ABSTRACT
Epstein-Barr virus (EBV) is an oncogenic virus that infects more than 90% of the world’s population. The proteins and miRNAs encoded by EBV are involved in multiple human malignancies. Recently R-resistance RNA-seq demonstrated that EBV-encoded circular RNAs. The current research aims to explore their functions in EBV-associated malignancies. Total 56 miRNAs were sponged by circRNAome. 24 and 9 in EBV host B and epithelial cells out of 56 miRNAs were detectable by miRNA-seq. 18 and 5 miRNAs were down-regulated in both types of host cells, respectively, after EBV infection. The network between five miRNAs and their targets included 1414 genes, 1419 nodes, and 2423 edges. These targets were enriched in multiple categories, and most of them were up-regulated in EBV-infected cells. These data represented the first report that EBV circRNAs could sponge the miRNAs to promote the up-regulated expression of their targets, involving in malignancies associated with EBV.
Graphical abstract
Introduction
Epstein-Barr virus (human herpesvirus 4, EBV/HHV4) is a human oncogenic gamma herpesvirus, which causes a variety of human malignancies including lymphoid and nonlymphoid malignancies [Citation1–Citation3]. The lymphoid malignancies associated with EBV include Burkitt’s lymphoma, Hodgkin’s disease and non-Hodgkin’s lymphoma, and T-cell lymphoma [Citation3]. In addition, EBV is also associated with nasopharyngeal carcinoma, leiomyocarcomas, and gastric carcinoma [Citation4–Citation7]. EBV may also cause lymphoproliferative disorders, such as posttransplant lymphoproliferative disease (PTLD) in solid organ or hematopoietic transplant patients [Citation8,Citation9]. Although studies indicated that EBV-encoded proteins and miRNAs played critical roles in the EBV associated with oncogenesis [Citation10], the mechanism is still not fully understood. Recently the R-resistance RNA-sequencing identified the circRNAs encoded by EBV genome in EBV positive PTLD, gastric cancer, and different latently infected B cell lines [Citation11,Citation12]. These circRNAs were related to EBV-encoded oncogenic genes such as LMP2, RPMS1, and BART [Citation11,Citation12]. Up to now, the functions of these circRNAs encoded by EBV genome have not been studied yet. Whether these circRNAs are involved in the oncogenesis mediated by EBV infection needs to be investigated.
The circRNAs are generated by covalently backsplicing of a downstream 5′ splice donor site onto an upstream 3′ splice acceptor site [Citation13,Citation14]. Currently, circRNAs have been demonstrated to exert their functions by acting as miRNA sponges [Citation15], competitively inhibiting their corresponding linear mRNAs [Citation16], and binding to the RNA binding proteins [Citation17]. Additionally, it has been found that some of circRNAs could be translated into proteins using cap-independent mechanism [Citation18–Citation20]. Considering miRNAs regulate multiple aspects of cancers including cell growth, invasion, angiogenesis, metastasis, and immune evasion [Citation21,Citation22], it is hypothesized that circRNAome composed of circRNAs encoded by EBV genome would sponge the miRNAs of host cells to regulate their behaviors, and promote the progression of EBV-associated malignancies. By creatively combining bioinformatics prediction, RNA-seq analysis, and database searching, it has been found that EBV infection caused the decrease in expression of several host miRNAs, and the increase in expression of their targets. These targets are enriched in multiple categories associated with EBV related cancers such as herpes virus infection, cell cycle, and cancer pathway. This first evidence indicated that EBV circRNAome was involved in the EBV-associated malignancies by sponging miRNAs of host cells.
Methods and materials
Identification of miRNAs which interacted with circrnaome of EBV
According to the genome sequence of Mutu-EBV, the circRNAs sequences were acquired, which were identified by R-resistance RNA sequencing [Citation11]. The sequences were inputted to the miRDB database to identify the binding sites on circRNAs for miRNAs using MirTarget algorithm [Citation23,Citation24]. The binding sites for all human miRNAs were predicted and scored. The scores were weighed according to the numbers of miRNA binding sites and the abundance of circRNAs in circRNAome.
