ABSTRACT
L-Isoleucine dioxygenase (IDO) specifically converts L-isoleucine(L-Ile) to 4-hydroxyisoleucine(4-HIL). To obtain IDO with improved activity, a strategy was developed that is dependent on the restoration of succinate-minus E. coli cell growth by the coupling of L-Ile hydroxylation and the oxidation of α-ketoglutarate(α-KGA) to succinate. Five mutants were obtained with this strategy, and the characteristics of IDOM3, which exhibited the highest activity, were studied. The catalytic efficiency, thermal stability and catalytic rate of IDOM3 were significantly improved compared with those of wild-type IDO. Moreover, an efficient method for the biotransformation of 4-HIL by resting cells expressing IDOM3 was developed, with which 151.9 mmol of 4-HIL/L (22.4 g/L) was synthesized in 12 h while the substrates seldom exhibited additional consumption.
Introduction
4-Hydroxyisoleucine(4-HIL) is a natural nonproteinogenic amino acid that was first isolated from the seeds of Trigonella foenum-graecum and has been proven to exhibit glucose-dependent insulinotropic activity in rat models of type 2 diabetes mellitus(T2DM). Moreover, 4-HIL does not induce side effects such as hypoglycemia, which occurs during T2DM therapy.Citation1-Citation4 4-HIL has also been found to be an effective drug to control body weight gain, glycemia, insulinemia and decreased plasma triglyceride levels in rodents.Citation5 Furthermore, recent studies have shown that 4-HIL exhibits effective antidiabetic activity in a model of type1 diabetes mellitus without insulin.Citation6
The primary method for 4-HIL production is extraction from fenugreek seeds; however, the yield by this method is rather low (approximately only150 mg of 4-HIL can be extracted from 1 kg of fenugreek seeds).Citation2,Citation7 In addition to seed extraction, chemical and enzymatic synthesis methods have also been developed but were considered to result in lowefficiency, highcost and heavy pollutionCitation8-Citation10
4-HIL has at least 8 stereo configurations, but only (2S, 3R, 4S)-4-HIL exhibits biologic activity.Citation11 L-Isoleucine dioxygenase (IDO) from Bacillus thuringiensis 2e2 has been found to specifically convert L-isoleucine(L-Ile) to (2S, 3R, 4S)-4-HIL, which is a member of the α-ketoglutarate(α-KGA)-dependent hydroxylase family, and (2S, 3R, 4S)-4-HIL has been successfully synthesized by IDO.Citation11 In a previous study, we cloned ido(Accession KC884243) from isolated B. thuringiensis TCCC 11826, and IDO was used to synthesize 4-HIL via biotransformation.Citation12 However, the 4-HIL production was rather low(44.64 mM in 36 h), and the substrates(L-Ile and α-KGA) were also found to exhibit additional consumption during the process of biotransformation.
To further gain IDO with improved activity, a strategy dependent on the coupling of L-Ile hydroxylation, the oxidation (decarboxylation) of α-KGA to succinate and cell growth was developed. Five mutants were obtained via this strategy, and the characteristics of the mutant exhibiting the highest activity were studied. Moreover, a method for 4-HIL production by resting Escherichia coli cells overexpressing activity-improved IDO was developed.
Results and discussion
Restoration of E. coli ΔsucAΔaceA-ido growth by IDO via the coupling of L-Ile hydroxylation and the oxidation of α-KGA to succinate
One unique property of the α-KGA-dependent hydroxylase reaction is the coupling of substrate hydroxylation and the oxidation (decarboxylation) of α-KGA to succinate.Citation13 Thus, IDO may shunt the TCA cycle when succinate synthesis is blocked, thereby coupling L-Ile hydroxylation and cell growth.
