ABSTRACT
Schwann cells (SCs) have been reported as a possible source of neurotrophic support for spiral ganglion neurons (SGNs). This study was aimed to investigate whether S100A4 was contributed in the functional effects of SCs on SGNs. SCs were transfected with S100A4 vector or small interfering RNA (siRNA) against S100A4, and the transfection efficiency was verified by quantitative PCR (qPCR) and Western blot. The migration of transfected SCs was determined by Transwell assay, and the expression levels of vascular endothelial growth factor precursor (VEGF) and matrix metallopeptidase 9 (MMP-9) were measured by Western blot. Co-culture of either S100A4 overexpressed or suppressed SCs with SGNs, and the growth associated protein 43 (GAP43) expression in SGNs was detected by immunofluorescence (IF), qPCR and Western blot. The migration of SCs was significantly enhanced by S100A4 overexpression (P < 0.001), while was suppressed by S100A4 knockdown (P < 0.01). Further, the expressions of VEGF and MMP-9 were notably up-regulated by S100A4 overexpression, while were down-regulated by S100A4 knockdown. Moreover, co-culture with the S100A4 overexpressed SCs significantly increased the expression of GAP43 in SGNs (P < 0.01). As expected, co-culture with S100A4 knockdown SCs decreased GAP43 level (P < 0.05). S100A4 enhanced the migratory ability of SCs. SCs genetically modified to overexpress the S100A4 could up-regulate the GAP43 expression in SGNs.
Introduction
Schwann cells (SCs) originate from the neural crest are the predominant cell type constituting the structure of peripheral nerves.Citation1 SCs are implicated in many important aspects of peripheral nerve biology, including the conduction of nervous impulses, production of the nerve extracellular matrix, modulation of neuromuscular synaptic activity, and presentation of antigens to T-lymphocytes.Citation2-5 Besides, SCs can produce and secrete neurotrophic factors, especially brain-derived neurotrophic factor (BDNF) which supports and influences the growth and regenerative capacity of neurons.Citation6 Spiral ganglion neurons (SGNs) are located in Rosenthal's canal within the modiolus of the cochlea.Citation7 Maintenance of a robust SGNs population may improve the efficacy of the electrode-neural interface and enhance cochlear implant performance.Citation8 Previous studies have shown that BDNF secreted from SCs can improve SGNs survival and prevent degeneration in models of deafness.Citation9,10 In a word, SCs have an intimate relationship with SGNs.
S100A4, also known as mts1, a member of S100 family of transcription factors, locates in the 1q21 human chromosome region.Citation11 S100A4 has been reported as a vital regulator in modulating cell cycle, proliferation, apoptosis, migration of various types of cells through different mechanisms.Citation11,12 S100A4 is widely expressed in the nervous system, and it appears to be involved in the regulation of neuron survival plasticity, and responses to injury or disease.Citation13 Later studies have identified S100A4 as a neuroprotectant in peripheral nervous system.Citation14,15 However, little was known about the neuroprotective effects of S100A4 on SCs and SGNs.
In this study, rat SCs line RT4-D6P2T were employed and transfected with S100A4 vector or small interfering RNA (siRNA) against S100A4, to obtain S100A4 overexpressed or suppressed SCs. Further, the migratory ability of transfected cells was determined to reveal the effects of S100A4 on SCs migration. Next, SGNs were co-cultured either with S00A4 overexpressed or suppressed SCs, and the expression level of growth associated protein 43 (GAP43) in SGNs was detected, to investigate the role of S100A4 dysregulated SCs in SGNs. This study might add data on the molecular correlation of SCs with SGNs.