Database extraction
miRNA expression data were extracted from miRmine database which collected the expression profiles from different publicly available miRNA-seq datasets [Citation25]. These included miRNAs from LCLd3 cells infected with or without EBV infection, and normal HEK293T and HEK293 cells. The original data of the RNA sequencing were downloaded from SRA database which stores raw sequencing data for research community to enhance reproducibility and allows for new discoveries by comparing data sets [Citation26,Citation27].
Rna-seq data analysis
The RNA-seq data for mock (SRR5309459, https://www.ncbi.nlm.nih.gov/sra?term=SRX2609768) and EBV infection (SRR5309460, https://www.ncbi.nlm.nih.gov/sra?term=SRX2609769) were downloaded from SRA database. The original reads were trimmed using FASTX-toolkit (V 0.0.13, http://hannonlab.cshl.edu/fastx_toolkit/index.html) and aligned to reference genome using TopHat2 (V2.1.1, https://ccb.jhu.edu/software/tophat/index.shtml). The differential expression analysis was performed using Cuffdiff (V2.2.1, http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/). The different data were used to evaluate the alteration of the target expressions of miRNAs.
Network establishment
The network between miRNAs, of which the expression were down-regulated by EBV infection, suggesting these miRNAs were sponged by circRNAome of EBV, and their targets were established by using miRNet database, which integrated 11 miRNA databases including miRTarBase, TarBase, miRecords, miRDB, and miRanda, etc [Citation28]. These targets were determined by high throughput sequencing methods such as CLASH, PAR-CLIP, and HITS-CLIP which were designed to map the miRNA interactome. The targets were then employed to analyze the enrichment in the pathways based on GO or KEGG database by hypergeometric test algorithm. The statistical value was calculated by the embedded statistical method of miRNet.
Statistical analysis
The up-regulated targets of miRNAs after EBV infection were acquired from the RNA-seq data with or without EBV infection. The data were analyzed using Fisher exact test in R (V3.6.1, https://www.r-project.org/). When P value was less than 0.05, it was statistically significant.
Results
Circrnaome of EBV as a miRNA sponge
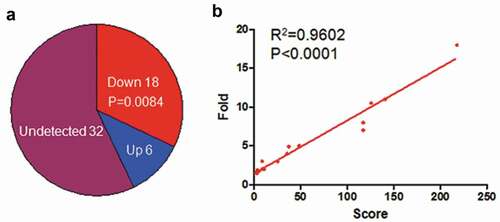
Recently two independent research groups identified the circRNAs from EBV genome by R-resistance RNA sequencing [Citation11,Citation12], and their location was shown in , but the functions of this circRNAs were not known yet. In the current research, we explored the possibility that the circRNAome constituted by all circRNAs from EBV transcripts as a sponge negatively regulated the functions of host cell miRNAs. Using miRDB screening, we identified 56 potential miRNAs in human which could interact with cicrRNAome of EBV (). They were ranked using the weighed score according to their binding ability and binding sites, and the abundance of circRNAs which interacted with the corresponding miRNAs (). 24 out of 56 miRNAs could be detected in LCLd3 cells, a B cell line (). 18 out of 24 miRNAs have decreased expression after EBV infection ( and ). lists all the decreased miRNA and their scores and decreased folds. The decreased fold of miRNAs was positively correlated with the weighed score (). This data indicated that circRNAome of EBV could inhibit the miRNA functions of host cells as a sponge.
Table 1. Down-regulated miRNAs after EBV infection in LCLd3 cells.
Figure 1. The potential miRNAs interacted with circRNAome of EBV. a. The circRNA location in the EBV genome. b. The potential miRNAs that interacted with circRNAome of EBV. The miRNAs were identified using miRDB database screening. The score was the weighed score according to the binding ability, binding sites of miRNAs and expression abundance of circRNAs in R-resistance RNA-sequencing data.