In E. coli, there are 4 pathways for succinate synthesis: (1) from α-KGA by α-ketoglutarate dehydrogenase via the TCA cycle under aerobic conditions, (2) from isocitrate by isocitrate lyase via the glyoxylate pathway, (3) from oxaloacetate through the reductive branch of the TCA cycle under anaerobic conditions, and (4) from L-glutamate and L-arginine via the γ-aminobutyrate catabolic pathway.Citation14 To block succinate synthesis in E. coli K-12 MG1655, both the sucA-encoding submit of α-ketoglutarate dehydrogenase and aceA-encoding isocitrate lyase were deleted, resulting in E. coli ΔsucAΔaceA. The plasmid pWSK-ido was subsequently introduced into E. coli ΔsucAΔaceA, and E.coliΔsucAΔaceA-ido was obtained. E. coli ΔsucAΔaceA could not grow in M9 medium without the addition of succinate during aerobic cultivation although the γ-aminobutyrate catabolic pathway was still retained (). The effect of succinate addition on the growth of E.coliΔsucAΔaceA was detected, and the biomass of E.coliΔsucAΔaceA was enhanced with the increased addition of succinate. Notably, E.coliΔsucAΔaceA-ido induced by IPTG restored growth in M9 medium supplemented with L-Ile and α-KGA, indicating that IDO activity shunts the blocked TCA cycle and restores cell growth coupling with L-Ile hydroxylation ().
Figure 1. Growth of E. coli cells under different conditions. (A) E. coli K-12 MG1655 grown on M9 medium(open squares) or M9 medium supplemented with succinate(solid squares); E. coli ΔsucAΔaceA grown on M9 medium(open triangles), M9 medium supplemented with 0.1 g succinate/L (gray solid triangles) or M9 medium supplemented with 0.5 g succinate/L (black solid triangles). (B) E. coli ΔsucAΔaceA harboring pWSK29(E. coli ΔsucAΔaceA-pWSK) grown on M9 medium supplemented with 1 g/L L-Ile and α-KGA(solid squares) or E. coli ΔsucAΔaceA harboring pWSK-ido(E. coli ΔsucAΔaceA-ido) grown on the same medium but under the induction of 0.01 mM(solid circles) or 0.05 mM(solid triangles) IPTG.
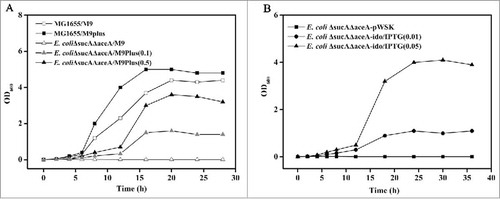
4-HIL accumulation was detected in culture broth during cell growth. The biomass () and 4-HIL accumulation(data not shown) were enhanced with the increase in IPTG addition. Because 4-HIL accumulation is coupled with succinate synthesis, it was deduced that the higher biomass was due to increased succinate synthesis catalyzed by the enhanced expression of ido induced by an increased IPTG concentration.
Screening the IDO variants with improved activities
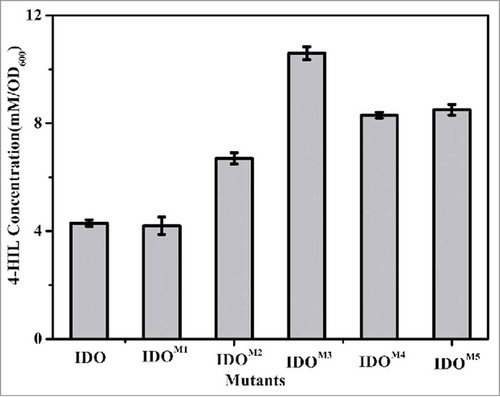
Because IDO can couple L-Ile hydroxylation and E.coliΔsucAΔaceA cell growth and because the biomass was dependent on the level of succinate synthesis, one would expect that a higher IDO activity would correspond to a greater supply of succinate and larger colonies of strains expressing ido on the plates. Error-prone PCR was performed with ido as a template, and the mutated gene products were subsequently ligated into the vector pWSK29. The recombinant plasmids were transformed into E.coliΔsucAΔaceA to obtain a mutant library. After the screening of approximately 9,000 clones, 5 were found to show considerably larger colonies than the wild type. The 5 clones were collected, and 4 clones repeatedly showed increased 4-HIL production compared with the strains with wild-type IDO, indicating improved IDO activity ().
