ABSTRACT
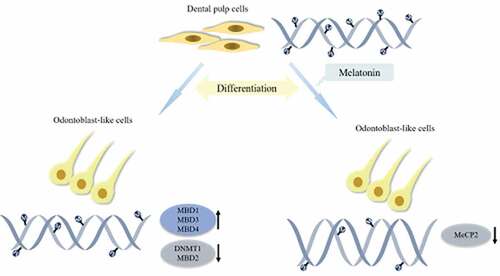
Differentiation potency of human dental pulp cells (hDPCs) is essential for dentin regeneration. DNA methylation is one of the major epigenetic mechanisms and is suggested to involve in differentiation of hDPCs, the machinery of which includes DNA methyltransferase enzymes (DNMTs) and methyl-CpG-binding domain proteins (MBDs). Our previous study has found that melatonin (MT) promoted hDPC differentiation, but its mechanism remains elusive. We aimed to investigate the role of DNA methylation in the promotion of MT to differentiation of hDPCs in vitro. hDPCs were cultured in basal growth medium (CO) or odontogenic medium (OM) exposed to MT at different concentrations (0, 10−12, 10−10, 10−8, 10−6, 10−4 M). The cell growth was analyzed using Cell Counting Kit-8 assay, and mineralized tissue formation was measured using Alizarin red staining. The expression of the 10 genes (DNMT1, DNMT3A, DNMT3B, MBD1-6, MeCP2) was determined using real-time qPCR and western blotting. The abundance of MeCP2 in the nuclei was evaluated using immunofluorescence analysis. Global methylation level was tested using ELISA. We found that mineralized tissue formation significantly increased in OM with MT at 10−4 M, while the levels of MeCP2 and global DNA methylation level declined. The expression of MBD1, MBD3, and MBD4 significantly increased in OM alone, and the expession of DNMT1 and MBD2 was decreased. These results indicate that MT promotes odontogenic differentiation of hDPCs in vitro by regulating the levels of DNMT1, MeCP2, and global DNA methylation, suggesting that MT-induced DNA methylation machinery may play an important role in tooth regeneration.
Introduction
Human dental pulp cells (hDPCs) are mesenchymal stem cells (MSCs) derived from the neural crest [Citation1]. These cells are capable of differentiating into odontoblast-like cells in response to exogenous stimuli or injury and subsequently generating reparative dentin [Citation2]. The differentiation potency of hDPCs plays a crucial role in homeostasis of dental pulp, which makes it possible to preserve pulp vitality in clinic. Moreover, hDPCs are easily obtained from extracted teeth and possess high proliferative ability [Citation3]. Emerging evidence has highlighted the critical role of epigenetic modifications in regulating MSC homeostasis between self-renewal and differentiation, and epigenetic processes involved in gene expression during differentiation of dental MSCs [Citation4,Citation5].
DNA methylation is the most extensively studied epigenetic modifications in mammalian genomics [Citation6]. This process depends on the activity of DNA methyltransferase (DNMTs) and methyl-CpG-binding domain proteins (MBDs) [Citation7]. DNMTs, including DNMT1, DNMT3A, and DNMT3B, mediate DNA methylation by adding the methyl group to the C5 position of cytosine (5-C) to form 5-methylcytosine (5-mC) [Citation8]. DNMT1 ensures the maintenance of DNA methylation during DNA replication, whereas DNMT3A and DNMT3B act on unmethylated or hemi-methylated sites as de novo DNA methyltransferases. The MBD family comprises MBD1-6 and MeCP2, which is thought to be linked to transcriptional repression by binding selectively to methylated DNA [Citation9].
There is direct evidence that hypermethylation helps maintain stemness of cells, while hypomethylation promotes stem cell differentiation [Citation10]. Li et al. [Citation11] have suggested that the DNA demethylation machinery was involved in the cytodifferentiation potential of hDPCs. In addition, DNA methyltransferase inhibitors, 5-aza-2ʹ-deoxycytidine (5-Aza) and RG108, have been demonstrated to promote hDPCs differentiation [Citation12,Citation13]. The first cloned MBD, MeCP2, has greater affinity for most of the methylated CpG sites than other MBDs, contributing its ability to bind to single CpG dinucleotides that are symmetrical and methylated [Citation14]. Downregulation of MeCP2 expression has been demonstrated during differentiation of human bone marrow MSCs [Citation15]. However, the expression profiles of these genes during odontogenic differentiation of hDPCs remain underexplored.
Melatonin (MT), N-acetyl-5-methoxytryptamine, is an endogenous hormone with pleiotropic bioactivities, including the control of circadian rhythms, oncostatic properties, immunomodulatory functions [Citation16]. Moreover, it has been shown to be involved in promoting MSC differentiation into osteogenic, adipogenic, chondrogenic and neurogenic lineages in recent studies [Citation12,Citation17]. There is mounting evidence implying the effect of MT on differentiation of MSCs and its epigenetic mechanism in gene expression [Citation18,Citation19]. Recent study on goat oocytes showed that MT downregulated the expression of DNMTs and global methylation level to promote differentiation [Citation20]. In addition, MT could change methylation levels to various degrees, which might function both as a gene inducer and silencer simultaneously without changing the DNA sequence [Citation21].