Materials and methods
SCs and primary SGNs culture
Rat SCs line RT4-D6P2T was purchased from the American Type Culture Collection (ATCC; Manassas, VA). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) high glucose (Life Technologies, Carlsbad), supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT) and were incubated at 37°C in a humidified atmosphere containing 5% CO2.Citation16
A total of 12 Sprague-Dawley rats (postnatal day 3–5) were provided by the Animal Center of the Academy of Military Science of the Chinese PLA. Cochleae were isolated after rats were decapitated, and SGNs were dissociated as previous described.Citation17 The SGNs were cultivated in DMEM high glucose (Life Technologies), 25 mM HEPES (Life Technologies), 30 U/mL penicillin (Grünenthal GmbH, Aachen), 3 μL/mL N2 supplement (Life Technologies) and 5 μg/mL insulin (Sigma-Aldrich, St. Louis).Citation18 SGNs were incubated in a humidified atmosphere with 5% CO2 at 37°C. The mere killing of rats for tissue analyses was approved by our local ethics committee. All procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Precautions were taken to minimize suffering and the number of animals used in each experiment.
Cell transfection
SCs were seeded in 6-well plates at a density of 2 × 10Citation5. After culture for 24 h, cells were divided into 3 groups, namely control, pc S100A4 and si S100A4. A S100A4 expression vector, i.e., pc S100A4, was constructed by sub-cloning the full-length S100A4 coding sequence into pcDNA3.1 (Sangon Biotech, Shanghai, China), and the vector was transfected into cells in pc S100A4 group. Cells in si S100A4 group were transfected with siRNA against S100A4 (RiboBio, Guangzhou, China). All transfections were performed by using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's protocol. At 48 h after transfection, the cells were prepared for further analyses.
Real-time quantitative PCR (qPCR)
After transfection, cells were collected and total RNA in cells were extracted by using TRIzol (Invitrogen). DNaseI (Invitrogen) was used for removing the DNA contamination. Afterward, 1 μg RNAs of each sample were used for cDNA synthesis by using Transcriptor First Strand cDNA Synthesis Kit (Roche, USA), according to the manufacturer's instructions. qPCR was carried out for a total of 20 μL reaction mixtures on an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using FastSTART Universal SYBR Green Master (ROX) (Roche, USA).Citation18 Data were analyzed according to 2−ΔΔCt method and were normalized to GAPDH expression in each sample. All primers were synthesized by GenePharma (Shanghai, China).
Western blotting
After transfection, cells were collected and cellular protein was extracted by RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China). Protein concentration was quantified using BCA™ Protein Assay Kit (Pierce, Appleton, WI, USA), according to the manufacturer's recommendations. Equal amounts of proteins were separated on a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% skim milk for 1 h, and then probed with primary antibodies: S100A4 (sc-292281), vascular endothelial growth factor precursor (VEGF; sc-7269), matrix metallopeptidase 9 (MMP-9; sc-21733), GAP43 (sc-17790) or GAPDH (sc-365062; Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C, overnight. Afterward, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies. The bands were visualized by the ECL Plus Western Blotting Substrate (Thermo Scientific).Citation19
Cell migration assay
Cell migration was evaluated by Transwell assay using the 8 μm-pore Transwell migration chambers (Corning, USA). Briefly, transfected cells were seeded in the upper portion of a chamber with serum-free medium. Complete medium filled in the lower chamber as an attractant. At 48 h after incubation, the non-migrated cells in the upper chamber were removed by cotton swabs. The migrated cells were stained with 0.1% crystal violet (Beyotime, Nantong, China) for 30 min, and then photographed and counted under optical microscope (Olympus, Tokyo, Japan).Citation19
Immunofluorescence (IF)
SGN and SCs were co-cultured as previous described.Citation8 Briefly, SGN were plated onto 8-well chamber slides (Nalge Nunc International, Rochester, NY, USA) at a density of 2 × 10Citation4 cells/well. The transfected SCs were collected and added to the SGN at 2 × 10Citation4 cells/well. After 24 h of incubation in DMEM high glucose with 10% FBS, cells were washed with phosphate-buffered saline (PBS) 3 times and fixed with 4% formaldehyde for 20 min on ice. Cells were then permeabilized with 1% Triton X-100 for 25 min, and blocked in 10% goat serum (Beyotime) for 30 min at 37°C. The cells were incubated with the primary antibody GAP43 (sc-17790, Santa Cruz Biotechnology) overnight at 4°C, and followed by incubation with fluorescent secondary antibody for 1 h at room temperature. Cell nuclei were stain for 5 min by using 4′, 6-diamidino-2-phenylindole (DAPI; Beyotime). The fluorescent images were obtained using an automated upright microscope system (Leica, DM4000B Leica Microsystems, Wetzlar, Germany).Citation20
Statistical analysis
Data in histograms were from 3 independent experiments in triplicate and were presented as the mean ± standard deviation (SD). All other data were displayed from one assaying only. Statistical analyses were performed using GraphPad Prism 5 software (GraphPad, San Diego, CA), and statistical significance between different groups was analyzed by Student's t tests. A P-value of < 0.05 was considered as statistical significance.