Figure 2. Production of 4-HIL by E. coli ΔsucAΔaceA harboring ido variants in 24-well microplates. The error bars represent the SD of the mean calculated for 3 replicates.
Sequence analysis of IDOM3, which exhibited the highest IDO activity, showed that the residues Leu27, Glu80, Gly169 and Ser182 were substituted for Ile27, Asp80, His169 and Asp182, respectively. The active site structures of α-KGA-dependent hydroxylase exhibit nearly identical arrangements of the 3 amino acid side chains, a His1-X-Asp/Glu-Xn-His2 motif.Citation13 As a member of the superfamily, IDO also has the only His159-X-Asp161-X50-His212 motif, which might be the active site. Thus, one could reason that the 2 mutated amino acid residues(Gly169 and Ser182) located in the motif resulted in the improved activity of IDOM3.
Expression and characterization of IDOM3
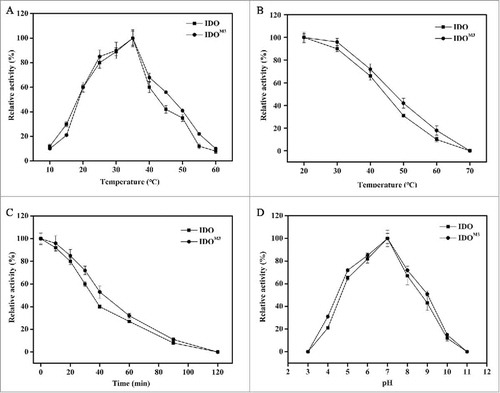
IdoM3 was expressed by E. coli BL-idoM3 under the induction of IPTG, and recombinant IDO M3 was purified by a Ni2+-NTA affinity column. The concentrations of the purified recombinant IDO and IDOM3 were 5.1 and 4.2 mg/mL, respectively. To calculate the Km, kcat and Vmax values of IDO and IDOM3, the activities of the 2 enzymes toward different concentrations of L-Ile were measured, and the data were plotted according to the Michaelis-Menten equation. As shown in , Km was lower, but Vmax and Kcat were higher for IDOM3 than IDO. The catalytic efficiency kcat/Km was approximately 1.5-fold higher for IDOM3 than for IDO under the measured conditions.
Table 1. Kinetic apparent constants determined for IDO and IDOM3.
The optimum temperature for the activity of IDOM3 and IDO was determined within a range of 10°C to 60°C. As shown in , the optimum temperature for both enzymes was approximately 35°C. Thermal stability assays showed that both IDO and IDOM3 were stable below 40°C, while the activities decreased dramatically when the temperature was above 40°C (). More than 60% of the maximum activity was observed after incubation at 60°C for 30 min (). Notably, IDOM3 exhibited a slightly higher stability than IDO. After incubation at 60°C for 40 min, 53% of IDOM3 activity was retained, compared with 40% of IDO activity (). The mutated amino acid residues at sites 27 and 80 may be responsible for the improved thermal stability. The optimum pH for the activity of IDOM3 and IDO was determined over a pH range of 3.0 to 11.0. The activity of the 2 enzymes rose with the increased pH until 7.0 and then decreased, with no difference in the optimum pH for the 2 enzymes ().
Figure 3. Effect of temperature and pH on enzyme activity and stability. (A) Effect of temperature on enzyme activity. The maximum activity at 35°C was taken as 100%. (B) Effect of temperature on enzyme stability. Enzymes were incubated for 1 h at the indicated temperatures. The samples were then measured under standard conditions. The activity without treatment was taken as 100%. (C) Time-course thermal stability. The enzyme activities were determined by evaluating residual activities after incubation for 0 min, 10 min, 20 min, 30 min, 40 min, 60 min, 90 min and 120 min at 60°C. The activity without treatment was taken as 100%. (D) Effect of pH on enzyme activity. The enzyme activity was measured at 35°C in different buffers with pH values ranging from 3 to 11. The maximum activity observed in pH 7.0 buffer was taken as 100%. The error bars represent the SD of the mean calculated for 3 replicates. The circles represent IDO, and the squares represent IDOM3.