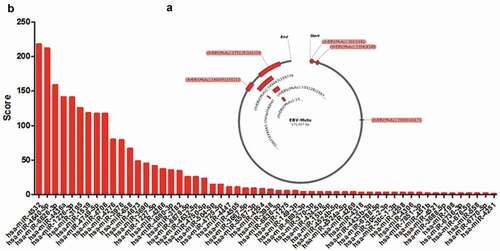
Figure 2. EBV infection decreased expression of miRNAs which were targeted by circRNAome of EBV. a. EBV infection decreased the expression of miRNAs that interacted with circRNAome of EBV. The miRNA expression in LCLd3 cell with or without EBV infection was from miRmine database. P-value was calculated using Fisher exact test in R. b. The positive correlation was between the decreased fold of miRNAs upon EBV infection and their scores targeted by EBV circRNAome.
The down-regulated miRNAs by EBV infection in lcld3 cell are highly expressed in epithelial cells, another type of EBV host cells
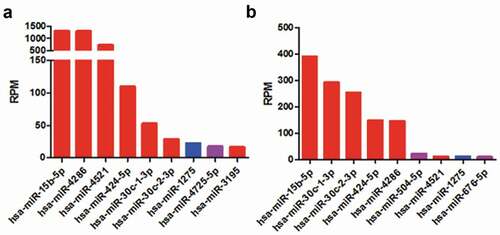
In addition to B cells, EBV could also infect epithelial cells [Citation29]. HEK293T and HEK293 cells, the epithelial cells derived from human kidney, were usually used as host cells to investigate EBV entry [Citation30,Citation31]. The miRNA expression data of HEK293T and HEK293 were acquired from miRmine [Citation25], showing that 9 out of 56 predicted miRNAs were detected in HEK293T or HEK293, respectively (). Interestingly, the down-regulated miRNAs such as has-miR-15b-5P, has-miR-4286 by EBV infection had the highest expression (red bars, ). This suggested that the sponge function of EBV circRNAome would essentially impact on the heavers of host cells.
Figure 3. The expression of miRNAs which potentially interacted with EBV circRNAome in HEK293T (a) and HEK293 (b) cells. The miRNA expression in HEK293T and HEK293 was from miRmine database. The red, purple and blue bars indicated miRNAs expression level that was decreased, increased or not altered after EBV infection.
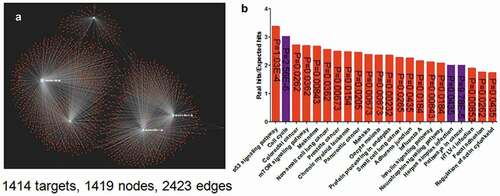
The network between miRNAs sponged by EBV circrnaome and their targets
Considering the high expression level of miR-15b-5p, miR-30c-1-3p, miR-30c-2-3p, miR-424-5p, and miR-4286 in both HEK 293T and HEK293 cells (), and their expression is decreased after EBV infection (), they were used to establish the network with their targets. Using miRNA database [Citation28], five miRNAs with their targets composed a comprehensive network with 1414 targets, 1419 nodes and 2423 edges (). The enrichment analysis indicated that their targets are enriched in cell cycle, cancer pathway and virus infection, etc., in KEGG pathway (). This was consistent with the fact that EBV infection was associated with multiple human cancers [Citation1–Citation3,Citation5,Citation9].
Figure 4. The network of the top five-decreased miRNAs associated with EBV circRNAome and their targets (a) and the enrichment of their targets (b). The network was generated using miRNet. The network was composed of 1414 targets, 1419 nodes, and 2423 edges. The enrichment assay indicated that the targets were enriched at herpes virus infection, cell cycle, cancer pathway, etc.