Results
Effects of transfection on SCs
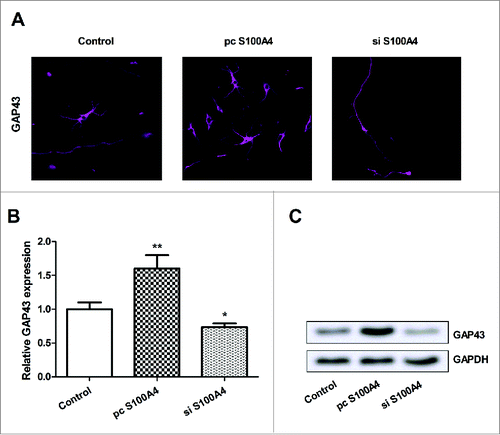
SCs were transfected with pc S100A4 or si S100A4, and then the transfection efficiency was verified in vitro. The morphology of transfected SCs observed from a light-microscope shown that (), there was no notable difference between the control group and transfection group. Subsequently, the protein and mRNA level expression of S100A4 in cells were detected by qPCR and Western blot analysis. As results showed in and 1C, both the protein and mRNA levels of S100A4 were elevated by pc S100A4 (P < 0.01), while were suppressed by si S100A4 (P < 0.01). Thus, the overexpressed and suppressed S100A4 SCs were successfully obtained, and were used for the forthcoming analyses.
Figure 1. Effects of transfection on SCs. SCs were transfected with S100A4 vector or siRNA against S100A4. (A) The morphology of the transfected SCs was observed and photographed under a light-microscope. (B) The mRNA level expression of S100A4 in transfected SCs was determined by qPCR. (C) The protein level of S100A4 was detected by Western blotting. SCs, Schwann cells; siRNA, small interfering RNA; qPCR, quantitative PCR; **, P < 0.01.
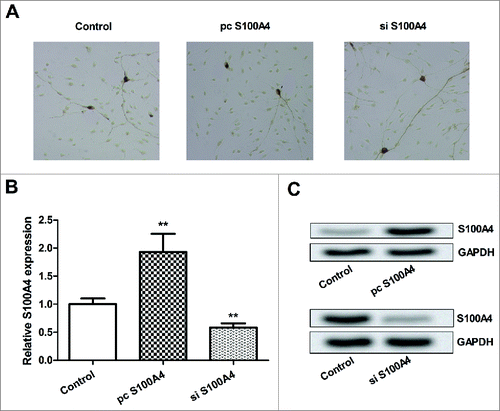
Effects of S100A4 on SCs migration
To explore the functional effects of S100A4 on SCs, the migration of transfected SCs was measured by Transwell assay. Results in showed that, cell migration was significantly enhanced by S100A4 overexpression (P < 0.001), while was inhibited by S100A4 suppression (P < 0.01). Furthermore, the protein expressions of VEGF and MMP-9 in transfected SCs were detected by Western blotting, to reveal the underling mechanism of S100A4 on SCs migration. We found that, both VEGF and MMP-9 were upregulated by S100A4 overexpression, whereas were downregulated by S100A4 suppression. Taken together, we inferred that S100A4 improved SCs migration might be via regulating VEGF and MMP-9.