4-HIL synthesis by resting cells expressing IDOM3
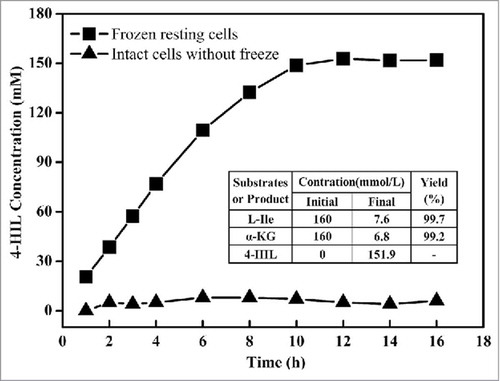
Under industrial application conditions, purified enzymes are not a suitable enzyme source. Biotransformation by E. coli harboring ido seems to be an appropriate method for 4-HIL synthesis. However, 50 nmol of glucose (150 nmol in total) was additionally consumed for cell growth rather than for 4-HIL synthesis during process biotransformation.Citation15 Moreover, additional consumption of L-Ile and α-KGA was also detected in our previous study(data not shown). One attractive solution would be to use resting cells, a strategy that has been successfully used in nitrile hydrolase biocatalysis.Citation16 However, the cell membrane seems to be the main limitation for the uptake of L-Ile and α-KGA and the export of 4-HIL. Thus, to improve the cell permeability of the substrate and products, harvested E. coli BL21(DE3) cells expressing IDOM3 were frozen at −80°C to obtain resting cells, and the strategy for 4-HIL synthesis was evaluated. As shown in , the resting cells could successfully transform L-Ile to 4-HIL, while intact cells that had not been frozen were not successful. Next, 151.9 mmol of 4-HIL/L (22.4 g/L) was synthesized in 12 h, with a yield of 0.997 mmol 4-HIL/mmol L-Ile and 0.992 mmol 4-HIL/mmol α-KGA, concentrations that are significantly higher than those obtained by the biotransformation method used previously.Citation12 According to the reaction stoichiometry, this process requires 151.9 mM L-Ile and α-KGA; the actual values were 152.4 mM and 153.2 mM, respectively, similar to the theoretical values. The results indicate that L-Ile and α-KGA were seldom subject to additional consumption during the process of biotransformation. Furthermore, the resting cells could be collected after the biotransformation and recycled at least 4 times while exhibiting almost the same yield (data not shown).
Figure 4. Production of 4-HIL by E. coli ΔsucAΔaceA-idoM cells subjected to freezing (squares) or not subjected to freezing (triangles). The final concentrations of L-Ile, α-KGA and 4-HIL as well as yield in the table were detected at 12 h of the process, when the 4-HIL production reached its peak value.
Materials and methods
Strains and plasmids
The strains and plasmids used in this work are listed in .
Table 2. Strains and plasmids.
Construction of E. coli ΔsucAΔaceA
Knockout of sucA and aceA was performed by PCR-based λ-red recombination as described previously.Citation17 A 1114-bp DNA fragment containing the chloramphenicol resistance gene cassette from pKD3 was amplified using primers sucA-1 and sucA-2.The PCR products were electroporated into E. coli K-12 MG1655 carrying the λ-red recombinase expression plasmid pKD46. Cells in which homologous recombination occurred were selected on an agar plate containing chloramphenicol and were identified by direct colony PCR using primers sucA-3 and sucA-4. The antibiotic marker was eliminated using the helper plasmid pCP20. The resulting strain was denoted E. coli ΔsucA. aceA in E. coli ΔsucA was deleted with the same method, resulting in E. coli ΔsucAΔaceA.
