biological remediation. However, there is still a great demand for better catalytic activity, stability and productivity of XDHs from the view-point of practical applications.
Future perspectives
As stated above, systematic research capable of linking discovery-engineering-production-application will promote the progress of XDHs. Some promising research aspects in the future include: 1) prospecting novel XDHs from extremophiles and metagenomes from various natural environments; 2) improving the existing enzymes by protein engineering, fusion protein or evolution approaches by combining the advanced high throughput technology; 3) designing and constructing engineered microbial cell factories for efficient XDH production with high activity and thermostability by overexpressing potential helper genes based on the complicated biosynthesis mechanism; 4) developing practical applications in diagnosis, detection, industrial biocatalysis and environmental remediation. The first future study will contribute to exploration of novel excellent enzymes more suitable for harsher operating environments with higher pH-stability, thermal stability and catalytic activity. This investigation will also help to understand the mechanisms responsible for the stability and activity of valuable XDHs in comparison with the 3 already well-characterized XDHs from bovine milk, R. capsulates and A. baumannii. The second research direction will evolve the already existing XDHs for improved stability, substrate affinity, activity and specificity to cater for the challenges to be solved by the first research direction. The commercially available XDHs and newly reported highly active AbXDH and RcXDH can be good starting materials. The third investigation may contribute to more efficient industrialized production of XDHs, which is an urgent task because the current inefficient extraction methods impede a variety of applications like industrial biocatalysis and environmental remediation due to the low efficiency and high cost. With these future researches on XDHs, it will be promising to expand their applications to in-situ enzymatic diagnosis and detection of target molecules such as purines and aldehydes in harsh environments using more sensitive but robust XDHs, and to detoxifying environmental pollutants like heterocyclic compounds including purines and aldehydes and synthesising nucleoside drugs like ribavirin in large scale.
Disclosure of potential conflicts of interest
No potential conflicts of interest are disclosed.
Supplementary_Materials_-_1206168.pdf
Download PDF (164.5 KB)Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant Number: 21406132), and the China Postdoctoral Science Foundation (Grant Numbers: 2014M550743 and 2015T80094).
References
- Schardinger F. Ueber das Verhalten der Kuhmilch gegen Methylenblau und seine Verwendung zur Unterscheidung von ungekochter und gekochter Milch. Z Unters Nahr Genussm 1902; 5:1113-21; http://dx.doi.org/10.1007/BF02506750
- Woolfolk CA, Downard JS. Distribution of Xanthine-oxidase and Xanthine dehydrogenase specificity types among bacteria. J Bacteriol 1977; 130:1175-91; PMID:863854
- Battelli MG, Lorenzoni E, Stripe F. Milk xanthine oxidase type D (dehydrogenase) and type O (oxidase). Purification, interconversion and some properties. Biochem J 1973; 131:191-8; PMID:4352904; http://dx.doi.org/10.1042/bj1310191
- Kalimuthu P, Leimkuhler S, Bernhardt PV. Low-potential amperometric enzyme biosensor for xanthine and hypoxanthine. Anal Chem 2012; 84:10359-65; PMID:23134312; http://dx.doi.org/10.1021/ac3025027