Figure 2. Effects S100A4 on SCs migration. SCs were transfected with S100A4 vector or siRNA against S100A4. (A) SCs migration was measured by Tranwell assay, and the migrated SCs were stained with crystal violet and photographed under a light-microscope. (B) Quantification of migrated SCs. (C) Protein expressions of VEGF and MMP-9 in transfected SCs were measured by Western blot. SCs, Schwann cells; siRNA, small interfering RNA; VEGF, vascular endothelial growth factor precursor; MMP-9, matrix metallopeptidase 9; **, P < 0.01; ***, P < 0.001.
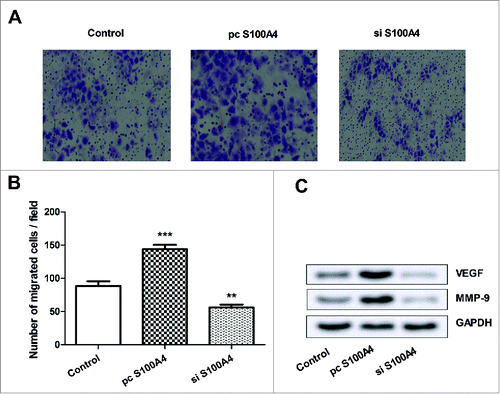
Effects of dysregulated S100A4 SCs on the expression of GAP43 in SGNs
To explore the effects of the dysregulated S100A4 SCs on SGNs, SCs were firstly transfected with pc S100A4 or si S100A4, and co-cultured with SGNs. GAP43 in SGNs was probed by IF and its expression was determined by qPCR and Western blot analysis. As results showed in , the expression of GAP43 in SGNs was remarkably elevated by S100A4 overexpressed SCs, while was decreased by S100A4 suppressed SCs. In line with the results in , showed that, both the protein and mRNA levels of GAP43 were up-regulated by S100A4 overexpressed SCs (P < 0.01), whereas were down-regulated by S100A4 suppressed SCs (P < 0.05). These data suggested that, the expression of S100A4 in SCs might play a pivotal role in GAP43 expression in SGNs.
Figure 3. Effects of dysregulated S100A4 SCs on the expression of GAP43 in SGNs. SGNs were co-cultured either with S100A4 overexpressed or suppressed SCs. (A) GAP43 in SGNs was probed by IF and observe under an automated upright microscope system. (B) The mRNA level expression of GAP43 in SGNs was detected by qPCR. (C) The protein expression of GAP43 was detected by Western blot. SCs, Schwann cells; SGNs, spiral ganglion neurons; GAP43, growth associated protein 43; IF, immunofluorescence; qPCR, quantitative PCR; *, P < 0.05; **, P < 0.01.
Discussion
SCs have an intimate relationship with SGNs with a number of important functions. S100A4 is widely expressed in the nervous system, and it appears to be involved in the regulation of neuron survival plasticity. However, few investigations have focus on the role of S100A4 in SCs and SGNs. In this study, SCs were genetically modified to overexpress or suppress S100A4. We found that S100A4 overexpression enhanced the migratory ability of SCs, while S100A4 suppression reduced SCs migration. Additionally, the expression levels of VEGF and MMP-9 proteins were upregulated by S100A4 overexpression, while were downregulated by S100A4 suppression. More important, co-culture of S100A4 overexpressed SCs with SGNs remarkably up-regulated the expression of GAP43 in SGNs. However, the level of GAP43 was down-regulated after SGNs were co-cultured with S100A4 suppressed SCs.
SCs migration is a pivotal step preceding myelination and remyelination in the peripheral nervous system.Citation21 Biochemical reports also demonstrated that SCs could gradually migrate to the peripheral nerve injured site and provide supportive effects to proximal axons which promotes successive neuro-regeneration by expressing a variety of trophic factors.Citation22 Nowadays, studies have illuminated that multiple factors, such as concentrated growth factor (CGF), NMDA receptors and Netrin-1, have the capacity of improving SCs migration.Citation23-25 However, our study provided the first evidence that S100A4 could enhance SCs migration. VEGF is a potent soluble growth factor, which plays a key role in regulating angiogenesis.Citation26 Accordingly, an increasing number of studies evidenced VEGF could modify the migration of neurons and glia.Citation27 MMP-9 is an intriguing MMP family member found in adult nerve only after injury and predominantly in SCs.Citation28 MMP-9 was also reported as a pivotal regulator in SCs migration.Citation29 In the current study, both VEGF and MMP-9 were upregulated by S100A4 overexpression in SCs, implying that S100A4 enhanced SCs migration at least partly via modulation these 2 factors.