Our previous study has shown that MT at physiological concentrations could promote odontogenic differentiation of hDPCs [Citation22]. It is proved that DNA methylation involved in the regulation of differentiation of hDPCs, and MT activity was modulated by the epigenetic factors in MSCs as mentioned before. However, what changes occur in DNA methylation during MT-induced differentiation of hDPCs, is poorly understood. Therefore, we investigated the effect of MT on DNA methylation during odontogenic differentiation of hDPCs in vitro and explored the underlying mechanisms. To validate this, we induced differentiation of hDPCs with MT exposure for 7 days, and then detected the expression of odontogenic markers, DNMTs and MBDs, the abundance of MeCP2 in the nuclei, and global methylation level.
Materials and methods
Ethics committee approval and patient consent
The entire study was approved by the Medical Ethics Review Committee of Guanghua School of Stomatology, Hospital of Stomatology, Institute of Stomatological Research, Sun Yat-sen University (Ethical Application Ref: KQEC-2019-10). Informed written consent was obtained from the donors. The investigation conformed to the principles outlined in the Declaration of Helsinki.
Cells isolation and culture
hDPCs were obtained from healthy premolars or third molars extracted from patients aged 18 to 25 at the Department of Oral and Maxillofacial Surgery, Guanghua School of Stomatology, Hospital of Stomatology, Sun Yat-sen University. Human dental pulp tissue isolation was performed according to an established protocol [Citation22]. After teeth were split with a chisel, pulp tissues were isolated and minced to pieces under sterile condition. The pieces were digested with collagenase type I and then cultured in 100 mm dishes with basal growth medium (CO), containing α-minimum essential medium (αMEM) supplemented with 20% fetal bovine serum (FBS) and antibiotics (10 U/L penicillin and 100 mg/L streptomycin). The medium was changed every 3 days in an atmosphere containing 95% O2 and 5% CO2 at 37°C. The cells were purified by passaging. The cells from passage 4 were used in subsequent experiments. All other reagents were purchased from Gibco (Grand Island, USA) unless otherwise stated.
Alizarin red staining and calcium concentration determination
A single-cell suspension was prepared from the fourth generation of hDPCs, and these processed cells were used for further experimentation. The cells were inoculated in 48-well plates at a density of 2 × 104 cells/well. Then, the cells were cultured in CO or odontogenic medium (OM) treated with MT (Sigma-Aldrich, St Louis, MO, USA) at different concentrations for 7 days: (a) CO+MT-, (b) OM+MT-, (c) OM+MT+, melatonin (10−12, 10−10, 10−8, 10−6, 10−4 M). OM contained α-MEM supplemented with 20% FBS and 2% antibiotics, 50 nM ascorbic acid, 100 nM dexamethasone, and 10 mM β-glycerol phosphate (Sigma-Aldrich, St Louis, MO, USA).
After a culture period of 7 days, the cells were washed thrice with phosphate-buffered saline (PBS, pH = 7.4), fixed in 4% paraformaldehyde solution (Sigma-Aldrich, St Louis, MO, USA) for 30 min, and then washed thrice with PBS again. The cells were then stained with 1% Alizarin red solution (Cyagen, Jiangsu, China) for 10 min. After washing thrice with PBS and drying at room temperature, the mineralized nodules were observed and photographed using an inversed microscope (Carl Zeiss Meditec AG, Jena, Germany). The amount of mineralization matrix was detected using 100 nM cetylpyridinium chloride (Sigma-Aldrich, St Louis, MO, USA) for 1 h. The concentration of the dye was determined by absorbance at 562 nm. The data were collected from three conditioned wells independently.
Cell viability assay
The proliferative viability of hDPCs was monitored using the Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Kumamoto, Japan) according to the manufacturer’s protocol. Briefly, 5000 cells were seeded in each well of the 96-well culture plates. After they were left to adhere for 24 h, cells were cultured in CO with MT at different concentrations: (a) CO+MT-, (b) CO+MT+, MT (10−12, 10−10, 10−8, 10−6, 10−4 M). After 1, 3, 5, and 7 days of culturing, the supernatant of each group was removed, and hDPCs were incubated in 100 μL/mL CCK-8 solution in serum-free medium for another 2 h at 37°C. The optical density value was measured using a spectrophotometer (Tecan, Grodig, Austria) at 450 nm. Experiments were repeated four times.
Real-time qPCR
The processed cells from the fourth passage were used for this experiment. They were inoculated in 6-well plates at a density of 2 × 105 cells/well. Then, the cells were cultured in CO or OM treated with MT at different concentrations for 7 days: (i) CO+MT-, (ii) CO+MT+, MT (10−12, 10−4 M), (iii) OM+MT-, (iv) OM+MT+, MT (10−12, 10−4 M).