Up-regulated expression of miRNAs’ targets is associated with herpes infection, cell cycle and cancer pathway in host cells after EBV infection
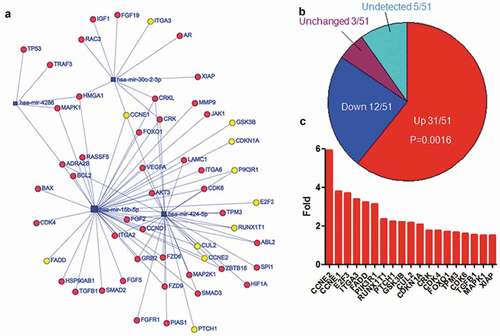
The subset networks (–) associated with herpes virus infection, cell cycle and cancer pathway were extracted from the entire network (). The expression of 12 out of 17 targets related to herpes virus infection was increased in EBV-infected cells, while 4 was decreased, and 1 was not altered (). Fisher exact test indicated that there was significant difference between the increased targets and others (p = 0.0381). The targets with twofold increase in expression were indicated in , including TAB2, IFNGR2, etc. The expression of 21 out of 31 targets linked to cell cycle was increased (), while 7 was decreased, 2 was not altered, and 1 was not detectable. Fisher exact test indicated there was significant difference between the increased targets and others (p = 0.0092). The expression of 15 out of 21 were more than twofold () in EBV-infected cells, including CCNE1/2, CDCD25A, E2F3, etc. There were 51 targets associated with cancer pathway and the expression of 31 targets was increased () after EBV infection, while 12 was decreased, 3 was not altered, and 5 was not detectable. Fisher exact test indicated that the increased targets were significantly different from others (p = 0.0016). The expression of 19 targets was greater than twofold (), including the well-known important genes such as CDK4/6, TGFB1, MAPK1.
Figure 5. Up-regulated expression of the top five-decreased was composed of miRNA targets are associated with herpes virus infection. a. The network of miRNAs and their targets which were enriched in herpes virus infection. The yellow circles indicated the genes of which expression were increased more than two-fold after EBV infection. b. The expression alteration of targets associated with herpes virus infection after EBV infection. P-value was calculated using Fisher exact test in R. c. The genes of which expression were increased more than twofold after EBV infection. The gene expression data were calculated using RNA-Seq data with or without EBV infection from SRA.
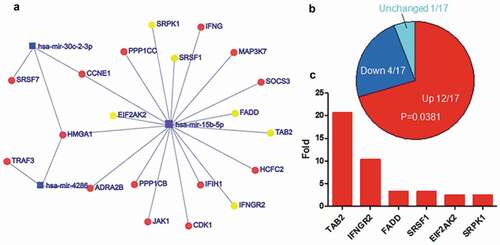
Figure 6. Upregulated expression of the top five-decreased miRNA targets associated with cell cycle. a. The network of miRNAs and their targets which were enriched in cell cycle. The yellow cycles indicated the genes of which expression were increased more than two-fold after EBV infection. b. The expression alteration of target genes associated with cell cycle after EBV infection. P-value was calculated using Fisher exact test in R. c. The genes of which expression were increased more than twofold after EBV infection. The gene expression data were calculated using RNA-Seq data with or without EBV infection from SRA.
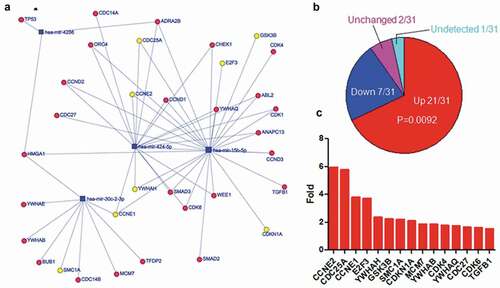
Figure 7. Upregulated expression of the top five-decreased miRNA targets associated with oncogenesis. a. The network of miRNAs and their targets which were enriched in cancer pathway. The yellow cycles indicated the genes of which expression were increased more than two-fold after EBV infection. b. The expression alteration of targets associated cell cycle after EBV infection. P-value was calculated using Fisher exact test in R. C. The genes of which expression were increased more than twofold after EBV infection. The gene expression data were calculated using RNA-Seq data with or without EBV infection from SRA.