GAP43, a protein kinase C substrate, is a member of the calmodulin-binding protein family and concentrates in the presynaptic membrane and growth cones.Citation30 GAP43 is enriched in neuronal growth cone and seems to play an important role in neurotransmitter vesicle fusion and recycling, long-term potentiation, spatial memory formation and learning.Citation31 Moreover, GAP43 is involved in the damage repair and regeneration of SGNs. In the rat, loss of spiral ganglion caused a substantial re-emergence of GAP43 immunoreactivity in varicose fibers of the ipsilateral ventral cochlear nucleus and cell bodies of the lateral superior olive.Citation32 Up-regulation of GAP43 in SGNs might indicate neuronal regeneration and plasticity.Citation7 Interestingly, in the present study, the expression of GAP43 in SGNs was elevated by co-culturing with S100A4 overexpressed SCs, indicating that S100A4 was implicated in the role of SCs in SGNs regeneration.
In summary, this study demonstrated that SCs genetically modified to overexpress the S100A4 could up-regulate the expression of GAP43 in SGNs in vitro. Transplantation of S100A4 overexpressed SCs into the cochlea may provide a clinically relevant means of preventing SGNs degeneration. Nevertheless, further research is still needed to fully confirming this hypothesis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The work is supported by a grant from Surface Project of Beijing Natural Science Foundation (7132109).
References
- Fu X, Tong Z, Li Q, Niu Q, Zhang Z, Tong X, Tong L, Zhang X. Induction of adipose-derived stem cells into Schwann-like cells and observation of Schwann-like cell proliferation. Mol Med Rep 2016; 14:1187-93; PMID:27279556
- Pinzon A, Calancie B, Oudega M, Noga BR. Conduction of impulses by axons regenerated in a Schwann cell graft in the transected adult rat thoracic spinal cord. J Neurosci Res 2001; 64:533-41; PMID:11391708; https://doi.org/10.1002/jnr.1105
- Chernousov MA, Carey DJ. Schwann cell extracellular matrix molecules and their receptors. Histol Histopathol 2000; 15:593-601; PMID:10809381
- Auld DS, Robitaille R. Perisynaptic Schwann cells at the neuromuscular junction: nerve- and activity-dependent contributions to synaptic efficacy, plasticity, and reinnervation. Neuroscientist 2003; 9:144-57; PMID:12708618; https://doi.org/10.1177/1073858403252229
- Argall KG, Armati PJ, Pollard JD, Watson E, Bonner J. Interactions between CD4+ T-cells and rat Schwann cells in vitro. One. Antigen presentation by Lewis rat Schwann cells to P2-specific CD4+ T-cell lines. J Neuroimmunol 1992; 40:1-18; PMID:1381378; https://doi.org/10.1016/0165-5728(92)90208-3
- Yi S, Yuan Y, Chen Q, Wang X, Gong L, Liu J, Gu X, Li S. Regulation of Schwann cell proliferation and migration by miR-1 targeting brain-derived neurotrophic factor after peripheral nerve injury. Sci Rep 2016; 6:29121; PMID:27381812; https://doi.org/10.1038/srep29121
- Dodson HC, Mohuiddin A. Response of spiral ganglion neurones to cochlear hair cell destruction in the guinea pig. J Neurocytol 2000; 29:525-37; PMID:11279367; https://doi.org/10.1023/A:1007201913730