Total RNA of hDPCs was isolated with RNA-Quick Purification Kit (Yishan Biotechnology, Shanghai, China) according to the manufacturer’s instructions. The quality of total RNA was monitored by capillary electrophoresis (NanoDrop Technologies, Wilmington USA). Next, cDNA was synthesized using PrimeScript️ RT Master Mix (TaKaRa, Shiga, Japan). QPCR was performed using SYBR Green I Master Mix (Roche Applied Science, Basel, Switzerland). The sequences of each primer are listed in . The qPCR conditions were as follows: 1 cycle at 95°C for 5 min, followed by 45 cycles at 95°C for 10 s, 60°C for 20 s, and 72°C for 20 s. Ct (cycle threshold) values were calculated using the 2−ΔΔCt method.
Table 1. List of primer pairs used for RT-qPCR analysis.
Western blotting
The processed cells from the fourth passage were used for this experiment. They were inoculated in 6-well plates at a density of 2 × 105 cells/well. Then, the cells were cultured in CO or OM treated with MT at different concentrations for 7 days: (i) CO+MT-, (ii) CO+MT+, MT (10−12, 10−4 M), (iii) OM+MT-, (iv) OM+MT+, MT (10−12, 10−4 M).
RIPA (radioimmunoprecipitation assay) buffer containing protease inhibitor was used to harvest proteins and the protein concentration was measured using the BCA (bicinchoninic acid) protein assay (Beyotime, Shanghai, China). Thirty micrograms of total protein were separated by 4–20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SurePAGE, Ubiotechnology, Guangdong, China), then transferred to polyvinylidene difluoride (PVDF) membrane (Merck Millipore, Billerica, MA, USA). The membrane was then blocked with tris-buffered saline with Tween-20 (TBST) containing 5% skim milk for 1 h at room temperature and incubated at 4°C overnight with the diluted primary antibodies shown in . After washing for 50 min, the membrane was blocked with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. Antibody binding was visualized using an enhanced chemiluminescence system (CW Biotech, Peking, China), and band densities were obtained and normalized to β-actin and the background using ImageJ software (National Institutes of Health, Bethesda, USA).
Table 2. List of various antibodies used for western blotting.
Global methylation level quantification
The processed cells from the fourth passage were used for this experiment. They were inoculated in 6-well plates at a density of 2 × 105 cells/well. Then, the cells were cultured in CO or OM treated with MT at different concentrations for 7 days: (i) CO+MT-, (ii) CO+MT+, MT (10−12, 10−4 M), (iii) OM+MT-, (iv) OM+MT+, MT (10−12, 10−4 M).
DNA was extracted from tissues using DNeasy kits (Omega Bio-tek, Guangzhou, China) following the manufacturer’s directions and then global methylation level was analyzed by MethylFlash Global DNA Methylation (5-mC) ELISA Easy Kit (EPIGENTEK, Farmingdale, USA) according to the manufacturer’s protocol. Hundred nanograms of sample DNA were added with binding solution into the wells and then incubated at 37°C for 30 min and washed thrice with washing buffer. Fifty microliters of the 5-mC detection complex solution were added to each well and then incubated at room temperature for 50 min. After washing five times, 50 μL of Developer Solution were added to each well and Stop Solution was added to stop the reaction. The absorbance was read on a microplate reader at 450 nm.
Immunofluorescence analysis
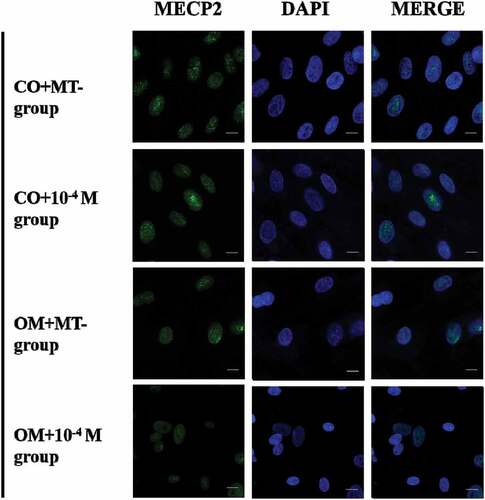
Cells were cultured on the confocal dishes at a density of 1 × 104 cells/well in CO or OM treated with MT at different concentrations for 7 days: (i) CO+MT-, (ii) CO+MT+, (10−4 M), (iii) OM+MT-, (iv) OM+MT+ (10−4 M). Then, the dishes were washed in PBS thrice and fixed in 4% paraformaldehyde for 20 min at room temperature, followed by washing in PBS. Permeabilization was performed with 0.5% Triton X-100 in PBS (v/v) for 20 min. The cells were then incubated for 30 min in 5% FBS dissolved in PBS (w/v). The cells were incubated overnight with primary antibodies-MeCP2 in 1:200 dilution at 4°C. After three washes in PBS, the cells were incubated for 1 h with goat anti-rabbit Alexa Fluor 488 (Invitrogen, California, USA), diluted to 1:200, as a secondary antibody. After further washing in PBS, the nuclei were counterstained with DAPI (4ʹ,6-diamidino-2-phenylindole) in antifade mounting medium (Melonepharma, Dalian, China).