Construction of an ido mutant library and ido overexpression strain
A mutant library of the ido gene was constructed by error-prone PCR from pET-ido using an instant error-prone PCR kit(Tiandz, Inc.; 101005) and the primers ido-1 and ido-2. The mutated gene products were consequently ligated into the vector pWSK29, which had been digested by Xba I via the ClonExpressTM II One Step Cloning Kit(Vazyme Biotech Co., Ltd; C11201), using a homologous recombinase. The recombinant plasmids were transformed into E. coli ΔsucAΔaceA, and the cells were grown on plates containing M9 medium(glycerol as a carbon source) supplemented with L-Ile and α-KGA(1 g/L) as well as ampicillin(100 mg/L) and IPTG(0.05 mM).
The ido gene was amplified from pWSK-ido or pWSK-ido M3 by PCR using primers ido-3 and ido-4. The obtained PCR products were digested by BamH I and Hind III and were cloned into the expression vector pET-His. The recombinant plasmids(pET-IDO and pET-IDOM3) were further transformed into competent E. coli BL21(DE3) cells, and the cells were selected on LB plates containing 100 mg/L ampicillin, resulting in BL-ido and BL-idoM3.
Screening IDO mutants with improved activity
The clones exhibiting larger colonies than the wild type were transferred to 24-well microplates with 800 μL of M9 medium supplemented with L-Ile(1 g/L), α-KGA(1 g/L) and IPTG(0.05 mM). After incubation at 37°C for 24 h, 40 μL of the culture in each well was then inoculated into the corresponding well of another 24-well microplate with the same medium and was then cultured for another 24 h. The biomass(OD600)was detected, and variants exhibiting a higher biomass were chosen for further analysis.
Biotransformation of 4-HIL by E. coli ΔsucAΔaceA harboring ido variants and resting cells of BL- idoM3
E. coli ΔsucAΔaceA cells harboring ido variants were cultured in 24-well microplates with M9 medium supplemented with L-Ile(1 g/L), α-KGA(1 g/L) and IPTG(0.05 mM) at 37°C for 24 h, and then 0.1 mL of the culture broth was transferred to another 24-well microplate with the same medium, was cultured for another 24 h and was centrifuged at 4°C for 5 min. Five hundred microliters of supernatant was collected for a quantitative analysis of 4-HIL.
BL- idoM3 cells were harvested after induction by IPTG for 6 h and were frozen at −80°C for 2 h to obtainresting cells. The cells (20 g) were suspended in a 1-L shake flask with 200 mL of Tris-HCl (100 mM, pH 7.0) containing 160 mM L-Ile and α-KGA, 5 mM FeSO4 and 10 mM ascorbic acid; the cells were thenshaken at 37°C at 200 rpm for 16 h.
Protein expression and purification
IPTG (0.1 mM) was added when the BL-ido and BL-idoM3 cells were grown in LB medium(with 100 mg/L ampicillin) to the midexponential stage. The cells were harvested by centrifuging and broken with sonication after cultivation for another 4 h. The expression of recombinant IDOs was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The 6×His-tagged IDOs were purified using a Ni2+-NTA affinity column. The concentration of the purified proteins was determined by the Bradford method using bovine serum albumin as a standard.
Characterization of IDO and IDOM3
For a catalytic characterization of IDO and IDOM3, the reaction mixture composed of 10 mM α-KGA and L-Ile, 5 mM FeSO4, 10 mM ascorbic acid and 100 mM Tris-HCl(pH 7.0) was combined with 0.5 mg/mL of purified recombinant IDO, and the reaction was performed at 30°C for 30 min. The enzymatic activity was determined by the production of 4-HIL, as measured by high-performance liquid chromatography(HPLC).