- Hille R. The mononuclear molybdenum enzymes. Chem Rev 1996; 96:2757-816; PMID:11848841; http://dx.doi.org/10.1021/cr950061t
- Zarepour M, Kaspari K, Stagge S, Rethmeier R, Mendel RR, Bittner F. Xanthine dehydrogenase AtXDH1 from Arabidopsis thaliana is a potent producer of superoxide anions via its NADH oxidase activity. Plant Mol Biol 2010; 72:301-10; PMID:19915948; http://dx.doi.org/10.1007/s11103-009-9570-2
- Nishino T, Okamoto K, Kawaguchi Y, Hori H, Matsumura T, Eger BT, Pai EF, Nishino T. Mechanism of the conversion of xanthine dehydrogenase to xanthine oxidase: identification of the two cysteine disulfide bonds and crystal structure of a non-convertible rat liver xanthine dehydrogenase mutant. J Biol Chem 2005; 280:24888-94; PMID:15878860; http://dx.doi.org/10.1074/jbc.M501830200
- Sato A, Nishino T, Noda K, Amaya Y, Nishino T. The structure of chicken liver xanthine dehydrogenase. cDNA cloning and the domain structure. J Biol Chem 1995; 270:2818-26; PMID:7852355; http://dx.doi.org/10.1074/jbc.270.6.2818
- Amaya Y, Yamazaki K, Sato M, Noda K, Nishino T, Nishino T. Proteolytic conversion of xanthine dehydrogenase from the NAD-dependent type to the O2-dependent type. Amino acid sequence of rat liver xanthine dehydrogenase and identification of the cleavage sites of the enzyme protein during irreversible conversion by trypsin. J Biol Chem 1990; 265:14170-5; PMID:2387845
- Gremer L, Meyer O. Characterization of xanthine dehydrogenase from the anaerobic bacterium Veillonella atypica and identification of a molybdopterin-cytosine-dinucleotide-containing molybdenum cofactor. Eur J Biochem / FEBS 1996; 238:862-6; PMID:8706691; http://dx.doi.org/10.1111/j.1432-1033.1996.0862w.x
- Aretz W, Kaspari H, Klemme JH. Molekulare und kinetische Charakterisierung der Xanthin-Dehydrogenase aus dem phototrophen Bakterium Rhodopseudomonas capsulata. Z Naturforsch 1981; 36:933-41.
- Ohe T, Watanabe Y. Purification and properties of xanthine dehydrogenase from Streptomyces cyanogenus. J Biochem 1979; 86:45-53; PMID:479130
- Sin IL. Purification and properties of xanthine dehydrogenase from Pseudomonas acidovorans. Biochimica et biophysica acta 1975; 410:12-20; PMID:1191668; http://dx.doi.org/10.1016/0005-2744(75)90203-X
- Hettrich D, Peschke B, Tshisuaka B, Lingens F. Microbial metabolism of quinoline and related compounds. X. The molybdopterin cofactors of quinoline oxidoreductases from Pseudomonas putida 86 and Rhodococcus spec. B1 and of xanthine dehydrogenase from Pseudomonas putida 86. Biol Chem Hoppe-Seyler 1991; 372:513-7; PMID:1657036; http://dx.doi.org/10.1515/bchm3.1991.372.2.513
- Enroth C, Eger BT, Okamoto K, Nishino T, Nishino T, Pai EF. Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion. Proc Natl Acad Sci U S A 2000; 97:10723-8; PMID:11005854; http://dx.doi.org/10.1073/pnas.97.20.10723
- Dietzel U, Kuper J, Doebbler JA, Schulte A, Truglio JJ, Leimkuhler S, Kisker C. Mechanism of Substrate and Inhibitor Binding of Rhodobacter capsulatus Xanthine Dehydrogenase. J Biol Chem 2009; 284:8768-76; PMID:19109249; http://dx.doi.org/10.1074/jbc.M808114200
- Leimkuhler S, Hodson R, George GN, Rajagopalan KV. Recombinant Rhodobacter capsulatus xanthine dehydrogenase, a useful model system for the characterization of protein variants leading to xanthinuria I in humans. J Biol Chem 2003; 278:20802-11; PMID:12670960; http://dx.doi.org/10.1074/jbc.M303091200
- Chen CG, Cheng GY, Hao HH, Dai MH, Wang X, Huang LL, Liu Z, Yuan Z. Mechanism of porcine liver Xanthine oxidoreductase mediated N-oxide reduction of cyadox as revealed by docking and mutagenesis studies. Plos One 2013; 8.