The cells were examined under Leica TCS SP8 MP confocal microscope (Leica Microsystems, Buffalo Grove, USA). The images for immunofluorescent staining were acquired with a 63×/1.4NA Plan-Apochromat oil objective. The pixel size was 187.25 × 187.25 μm (1024 × 1024). The images were acquired by lasers with two excitation lines: 488 nm for Alexa Fluor 488 and 405 nm for DAPI. The objectives, filters, pinhole, and gain were kept constant as all slides were examined.
Statistical analysis
GraphPad Prism 5.0 (GraphPad Software, Inc.) was employed to perform all of the statistical analyzes. All of the data were analyzed as the mean ± SEM (standard error of mean) from at least three independent experiments. Statistical comparisons of the data were analyzed using one‐way analysis of variance (ANOVA), whereas differences between groups were compared using Bonferroni test using SPSS v. 20.0 software (SPSS Inc, Chicago, USA).
P < 0.05 was considered to indicate statistical significance.
Results
Effect of MT on proliferation of hDPCs
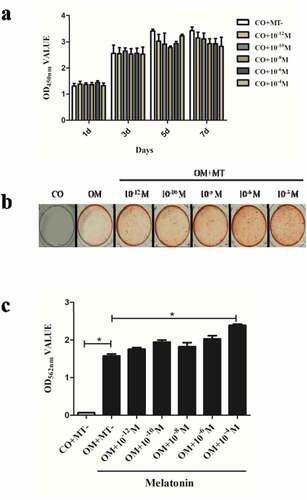
To investigate the effect of MT at various concentrations (10−12, 10−10, 10−8, 10−6, 10−4 M) on the growth rates of hDPCs, cell proliferation was measured with the CCK-8 assay on days 1, 3, 5, and 7. The results showed that there was no significant difference in the growth rates between any of the groups on days 1, 3, 5 and 7 ().
Figure 1. The effect of melatonin on proliferation and odontogenic differentiation of hDPCs.
Effect of MT on the mineralization potential of hDPCs
To determine the effect of MT on the mineralization potential of hDPCs, the formation of mineralized tissue was observed and measured using Alizarin red staining.
The greatest mineralized tissue formation of hDPCs was observed in OM with MT at 10−4 M (OM+10−4 M group) with statistical significance (P < 0.05). The lowest mineralized tissue formation was found in OM+MT- group, but the groups with MT at 10−12, 10−10, 10−8, and 10−6 M showed no significant increase in mineralized tissue formation compared with OM+MT- group. There was a statistically significant difference between OM+10−4 M and OM+10−12 M groups (). Therefore, the concentration of 10−4 M was selected for subsequent experiments.
Expression features of DNMTs during odontogenic differentiation of hDPCs treated with MT
To identify the expression of DNMTs in regulating odontogenic differentiation potential of hDPCs, the expression of the odontogenic markers, DSPP (dentin sialophosphoprotein) and DMP-1 (dentin matrix protein-1), and the genes of DNMTs, DNMT1, DNMT3A, and DNMT3B were assessed by qPCR and western blotting on day 7.
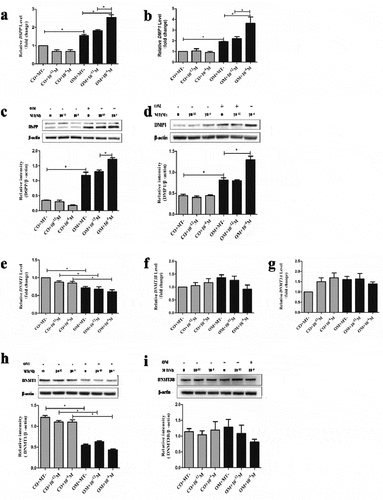
The mRNA and protein expression of DSPP and DMP-1 showed the highest level in OM+10−4 M group with statistical significance (P < 0.05, ). The expression of DNMT1 was significantly lower in all OM groups than that in all CO groups at the same MT concentration (P < 0.05), while there was no significant difference among MT-treated groups (e, h). The expression of DNMT3A and DNMT3B remained unchanged ().
Figure 2. The effect of melatonin on the expression of odontogenic markers and DNMTs during odontogenic differentiation of hDPCs.
Expression features of MBDs during odontogenic differentiation of hDPCs treated with MT
The results indicated that mRNA levels of MBD genes (MBD1, MBD3, and MBD4) were higher in all OM groups, but MBD2 expression was contrary (P < 0.05, ). The mRNA levels of MBD5 and MBD6 were not significantly altered during odontogenesis ().
Figure 3. Transcriptional profiles of methyl-CpG-binding domain proteins (MBDs) genes during odontogenic differentiation of hDPCs.
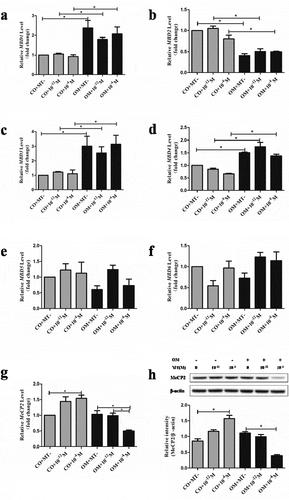
The expression of MeCP2 was significantly lower in OM+10−4 M group than OM+10−12 M and OM+MT- groups. There was no significant increase in OM+MT- group compared with CO+MT- group. The expression of MeCP2 increased in CO+10−12 M and CO+10−4 M groups, compared with CO+MT- group (). The abundance of MeCP2 in the nuclei was also proved to decrease in OM+10−4 M group ().