The network between the top five-deceased miRNAs through interaction with circrnaome of EBV and small compounds
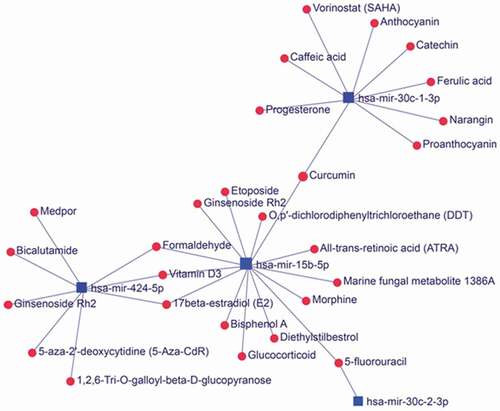
The expression of some miRNAs was decreased after EBV infection including miRNAs sponged by EBV circRNAome. According to the miRNet database, these miRNAs including has-miR-30c-1-3p, has-miR15b-5p and has-miR-424-5p were associated with 27 small compounds forming a miRNA-compound network (). Interestingly, some of the compounds such as etoposide, 5-fluororacil in the network were extensively used to treat cancers including EBV-associated non-Hodgkin’s lymphoma and nasopharyngeal carcinoma [Citation32–Citation35]. These data suggested that these chemotherapeutical compounds might exert their treatment by interfering the miRNAs sponged by EBV circRNAome.
Discussion
Recently, R-resistance RNA sequencing identified the circRNAs encoded by EBV genome, which composed the EBV circRNAome [Citation11,Citation12]. The functions of the circRNAs are not well studied yet. In the current research, we tried to investigate the function of cicrRNAs as a sponge of host miRNAs using miRNA database, miRNA-seq data, and bulk RNA-seq data from EBV-infected cells.
Circrnaome of EBV sponged mirnas of host cells involved in malignancies
CircRNAs could bind to miRNA so that circRNAs competitively inhibit miRNAs to bind to their targets which prevented the function of miRNAs. Through miRNA binding prediction, miRNA- and RNA-seq data analysis, miR-15b-5p, miR-30c-1-3p, miR-30c-2-3p, miR-424-5p, and miR-4286 were identified as top possible candidates sponged by EBV circRNAome. The overexpression of miR-15b promoted apoptosis and arrested cell cycle of glioma by targeting cyclin D1 [Citation36] while the knockout of miR-15b promoted B-cell malignancies by modulating the expression of cyclin D1 and D2 [Citation37]. EBV was associated with B lymphoma such as Burkitt, Hodgkin and Diffuse Large B cell Lymphomas (DLBCL) [Citation3]. The expression of miR-30c was positively correlated to survival of breast cancer patients [Citation38,Citation39]. Loss of miR-424 was associated with aggressiveness of breast cancers in human. Knockout of miR-424 promoted breast tumorigenesis in mice [Citation40]. miR-4286 was also highly expressed in chemo-sensitive tumors [Citation16]. Since these miRNAs had such important functions in the multiple aspects of tumor biology, the inhibition of these miRNAs by EBV circRNAs would promote tumorigenesis, cancer aggressiveness and chemotherapy resistance.
The targets of miRNAs sponged by EBV circrnaome regulated herpes virus infection
CircRNAs sponged miRNAs to increase the expression of miRNAs’ target genes. Most of the targets associated with herpes virus infection were from miR-15 such as FADD, TAB2, SRSF1, EIF2AK2 and SRPK1. Their expressions elevated more than twofold in EBV-infected cells. Kaposi’s-sarcoma-associated human herpesvirus-8 encoded v-FLIP could interact with FADD through death-effector domains and inhibit apoptosis signaling by death receptors [Citation41]. The protein complex TAB2/TAK1/TRAF6 was recruited by N-terminal of Epstein-Barr virus latent membrane protein 1 (LMP1) to promote LMP1-mediated activation of JNK [Citation42]. Herpes simplex virus (HSV) infected cell culture polypeptide 27 (ICP27) interacted with SRSF1 or SRPK1 to regulate host pre-mRNA aberrant splicing, which contributed to virus-induced host shutoff required for efficient viral growth [Citation43,Citation44]. HSV-1 protein US11 physically interacted with EIF2AK2 to negatively regulate the autophagy-constrained virus infection [Citation45]. These evidences supported that the up-regulated targets of miRNAs sponged by circRNAs could regulate a variety of functions associated with virus infection including viral growth, avoidance of limitation by host cells, host cell activation and survival.