The optimal temperatures for IDO and IDOM3 were determined by evaluating the activities at temperatures ranging from 10°C to 70°C. The thermal stability of the enzymes was determined by assessing their residual activities after incubation at temperatures from 10°C to 60°C for 1 h. The time-course thermal stability of the enzymes at 60°C was determined by evaluating their residual activities after incubation for 0 min, 10 min, 20 min, 30 min, 40 min, 60 min, 90 min and 120 min.
To determine the Km values, L-Ile was used at concentrations of 0.02 to 5 mM. To examine the pH dependency of the reaction, citric acid sodium-citrate buffer (pH 3.0–5.0), phosphate-buffered saline (pH 6.0–8.0), Tris-HCl (pH 9.0) and sodium bicarbonate-sodium carbonate buffer (pH 10.0–11.0) were used.
Analytical determination of 4-HIL concentration by HPLC
Four-HIL was analyzed by precolumn derivatization using 2, 4-fluoro-dinitrobenzene and was detected by HPLC using an Agilent C18 column (150 mm × 4.6 mm, 3.5 μm). Elution was performed using a gradient of 50% acetonitrile (v/v)/50mM (CH3COONa), fed at a constant flow rate of 1.0 mL/min. UV absorption was measured at 360 nm, and the column temperature was maintained at 33°C.
Conclusions
A strategy for the selection of IDO with improved activity was developed based on the coupling of L-Ile hydroxylation, the oxidation of α-KGA to succinate and cell growth. Five mutants were obtained using this strategy. The catalytic efficiency, thermal stability, and catalytic rate of the IDOM3 variant selected by the strategy were significantly improved compared with those of wild-type IDO. A method for 4-HIL biotransformation by resting cells expressing IDOM3 was developedand was shown to be suitable for 4-HIL synthesis with substrates only seldom being subjected to additional consumption.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Natural Science Foundation of China under Grant 31300069, the Tianjin Municipal Science and Technology Commission under Grant 15JCTPJC62800, the Innovation Fund for Youth of Tianjin University of Science and Technology under Grant 2016LG07 and the Training Program of Innovation and Entrepreneurship for Undergraduates under Grant 201510057063.
References
- Fowden L, Pratt HM, Smith A. 4-Hydroxyisoleucine from seed of Trigonella foenum-graecum. Phytochemistry 1973; 12(7):1707-11; http://dx.doi.org/10.1016/0031-9422(73)80391-7
- Narender T, Puri A, Shweta KT, Khaliq T, Saxena R, Bhatia G, Chandra R. 4-hydroxyisoleucine an unusual amino acid as antidyslipidemic and antihyperglycemic agent. Bioorg Med Chem Lett 2006; 16(2):293-6; PMID:16246556; http://dx.doi.org/10.1016/j.bmcl.2005.10.003
- Eidi A, Eidi M, Sokhteh M. Effect of fenugreek (Trigonella foenum-graecum L) seeds on serum parameters in normal andstreptozotocin-induced diabetic rats. Nutr Res 2007; 27(11):728-33; http://dx.doi.org/10.1016/j.nutres.2007.09.006
- Singh AB, Tamarkar AK, Narender T, Srivastava AK, Srivastava AK. Antihyperglycaemic effect of an unusual amino acid (4-Hydroxyisoleucine) in C57BL/KsJ-db/db mice. Nat Prod Res 2010; 24(3):258-65; PMID:20140804; http://dx.doi.org/10.1080/14786410902836693
- Jette L, Harvey L, Eugeny K, Levens N. 4-Hydroxyisoleucine: a plant-derived treatment for metabolic syndrome. Curr Opininon Investig Drugs 2009; 10(4):353-8.