Global methylation level mediated by MT during odontogenesis
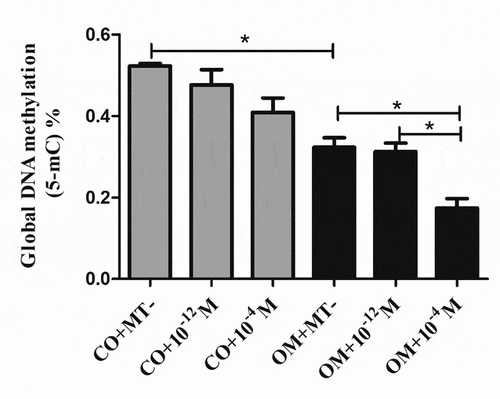
To further explore the change of global methylation level (5-mC) during odontogenesis in hDPCs, the level of 5-mC was analyzed on day 7. The results revealed that global methylation level was significantly lower in OM+MT- group, compared with CO+MT- group. Moreover, the level was further declined in OM+10−4 M group ().
Figure 5. The effect of melatonin on global methylation level in odontogenic differentiation of hDPCs. Global methylation level was detected using MethylFlash Global DNA Methylation (5-mC) ELISA on day 7. All results are presented as the mean ± SEM (standard error of mean) from at least three independent experiments. Procedures were performed as described in the text. *P < 0.05 indicates a statistically significant difference.
Discussion
hDPCs derived from dental pulp tissue have been previously characterized as a subpopulation of MSCs migrated from neural crest cells with spindle-shaped appearance and rapid expansion property [Citation1]. The potency of odontogenic differentiation of hDPCs is crucial for pulp and tooth regeneration, involving multiple and overlapping molecular factors [Citation23]. Epigenetic mechanisms, including DNA methylation and histone modification, play crucial roles in differentiation of dental MSCs [Citation5,Citation24]. DNA methylation is one of the major epigenetic modification systems. The major components of the DNA methylation machinery responsible for the process, include DNMTs and MBDs [Citation7]. A study reported that methylation status has an effect on a network of interactive signaling pathways involved in tooth development [Citation25]. Nakatsuka et al. [Citation26] found that DNA demethylation by 5-Aza treatment-induced skeletal myogenic differentiation of dental pulp stem cells. More studies have been reported that demethylation status promoted odontogenesis of hDPCs [Citation11,Citation27], and suppression of DNA methylation facilitated osteogenic differentiation capacity of human periodontal ligament stem cells exposed to high glucose [Citation28]. So far, little is known about the transcriptional profiles of DNMTs and MBDs genes during odontogenesis in hDPCs. Hence, the expression of these 10 genes and genomic methylation levels were comprehensively analyzed in the present study.
In this study, the effect of MT at different concentrations on odontogenic differentiation and proliferation of hDPCs was analyzed. We examined the expression of odontogenic markers DSPP and DMP1, the formation of mineralized tissue, and the cellular growth rates. The results showed that MT at pharmacological concentration level (10−4 M) upregulated the expression levels of DSPP and DMP-1, and increased mineralized nodules formation without showing any inhibiting effect on proliferation. These data confirmed that MT promoted odontogenic differentiation of hDPCs.
We found that DNMT1expression decreased during differentiation of hDPCs in the present study. Furthermore, the odontogenic induction, not the effect of MT, affected DNMT1 expression. This result was consistent with a previous report that the suppression of DNMT1 resulted in elevating the efficiency of odontoblastic differentiation of mouse dental papilla cells [Citation13]. In human MSCs, induction of osteogenic differentiation has been found to be associate with the decline of DNMT1 [Citation29]. Gasiuniene et al. [Citation30] reported that osteogenic differentiation of MSCs was related to the gradual decrease in the level of DNMT1. Additionally, high level of DNMT1 expression was noted in proliferating cells [Citation31]. It is possible that DNMT1 is important for odontogenesis.
After odontogenic induction and MT treatment, no significant changes on the expression of DNMT3A and DNMT3B were observed in this study, implying that these two DNMTs may not contribute to the suppression of DNA methylation on odontogenic differentiation of hDPCs. This is in line with the finding that fiber cell differentiation occurred in the absence of DNMT3A and DNMT3B, whose de novo DNA methylation may be compensated by DNMT1 [Citation32]. Emerging evidence has highlighted the critical roles of DNMT3A and DNMT3B in differentiation of embryonic stem cells during early development [Citation33]. High expression of DNMT3A and DNMT3B in early embryos facilitates de novo methylation, while the expression levels decreased during the differentiation of embryonic stem cells and somatic tissue [Citation31]. Studies of DNMT3A and DNMT3B in differentiation have yielded differing results [Citation34–Citation36], which is likely to depend on the MSC fate. These observations indicated that DNMT1 may have a major role in odontogenesis of hDPCs.