The targets of miRNAs sponged by EBV circrnaome regulated cell cycle
Except for miR-30c-1, the targets of the other four miRNAs () were involved in cell cycle regulation. These targets fall into the following categories: 1) cyclin and cyclin-dependent kinase such as CCNE1/2, CDK4/6, 2) cell division cycle phosphatase such as CDC25A, CDC27, 3) minichromosome maintenance protein complex such as MCM7, 4)YWHA family such as YWHAB/H/Q, 5) the others such E2F3, GSK3B, SMC1A. Cyckin E controlled G1-S phase of cell cycle in complex with CDK2 [Citation46]. CDK4/6 controlled G1 phase of cell cycle in complex with cyclin D [Citation46]. Both CDC25A and CDC27 promoted G1/S-phase transition [Citation47,Citation48]. MCMs primed chromatin for DNA replication through binding origins of DNA replication during the late M to early G1 phase transition of the cell cycle [Citation49]. 14-3-3 proteins (YWHA family) functioned at several key points in G1-S and G2-M transition by binding to regulatory proteins and modulating their functions [Citation50]. E2F3 controlled the expression of genes which rately limited the initiation of DNA replication [Citation51]. GSK3B regulated the expression of cyclin D, which was involved in controlling G1 phase cell cycle in CDK4/6 complex [Citation52]. Knockdown of SMC1A led to arresting at G0/G1 phase [Citation53]. These studies suggested that the up-regulated targets of miRNAs which were sponged by circRNAs promoted the cell cycle progress. In contrast, the negative regulators of cell cycle such as CDKN1A and TGFB1were up-regulated. This might be the negative feedback to balance the over-activation of cell cycle.
The targets of mirnas sponged by EBV circrnaome regulated cancer pathway
Except for miR-30c-2, the targets of other four miRNAs were involved in oncogenesis. Since cell cycle are tightly associated with cancers, a lot of targets in cancer pathway overlapped with the targets in cell cycle including cyclin and cyclin-dependent kinases. In addition to the targets associated with cell cycle, the other targets including ITGA3, PTCH1, CUL2, CRK, FOXO1, were involved in other aspects of cancers. Knockout of ITGA3 prevented skin tumor formation by promoting epidermal turnover and depletion of slow-cycling cells [Citation54]. Overexpression PTCH1 drove skin carcinogenesis [Citation55]. Overexpression of CUL2 drove cervical carcinogenesis which might be associated with miR424 [Citation55]. Overexpression of CRK induced the prominent tyrosine phosphorylations of scaffolding molecules such as p130Cas and paxillin, ultimately contributing to the increased motility and aggressive behavior of cancer cells [Citation56]. FOXO1 could promote tumor metastasis by up-regulating phosphorylation and MMP expression [Citation57]. Thus, in addition to targets that regulate cell cycle, lots of targets also regulated other aspects of cancers ranging from carcinogenesis to migration, invasion and metastasis.
Applications and perspective
Currently, tons of data from next generation sequencing (NGS) were deposited or collected by various databases such SRA to be used for free by the research community. It is a huge challenge on how to utilize these data to dig out useful information so as to aid the biomedical research. In the current research, integrative data from R-resistance RNA-seq, miRNA-seq, and RNA-seq using EBV-associated biomaterials found the miRNAs sponged by cicrRNAome of EBV as well as the target genes that regulated these miRNAs. This research provided an example of how to use the NGS data stored in the databases to discover the real biomedical project. The network among EBV circRNAome, host miRNAs, and miRNAs’ targets was beneficial for the investigation of malignancies associated with EBV.
Conclusion
R-resistance RNA-seq defined the circRNAome of EBV. The miRNA-seq and RNA-seq data supported that circRNAome of EBV as miRNA sponge regulated miRNA functions to increase gene expression which was involved in carcinogenesis and cancer progression.
Highlights
EBV cicrRNAome was involved in EBV-associated malignancies.
EBV circRNAome was a sponge of host cells’ miRNA.
The targets of EBV circRNAome sponged miRNAs constituted to the comprehensive network.