- Pettingill LN, Minter RL, Shepherd RK. Schwann cells genetically modified to express neurotrophins promote spiral ganglion neuron survival in vitro. Neuroscience 2008; 152:821-8; PMID:18304740; https://doi.org/10.1016/j.neuroscience.2007.11.057
- Gillespie LN, Clark GM, Bartlett PF, Marzella PL. BDNF-induced survival of auditory neurons in vivo: Cessation of treatment leads to accelerated loss of survival effects. J Neurosci Res 2003; 71:785-90; PMID:12605404; https://doi.org/10.1002/jnr.10542
- Miller JM, Chi DH, O'Keeffe LJ, Kruszka P, Raphael Y, Altschuler RA. Neurotrophins can enhance spiral ganglion cell survival after inner hair cell loss. Int J Dev Neurosci 1997; 15:631-43; PMID:9263039; https://doi.org/10.1016/S0736-5748(96)00117-7
- Qi R, Qiao T, Zhuang X. Small interfering RNA targeting S100A4 sensitizes non-small-cell lung cancer cells (A549) to radiation treatment. Onco Targets Ther 2016; 9:3753-62; PMID:27382312; https://doi.org/10.2147/OTT.S106557
- Tahara S, Nojima S, Ohshima K, Hori Y, Kurashige M, Wada N, Ikeda JI, Morii E. S100A4 accelerates the proliferation and invasion of endometrioid carcinoma and is associated with the ‘MELF’ pattern. Cancer Sci 2016; 107(9):1345-52
- Sandelin M, Zabihi S, Liu L, Wicher G, Kozlova EN. Metastasis-associated S100A4 (Mts1) protein is expressed in subpopulations of sensory and autonomic neurons and in Schwann cells of the adult rat. J Comp Neurol 2004; 473:233-43; PMID:15101091; https://doi.org/10.1002/cne.20115
- Dmytriyeva O, Pankratova S, Owczarek S, Sonn K, Soroka V, Ridley CM, Marsolais A, Lopez-Hoyos M, Ambartsumian N, Lukanidin E, et al. The metastasis-promoting S100A4 protein confers neuroprotection in brain injury. Nat Commun 2012; 3:1197; PMID:23149742; https://doi.org/10.1038/ncomms2202
- Moldovan M, Pinchenko V, Dmytriyeva O, Pankratova S, Fugleholm K, Klingelhofer J, Bock E, Berezin V, Krarup C, Kiryushko D. Peptide mimetic of the S100A4 protein modulates peripheral nerve regeneration and attenuates the progression of neuropathy in myelin protein P0 null mice. Mol Med 2013; 19:43-53; PMID:23508572
- Gupta D, Venugopal J, Prabhakaran MP, Dev VR, Low S, Choon AT, Ramakrishna S. Aligned and random nanofibrous substrate for the in vitro culture of Schwann cells for neural tissue engineering. Acta Biomater 2009; 5:2560-9; PMID:19269270; https://doi.org/10.1016/j.actbio.2009.01.039
- Warnecke A, Sasse S, Wenzel GI, Hoffmann A, Gross G, Paasche G, Scheper V, Reich U, Esser KH, Lenarz T, et al. Stable release of BDNF from the fibroblast cell line NIH3T3 grown on silicone elastomers enhances survival of spiral ganglion cells in vitro and in vivo. Hearing Res 2012; 289:86-97; PMID:22564255; https://doi.org/10.1016/j.heares.2012.04.007
- Bird CD, Emery NJ. Insightful problem solving and creative tool modification by captive nontool-using rooks. Proc Natl Acad Sci U S A 2009; 106:10370-5; PMID:19478068; https://doi.org/10.1073/pnas.0901008106
- Chiappetta G, Valentino T, Vitiello M, Pasquinelli R, Monaco M, Palma G, Sepe R, Luciano A, Pallante P, Palmieri D, et al. PATZ1 acts as a tumor suppressor in thyroid cancer via targeting p53-dependent genes involved in EMT and cell migration. Oncotarget 2015; 6:5310-23; PMID:25595894; https://doi.org/10.18632/oncotarget.2776