It was previously suggested the scale of genomic methylation generally also related to the number of MBD proteins [Citation37]. So far, knowledge of the roles of MBDs during MSC differentiation is limited. Therefore, we first detected the expression changes of MBDs in hDPCs during differentiation. We observed the differentiation-dependent instead of MT-dependent changes in expression levels of MBDs, except for MBD5 and MBD6. A similar result was reported in porcine adipocyte differentiation of bone marrow MSCs [Citation15]. However, this has not yet been studied in odontogenesis. The differences in their expression during odontogenesis may reflect their diverse functions.
The roles of MBDs have been described in reviews [Citation37–Citation39]. For example, MBD1 is crucial for gene silencing in cell division and differentiation, and MBD2 has a role in the self-renewing state of stem cells, enriched at promoter of two master pluripotency regulators, OCT4 and NANOG. MBD3 is known to play an important role in transcription factor-induced somatic cell reprogramming, pluripotent stem cell differentiation and embryonic development [Citation40]. Additionally, MBD4 is involved in promoter and genome-wide demethylation [Citation41]. The functions of MBD5 and MBD6 have not been fully described. MBD5 has been shown to be associated with heterochromatin by binding to DNA in a complex, and to be of importance in development, neurogenesis, and neuronal gene regulation [Citation42]. Similarly, MBD6 has been shown to be critical for cell differentiation into mesodermal and neural lineages and proliferation as an OCT4 regulatory gene [Citation43]. Thus, based on the observation, we concluded that MBD1-4 has major roles in odontogenesis instead of MBD5 and MBD6.
In the present study, the expression level of MeCP2 decreased after treatment with MT during odontogenesis, in contrast to alterations in the control group in which there was a mild increase in MT expression. On one hand, MeCP2 directly or indirectly represses transcription by specifically binding to methylated DNA. Recent study has found that it is important at the organ differentiation stage, instead of expressing during proliferation or differentiation of embryonic stem cells [Citation39]. The overexpression of MeCP2 was suggested to suppress myofibroblast differentiation in normal fibroblasts [Citation44], and correspondingly the level of MeCP2 declined significantly during differentiation of smooth muscle cells [Citation45]. In the neural cell lineage, the absence of MeCP2 was demonstrated to promote astrocyte-specific differentiation of neural stem cells [Citation46]. On the other hand, Tao et al. [Citation47] have reported that MeCP2 knockdown led to downregulation of vascular smooth muscle cell proliferation. Besides, similar change that MT increased the MeCP2 expression during proliferation occurred in neural stem cells in the adult hippocampus under sleep-deprived conditions [Citation48]. Therefore, we suggest that MeCP2 plays an essential role in DNA methylation in hDPCs.
Our findings showed a decline in the global methylation level in odontogenic induction alone, consistent with the DNMT1 level. This agrees with the result that another DNMT inhibitor, Zebularine promoted differentiation by downregulating the global methylation status of the cells, thus inducing the transcription of differentiation-specific genes [Citation49]. Furthermore, MT exposure in differentiated cells intensified this decline in accordance with reduction of MeCP2. Mammalian genomes have a tendency to be methylated genome-wide with the exception of CpG islands. The DNA methylation patterns in the mammalian genome are mainly maintained by DNMT1 because of its maintenance DNA methyltransferase activity. Loss of DNMT1 results in a loss of DNA demethylation globally and can be lethal [Citation50]. During DNA duplication, DNMT1 methylates hemi-methylated DNA by recruiting MeCP2, even though it does not possess a methyl-CpG binding domain that helps to bind to the methylated cytosine of the template DNA. The interaction of MeCP2-DNMT1 complex maintained the methyltransferase activity. The similar cooperation of MeCP2 and DNMTs was also confirmed in GABAergic neurons of mice and prostate cancer cells [Citation51,Citation52]. This direct interplay of MeCP2-DNMT1 complex was suggested to be responsible for genome-wide DNA methylation [Citation53]. The expression of DNMT1 was induced as DNA synthesis complete in proliferating cells [Citation54], which may also induce the expression of MeCP2. This might explain the cause of increase in MeCP2 expression in the control group with or without MT treatment in this study. Therefore, global methylation level in hDPCs was not affected by MT alone, considering that the upregulation of DNMT1 was inhibited and the mild increase of MeCP2 was not enough to increase global methylation. Moreover, DNMT1 activity was inhibited during odontogenic differentiation, and the 5-mC level correspondingly decreased. Furthermore, MeCP2 expression was deregulated and then global methylation level was declined during MT-induced differentiation, suggesting that MT may have effects on MeCP2 or MeCP2-DNMT1 complex.
To our knowledge, these findings provide the first indication that DNMTs and MBDs involved in the process of MT-induced differentiation in hDPCs in vitro. In conclusion, the present study shows that the expression of DNMT1 and MBD2 is downregulated in odontogenically differentiated hDPCs, while the expression of MBD1, MBD3, and MBD4 is upregulated. In addition, during the investigation into MT promotion on odontogenic differentiation of hDPCs, the levels of MeCP2 and global methylation are further downregulated. These results indicate that the positive effect of MT on hDPCs may be related to the interaction of DNMT1 and MeCP2, which contributes to global hypomethylation, expanding our knowledge on the understanding of epigenetic mechanisms of odontogenic differentiation in hDPCs.