The targets of EBV circRNAome were involved in multiple categories including virus infection, cell cycle, and oncogenesis.
CCNE1/2, CDC25A, E2F3, and TAB2 genes were the top-ranked targets of miRNAs which were sponged by EBV circRNAome.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author’s contributions
WY contributed to the study design; all authors collected the data and performed the data analysis; all authors prepared the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer. 2016;16(12):789–802.
- Rowlands DC, Ito M, Mangham DC, et al. Epstein-Barr virus and carcinomas: rare association of the virus with gastric adenocarcinomas. Br J Cancer. 2016;114(12):e15.
- Shannon-Lowe C, Rickinson AB, Bell AI. Epstein-Barr virus-associated lymphomas. Philos Trans R Soc Lond B Biol Sci. 2017;372:1732.
- Hsu JL, Glaser SL. Epstein-barr virus-associated malignancies: epidemiologic patterns and etiologic implications. Crit Rev Oncol Hematol. 2000;34(1):27–53.
- Tsao SW, Tsang CM, Lo KW. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos Trans R Soc Lond B Biol Sci. 2017;372:1732.
- Thompson MP, Kurzrock R. Epstein-Barr virus and cancer. Clin Cancer Res. 2004;10(3):803–821.
- Galetsky SA, Tsvetnov VV, Land CE, et al. Epstein-Barr-virus-associated gastric cancer in Russia. Int J Cancer. 1997;73(6):786–789.
- Dierickx D, Habermann TM. Post-transplantation lymphoproliferative disorders in adults. N Engl J Med. 2018;378(6):549–562.
- Dharnidharka VR, Webster AC, Martinez OM, et al. Post-transplant lymphoproliferative disorders. Nat Rev Dis Primers. 2016;2:15088.
- Yin H, Qu J, Peng Q, et al. Molecular mechanisms of EBV-driven cell cycle progression and oncogenesis. Med Microbiol Immunol. 2019;208(5):573–583.
- Toptan T, Abere B, Nalesnik MA, et al. Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci U S A. 2018;115(37):E8737–E45.
- Ungerleider N, Concha M, Lin Z, et al. The Epstein Barr virus circRNAome. PLoS Pathog. 2018;14(8):e1007206.
- Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264.
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442.
- Li LJ, Leng RX, Fan YG, et al. Translation of noncoding RNAs: focus on lncRNAs, pri-miRNAs, and circRNAs. Exp Cell Res. 2017;361(1):1–8.
- Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell. 2017;66(1):9–21 e7.
- Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859(10):1245–1251.
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469.
- Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561.
- Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18.
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Databaseissue):D146–52.
- Panwar B, Omenn GS, Guan Y. miRmine: a database of human miRNA expression profiles. Bioinformatics. 2017;33(10):1554–1560.
- International Nucleotide Sequence Database C, Kodama Y, Shumway M, Leinonen R. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40(Databaseissue):D54–6.
- International Nucleotide Sequence Database C, Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res. 2011;39(Databaseissue):D19–21.
- Fan Y, Siklenka K, Arora SK, et al. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44(W1):W135–41.
- Chesnokova LS, Hutt-Fletcher LM. Epstein-Barr virus infection mechanisms. Chin J Cancer. 2014;33(11):545–548.
- Chen J, Sathiyamoorthy K, Zhang X, et al. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat Microbiol. 2018;3(2):172–180.
- Chen J, Jardetzky TS, Longnecker R. The large groove found in the gH/gL structure is an important functional domain for Epstein-Barr virus fusion. J Virol. 2013;87(7):3620–3627.
- Polee MB, Kok TC, Siersema PD, et al. Phase II study of the combination cisplatin, etoposide, 5-fluorouracil and folinic acid in patients with advanced squamous cell carcinoma of the esophagus. Anticancer Drugs. 2001;12(6):513–517.
- Kok TC, Van der Gaast A, Dees J, et al. Cisplatin and etoposide in oesophageal cancer: a phase II study. rotterdam oesophageal tumour study group. Br J Cancer. 1996;74(6):980–984.
- Young RC. Etoposide in the treatment of non-Hodgkin’s lymphomas. Semin Oncol. 1992;19(6 Suppl 13):19–25.