- Mani NL, Schalper KA, Hatzis C, Saglam O, Tavassoli F, Butler M, Chagpar AB, Pusztai L, Rimm DL. Quantitative assessment of the spatial heterogeneity of tumor-infiltrating lymphocytes in breast cancer. Breast Cancer Res 2016; 18:78; PMID:27473061; https://doi.org/10.1186/s13058-016-0737-x
- Anliker B, Choi JW, Lin ME, Gardell SE, Rivera RR, Kennedy G, Chun J. Lysophosphatidic acid (LPA) and its receptor, LPA1, influence embryonic schwann cell migration, myelination, and cell-to-axon segregation. Glia 2013; 61:2009-22; PMID:24115248; https://doi.org/10.1002/glia.22572
- Chang HM, Shyu MK, Tseng GF, Liu CH, Chang HS, Lan CT, Hsu WM, Liao WC. Neuregulin facilitates nerve regeneration by speeding Schwann cell migration via ErbB2/3-dependent FAK pathway. PLoS One 2013; 8:e53444; PMID:23301073; https://doi.org/10.1371/journal.pone.0053444
- Qin J, Wang L, Zheng L, Zhou X, Zhang Y, Yang T, Zhou Y. Concentrated growth factor promotes Schwann cell migration partly through the integrin beta1-mediated activation of the focal adhesion kinase pathway. Int J Mol Med 2016; 37:1363-70; PMID:26986804
- Mantuano E, Lam MS, Shibayama M, Campana WM, Gonias SL. The NMDA receptor functions independently and as an LRP1 co-receptor to promote Schwann cell survival and migration. J Cell Sci 2015; 128:3478-88; PMID:26272917; https://doi.org/10.1242/jcs.173765
- Lv J, Sun X, Ma J, Ma X, Zhang Y, Li F, Li Y, Zhao Z. Netrin-1 induces the migration of Schwann cells via p38 MAPK and PI3K-Akt signaling pathway mediated by the UNC5B receptor. Biochem Biophys Res Commun 2015; 464:263-8; PMID:26116534; https://doi.org/10.1016/j.bbrc.2015.06.140
- Dimmeler S, Dernbach E, Zeiher AM. Phosphorylation of the endothelial nitric oxide synthase at ser-1177 is required for VEGF-induced endothelial cell migration. FEBS Lett 2000; 477:258-62; PMID:10908731; https://doi.org/10.1016/S0014-5793(00)01657-4
- Rosenstein JM, Krum JM, Ruhrberg C. VEGF in the nervous system. Organogenesis 2010; 6:107-14; PMID:20885857; https://doi.org/10.4161/org.6.2.11687
- Chattopadhyay S, Shubayev VI. MMP-9 controls Schwann cell proliferation and phenotypic remodeling via IGF-1 and ErbB receptor-mediated activation of MEK/ERK pathway. Glia 2009; 57:1316-25; PMID:19229995; https://doi.org/10.1002/glia.20851
- Mantuano E, Inoue G, Li X, Takahashi K, Gaultier A, Gonias SL, Campana WM. The hemopexin domain of matrix metalloproteinase-9 activates cell signaling and promotes migration of schwann cells by binding to low-density lipoprotein receptor-related protein. J Neurosci 2008; 28:11571-82; PMID:18987193; https://doi.org/10.1523/JNEUROSCI.3053-08.2008
- Basi GS, Jacobson RD, Virag I, Schilling J, Skene JH. Primary structure and transcriptional regulation of GAP-43, a protein associated with nerve growth. Cell 1987; 49:785-91; PMID:3581170; https://doi.org/10.1016/0092-8674(87)90616-7
- Flamm AG, Zerko S, Zawadzka-Kazimierczuk A, Kozminski W, Konrat R, Coudevylle N. 1H, 15N, 13C resonance assignment of human GAP-43. Biomol NMR Assign 2016; 10:171-4; PMID:26748655; https://doi.org/10.1007/s12104-015-9660-9
- Illing RB, Horvath M. Re-emergence of GAP-43 in cochlear nucleus and superior olive following cochlear ablation in the rat. Neurosci Lett 1995; 194:9-12; PMID:7478222; https://doi.org/10.1016/0304-3940(95)11706-3