- Wang CH, Zhao TX, Li M, Zhang C, Xing XH. Characterization of a novel Acinetobacter baumannii xanthine dehydrogenase expressed in Escherichia coli. Biotechnol Lett 2016; 38:337-44; PMID:26543035; http://dx.doi.org/10.1007/s10529-015-1986-y
- Corran HS, Dewan JG, Gordon AH, Green DE. Xanthine oxidase and milk flavoprotein: With an Addendum by J. St L. Philpot. Biochem J 1939; 33:1694-708; PMID:16747086; http://dx.doi.org/10.1042/bj0331694
- Harrison R. Structure and function of xanthine oxidoreductase: where are we now? Free Radic Biol Med 2002; 33:774-97; PMID:12208366; http://dx.doi.org/10.1016/S0891-5849(02)00956-5
- Hughes RK, Bennett B, Bray RC. Xanthine dehydrogenase from Drosophila-melanogaster - purification and properties of the wild-type enzyme and of a variant lacking iron sulfur centers. Biochemistry 1992; 31:3073-83; PMID:1313286; http://dx.doi.org/10.1021/bi00127a007
- Smith ST, Rajagopa Kv, Handler P. Purification and properties of Xanthine dehydrogenase from micrococcus lactilytkicus. J Biol Chem 1967; 242:4108-&; PMID:6061702
- Wagner R, Cammack R, Andreesen JR. Purification and characterization of Xanthine dehydrogenase from clostridium-acidiurici grown in the presence of selenium. Biochimica et biophysica acta 1984; 791:63-74; http://dx.doi.org/10.1016/0167-4838(84)90282-6
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res 2000; 28:235-42; PMID:10592235; http://dx.doi.org/10.1093/nar/28.1.235
- Ishikita H, Eger BT, Okamoto K, Nishino T, Pai EF. Protein conformational gating of enzymatic activity in xanthine oxidoreductase. J Am Chem Soc 2012; 134:999-1009; PMID:22145797; http://dx.doi.org/10.1021/ja207173p
- Hille R. Structure and function of mononuclear molybdenum enzymes. J Biol Inorg Chem 1996; 1:397-404; http://dx.doi.org/10.1007/s007750050071
- Schumann S, Saggu M, Moller N, Anker SD, Lendzian F, Hildebrandt P, Leimkühler S. The mechanism of assembly and cofactor insertion into Rhodobacter capsulatus xanthine dehydrogenase. J Biol Chem 2008; 283:16602-11; PMID:18390908; http://dx.doi.org/10.1074/jbc.M709894200
- Neumann M, Stocklein W, Walburger A, Magalon A, Leimkuhler S. Identification of a Rhodobacter capsulatus L-cysteine desulfurase that sulfurates the molybdenum cofactor when bound to XdhC and before its insertion into xanthine dehydrogenase. Biochemistry 2007; 46:9586-95; PMID:17649978; http://dx.doi.org/10.1021/bi700630p
- Waud WR, Rajagopalan KV. The mechanism of conversion of rat liver xanthine dehydrogenase from an NAD+-dependent form (type D) to an O2-dependent form (type O). Arch Biochem Biophy 1976; 172:365-79; PMID:176940; http://dx.doi.org/10.1016/0003-9861(76)90088-6
- Kontos HA. Role of products of univalent reduction of oxygen in hypertensive vascular injury. Hypertension: pathophysiology, diagnosis and management. New York: Raven Press Ltd., 1990:667-75.
- World-Health-Organization. WHO model list of essential medicines 18th list [Internet]. 2013. Available from: http://www.who.int/medicines/publications/essentialmedicines/en/
- Hori N, Uehara K, Mikami Y. Effects of xanthine oxidase on synthesis of 5-methyluridine by the ribosyl transfer reaction. Agric Biol Chem 1991; 55:1071-4.
- Zikakis JP, Townsend D. Preparation of high purity xanthine oxidase from bovine milk. US: University of Delaware, 1979.
- Nakanishi T, Machida Y. Method and test composition for the determination of the substrate for xanthine oxidase. Japan: Kyowa Hakko Kogyo Co., Ltd., 1982.
- Tanigaki N, Furukawa K, Sogabe Y, Emi S. Thermostable xanthine oxidase from Arthrobacter luteus. Japan: Toyo Boseki Kabushiki Kaisha, 1993.