- Comella P, Abate G, Di Finizio G, et al. Treatment of resistant non-Hodgkin’s lymphomas with cisplatin, etoposide, and bleomycin. Cancer Chemother Pharmacol. 1990;26(4):306–309.
- Sun G, Shi L, Yan S, et al. MiR-15b targets cyclin D1 to regulate proliferation and apoptosis in glioma cells. Biomed Res Int. 2014;2014:687826.
- Lovat F, Fassan M, Gasparini P, et al. miR-15b/16-2 deletion promotes B-cell malignancies. Proc Natl Acad Sci U S A. 2015;112(37):11636–11641.
- McCann JV, Xiao L, Kim DJ, et al. Endothelial miR-30c suppresses tumor growth via inhibition of TGF-beta-induced Serpine1. J Clin Invest. 2019;130:1654–1670.
- Kawaguchi T, Yan L, Qi Q, et al. Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients. Sci Rep. 2017;7(1):15945.
- Rodriguez-Barrueco R, Nekritz EA, Bertucci F, et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31(6):553–566.
- Thome M, Schneider P, Hofmann K, et al. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386(6624):517–521.
- Uemura N, Kajino T, Sanjo H, et al. TAK1 is a component of the Epstein-Barr virus LMP1 complex and is essential for activation of JNK but not of NF-kappaB. J Biol Chem. 2006;281(12):7863–7872.
- Tang S, Patel A, Krause PR. Herpes simplex virus ICP27 regulates alternative pre-mRNA polyadenylation and splicing in a sequence-dependent manner. Proc Natl Acad Sci U S A. 2016;113(43):12256–12261.
- Sciabica KS, Dai QJ, Sandri-Goldin RM. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. Embo J. 2003;22(7):1608–1619.
- O’Connell D, Liang C. Autophagy interaction with herpes simplex virus type-1 infection. Autophagy. 2016;12(3):451–459.
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079–3093.
- Nilsson I, Hoffmann I. Cell cycle regulation by the Cdc25 phosphatase family. Prog Cell Cycle Res. 2000;4:107–114.
- Qiu L, Wu J, Pan C, et al. Downregulation of CDC27 inhibits the proliferation of colorectal cancer cells via the accumulation of p21Cip1/Waf1. Cell Death Dis. 2016;7:e2074.
- Lei M. The MCM complex: its role in DNA replication and implications for cancer therapy. Curr Cancer Drug Targets. 2005;5(5):365–380.
- Hermeking H, Benzinger A. 14-3-3 proteins in cell cycle regulation. Semin Cancer Biol. 2006;16(3):183–192.
- Leone G, DeGregori J, Yan Z, et al. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12(14):2120–2130.
- Takahashi-Yanaga F, Sasaguri T. GSK-3beta regulates cyclin D1 expression: a new target for chemotherapy. Cell Signal. 2008;20(4):581–589.
- Zhang YF, Jiang R, Li JD, et al. SMC1A knockdown induces growth suppression of human lung adenocarcinoma cells through G1/S cell cycle phase arrest and apoptosis pathways in vitro. Oncol Lett. 2013;5(3):749–755.
- Sachs N, Secades P, van Hulst L, et al. Loss of integrin alpha3 prevents skin tumor formation by promoting epidermal turnover and depletion of slow-cycling cells. Proc Natl Acad Sci U S A. 2012;109(52):21468–21473.
- Kang HC, Wakabayashi Y, Jen KY, et al. Ptch1 overexpression drives skin carcinogenesis and developmental defects in K14Ptch(FVB) mice. J Invest Dermatol. 2013;133(5):1311–1320.
- Xu J, Fang Y, Wang X, et al. CUL2 overexpression driven by CUL2/E2F1/miR-424 regulatory loop promotes HPV16 E7 induced cervical carcinogenesis. Oncotarget. 2016;7(21):31520–31533.
- Hornsveld M, Dansen TB, Derksen PW, et al. Re-evaluating the role of FOXOs in cancer. Semin Cancer Biol. 2018;50:90–100.