- Haeri MR, Limaki HK, White CJ, White KN. Non-insulin dependent anti-diabet activity of (2S, 3R, 4S)-4-Hydroxyisoleucine of fenugreek (Trigonella foenum graecum) in streptozotocin- induced type I diabetic rats. Phytomedicine 2012; 19(7):571-4; PMID:22397995; http://dx.doi.org/10.1016/j.phymed.2012.01.004
- Alcock NW, Crout DHG, Gregorio MVM, Lee E, Pike G, Samuel CJ. Stereochemistry of the 4-hydroxyisoleucine from Trigonella foenum-graecum. Phytochemistry 1989; 28(7):1835-41; http://dx.doi.org/10.1016/S0031-9422(00)97870-1
- Ogawa J, Yamanaka H, Mano J, Doi Y, Horinouchi N, Kodera T, Nio N, Smirnov SV, Samsonova NN, Kozlov YI. Synthesis of 4-hydroxyisoleucine by the aldolase-transaminase coupling reaction and basic characterization of the aldolase from Arthrobacter simplex AKU 626. Biosci Biotech and Bioch 2007; 71(7):1607-15; http://dx.doi.org/10.1271/bbb.60655
- Smirnov SV, Samsonova NN, Novikova AE, Matrosov NG, Rushkevich NY, Kodera T, Ogawa J, Yamanaka H, Shimizu S. A novel strategy for enzymatic synthesis of 4-hydroxyisoleucine: identification of an enzyme possessing HMKP (4-hydroxy-3-methyl-2-keto-pentanoate) aldolase activity. FEMS Microbiol Lett 2007; 273(1):70-7; PMID:17559390; http://dx.doi.org/10.1111/j.1574-6968.2007.00783.x
- Aouadi K, Jeanneau E, Msaddek M, Praly JP. 1, 3-Dipolar cycloaddition of a chiral nitrone to (E)-1, 4-dichloro-2-butene: a new efficient synthesis of (2S, 3S, 4R)-4-hydroxyisoleucine. Tetrahedron Lett 2012; 53(23):2817-21; http://dx.doi.org/10.1016/j.tetlet.2012.03.110
- Ogawa J, Kodera T, Smirnov SV, Hibi M, Samsonova NN, Koyama R, Yamanaka H, Mano J, Kawashima T, Yokozeki K, et al. A novel L-isoleucine metabolism in Bacillus thuringiensis generating (2S, 3R, 4S)-4-hydroxyisoleucine, a potential insulinotropic and anti-obesity amino acid. Appl Microbiol Biotechnol 2001; 89(6):1929-38; http://dx.doi.org/10.1007/s00253-010-2983-7
- Zhang CL, Liu Y, Xue N, Wang XX, Xie XX, Xu QY, Chen N. Characterization of recombinant L-isoleucine-4-hydroxylase from Bacillus thuringiensis and its application in 4-hydroxyisoleucine biosynthesis. Acta Microbiol Sin 2014; 54(8):889-96; http://dx.doi.org/10.1007/s10409-014-0119-5
- Hausinger RP. Fe(II)/α-Ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol 2004; 39(1):21-68; PMID:15121720; http://dx.doi.org/10.1080/10409230490440541
- Samsonova NN, Smirnov SV, Novikova AE, Ptitsyn LR. Identification of Escherichia coli K12 YdcW protein as a γ-aminobutyraldehyde dehydrogenase. FEBS Lett 2005; 579(19):4107-12; PMID:16023116; http://dx.doi.org/10.1016/j.febslet.2005.06.038
- Smirnov SV, Kodera T, Samsonova NN, Kotlyarova VA, Rushkevich NY, Kivero AD, Sokolov PM, Hibi M, Ogawa J, Shimizu S. Metabolic engineering of Escherichia coli to produce (2S, 3R, 4S)-4-hydroxyisoleucine. Appl Microbiol Biotechnol 2010; 88(3):719-26; PMID:20665018; http://dx.doi.org/10.1007/s00253-010-2772-3
- Kobayashi M, Shimizu S. Nitrile Hydrolases. Curr Opin Biol 2000; 4(1):95-102; http://dx.doi.org/10.1016/S1367-5931(99)00058-7
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 2000; 97(12):6640-5; PMID:10829079; http://dx.doi.org/10.1073/pnas.120163297