Research highlights
Changes in DNMTs and MBDs expression during MT promoting hDPC differentiation were studied
The expression of MBD1, MBD3, and MBD4 increased, while the expression of DNMT1 and MBD2 decreased during mineralization induction
The level of global methylation and MeCP2 declined during MT-induced differentiation
Acknowledgements
We thank Taylor & Francis for their linguistic assistance during the preparation of this manuscript.
Disclosure statement
The authors declare no potential conflicts of interest.
Additional information
Funding
References
- Harpe PT. Dental mesenchymal stem cells. Development. 2016;143:2273–2280.
- Yen AH, Yelick PC. Dental tissue regeneration - a mini-review. Gerontology. 2011;57:85–94.
- Permuy M, López-Peña M, González-Cantalapiedra A, et al. Melatonin: a review of its potential functions and effects on dental diseases. Int J Mol Sci. 2017;18(4):865.
- Deng P, Chen QM, Hong C, et al. Histone methyltransferases and demethylases: regulators in balancing osteogenic and adipogenic differentiation of mesenchymal stem cells. Int J Oral Sci. 2015;7:197–204.
- Rodas-Junco BA, Canul-Chan M, Rojas-Herrera RA, et al. Stem cells from dental pulp: what epigenetics can do with your tooth. Front Physiol. 2017;8:999.
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38.
- Hamidi T, Singh AK, Chen T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015;7:247–265.
- Utsey K, Keener JP. A mathematical model for inheritance of DNA methylation patterns in somatic cells. Bull Math Biol. 2020;82(7):84.
- Rausch C, Hastert FD, Cardoso MC. DNA modification readers and writers and their interplay. J Mol Biol. 2019;0022-2836(19):30718–30719.
- Berdasco M, Melguizo C, Prados J, et al. DNA methylation plasticity of human adipose-derived stem cells in lineage commitment. Am J Pathol. 2012;181:2079–2093.
- Li Q, Yi B, Feng Z, et al. FAM20C could be targeted by TET1 to promote odontoblastic differentiation potential of human dental pulp cells. Cell Prolif. 2018;51:e12426.
- Sharma R, Ottenhof T, Rzeczkowska PA, et al. Epigenetic targets for melatonin: induction of histone H3 hyperacetylation and gene expression in C17.2 neural stem cells. J Pineal Res. 2008;45:277–284.
- Sun Z, Yu S, Chen S, et al. SP1 regulates KLF4 via SP1 binding motif governed by DNA methylation during odontoblastic differentiation of human dental pulp cells. J Cell Biochem. 2019;120:14688–14699.
- Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914.
- Stachecka J, Lemanska W, Noak M, et al. Expression of key genes involved in DNA methylation during in vitro differentiation of porcine mesenchymal stem cells (MSCs) into adipocytes. Biochem Biophys Res Commun. 2020;522:811–818.
- Varoni EM, Soru C, Pluchino R, et al. The impact of melatonin in research. Molecules. 2016;21:240.
- Wang B, Wen H, Smith W, et al. Regulation effects of melatonin on bone marrow mesenchymal stem cell differentiation. J Cell Physiol. 2019;234:1008–1015.
- Radio NM, Doctor JS, Witt-Enderby PA. Melatonin enhances alkaline phosphatase activity in differentiating human adult mesenchymal stem cells grown in osteogenic medium via MT2 melatonin receptors and the MEK/ERK (1/2) signaling cascade. J Pineal Res. 2006;40:332–342.
- Tain YL, Huang LT, Chan JY. Transcriptional regulation of programmed hypertension by melatonin: an epigenetic perspective. Int J Mol Sci. 2014;15:18484–18495.
- Saeedabadi S, Abazari-Kia AH, Rajabi H, et al. Melatonin improves the developmental competence of goat oocytes. Int J Fertil Steril. 2018;12:157–163.
- Korkmaz A, Reiter RJ. Epigenetic regulation: a new research area for melatonin? J Pineal Res. 2008;44:41–44.
- Liu Q, Fan W, He Y, et al. Effects of melatonin on the proliferation and differentiation of human dental pulp cells. Arch Oral Biol. 2017;83:33–39.
- Zhang X, Ning T, Wang H, et al. Stathmin regulates the proliferation and odontoblastic/osteogenic differentiation of human dental pulp stem cells through Wnt/beta-catenin signaling pathway. J Proteomics. 2019;202:103364.
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500.
- Wang J, Sun K, Shen Y, et al. DNA methylation is critical for tooth agenesis: implications for sporadic non-syndromic anodontia and hypodontia. Sci Rep. 2016;6:19162.
- Nakatsuka R, Nozaki T, Uemura Y, et al. 5-Aza-2ʹ-deoxycytidine treatment induces skeletal myogenic differentiation of mouse dental pulp stem cells. Arch Oral Biol. 2010;55:350–357.
- Zhang D, Li Q, Rao L, et al. Effect of 5-Aza-2ʹ-deoxycytidine on odontogenic differentiation of human dental pulp cells. J Endod. 2015;41:640–645.
- Liu Z, Chen T, Sun W, et al. DNA demethylation rescues the impaired osteogenic differentiation ability of human periodontal ligament stem cells in high glucose. Sci Rep. 2016;6:27447.
- Zentelyte A, Gasiuniene M, Treigyte G, et al. Epigenetic regulation of amniotic fluid mesenchymal stem cell differentiation to the mesodermal lineages at normal and fetus-diseased gestation. J Cell Biochem. 2020;121:1811–1822.
- Gasiuniene M, Zentelyte A, Treigyte G, et al. Epigenetic alterations in amniotic fluid mesenchymal stem cells derived from normal and fetus-affected gestations: A focus on myogenic and neural differentiations. Cell Biol Int. 2019;43:299–312.
- Dan J, Chen T. Genetic studies on mammalian DNA methyltransferases. Adv Exp Med Biol. 2016;945:123–150.
- Hoang TV, Horowitz ER, Chaffee BR, et al. Lens development requires DNMT1 but takes place normally in the absence of both DNMT3A and DNMT3B activity. Epigenetics. 2017;12:27–40.
- Gifford CA, Ziller MJ, Gu H, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163.
- Song N, Endo D, Song B, et al. 5-aza-2ʹ-deoxycytidine impairs mouse spermatogenesis at multiple stages through different usage of DNA methyltransferases. Toxicology. 2016;361-362:62–72.
- Nomura Y, Hara ES, Yoshioka Y, et al. DNA methylation-based regulation of human bone marrow-derived mesenchymal stem/progenitor cell chondrogenic differentiation. Cells Tissues Organs. 2019;207:115–126.
- Li L, Ling Z, Dong W, et al. Dnmt3a-mediated DNA methylation changes regulate osteogenic differentiation of hMSCs cultivated in the 3D scaffolds under oxidative stress. Oxid Med Cell Longev. 2019;2019:4824209.
- Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277.
- Wade PA. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene. 2001;20:3166–3173.
- Gigek CO, Chen ES, Smith MA. Methyl-CpG-Binding Protein (MBD) family: epigenomic read-outs functions and roles in tumorigenesis and psychiatric diseases. J Cell Biochem. 2016;117:29–38.
- Wang X, Shi J, Cai G, et al. Overexpression of MBD3 improves reprogramming of cloned pig embryos. Cell Reprogram. 2019;21:221–228.
- Defossez PA, Stancheva I. Biological functions of methyl-CpG-binding proteins. Prog Mol Biol Transl Sci. 2011;101:377–398.
- Li H, Radford JC, Ragusa MJ, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A. 2008;105:9397–9402.
- Jung JS, Jee MK, Cho HT, et al. MBD6 is a direct target of Oct4 and controls the stemness and differentiation of adipose tissue-derived stem cells. Cell Mol Life Sci. 2013;70:711–728.
- He Y, Tsou PS, Khanna D, et al. Methyl-CpG-binding protein 2 mediates antifibrotic effects in scleroderma fibroblasts. Ann Rheum Dis. 2018;77:1208–1218.
- Zhao H, Wen G, Huang Y, et al. MicroRNA-22 regulates smooth muscle cell differentiation from stem cells by targeting methyl CpG-binding protein 2. Arterioscler Thromb Vasc Biol. 2015;35:918–929.
- Andoh-Noda T, Akamatsu W, Miyake K, et al. Differentiation of multipotent neural stem cells derived from Rett syndrome patients is biased toward the astrocytic lineage. Mol Brain. 2015;8:31.
- Tao H, Yang JJ, Hu W, et al. MeCP2 regulation of cardiac fibroblast proliferation and fibrosis by down-regulation of DUSP5. Int J Biol Macromol. 2016;82:68–75.
- Hinojosa-Godinez A, Jave-Suarez LF, Flores-Soto M, et al. Melatonin modifies SOX2(+) cell proliferation in dentate gyrus and modulates SIRT1 and MECP2 in long-term sleep deprivation. Neural Regen Res. 2019;14:1787–1795.
- Luo YH, Chen J, Xiao EH, et al. Zebularine Promotes Hepatic Differentiation of Rabbit Bone Marrow Mesenchymal Stem Cells by Interfering with p38 MAPK Signaling. Stem Cells Int. 2018;2018:9612512.
- Liao J, Karnik R, Gu H, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet. 2015;47:469–478.
- Matrisciano F, Tueting P, Dalal I, et al. Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice. Neuropharmacology. 2013;68:184–194.
- Guan Y, Guan X, An H, et al. Epigenetic silencing of miR-137 induces resistance to bicalutamide by targeting TRIM24 in prostate cancer cells. Am J Transl Res. 2019;11:3226–3237.
- Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–4812.
- Szyf M, Bozovic V, Tanigawa G. Growth regulation of mouse DNA methyltransferase gene expression. J Biol Chem. 1991;266:10027–10030.