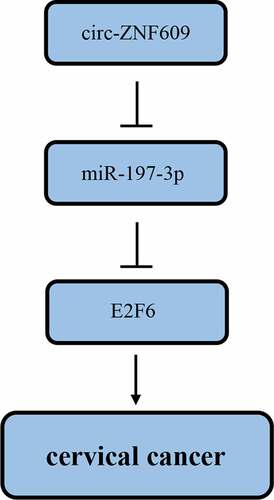
ABSTRACT
Countless studies have demonstrated that Circular RNAs (circRNAs) exert vital effects in regulating tumorigenesis of various cancers. CircRNA ZNF609 (circ-ZNF609) has been reported as an oncogene in various human cancers. Nevertheless, its regulating effect in cervical cancer (CC) remains to be further explored. RT-qPCR was adopted to measure circ-ZNF609, miR-197-3p and E2F6 levels. CC cell proliferation, migration and invasion were analyzed via CCK-8 and transwell assays. Dual-luciferase reporter assay was adopted to confirm the interaction between miR-197-3p and circ-ZNF609 or E2F6. In the present study, it was found that circ-ZNF609 was elevated in CC tissues and cell lines, and circ-ZNF609 deletion repressed cell viability, migration and invasion in CC. Moreover, circ-ZNF609 was identified to negatively regulate miR-197-3p expression in CC cells. The inhibition of miR-197-3p abrogated the inhibitory effect on CC cell proliferation, migration and invasion induced by circ-ZNF609 knockdown. Additionally, we further demonstrated that circ-ZNF609 upregulated E2F6 by interacting with miR-197-3p. Finally, rescue assays indicated that E2F6 overexpression upended the suppression of CC progression induced by circ-ZNF609 deletion. In conclusion, circ-ZNF609 promoted CC progression through modulating the miR-197-3p/E2F6 axis as an oncogene. This finding offers a unique insight into CC molecular mechanism and suggests a potential target for CC therapy.
KEYWORDS:
Introduction
Cervical cancer (CC), one of the most commonly diagnosed carcinomas in women, makes up a large proportion of cancer-relevant death in females around the world. Each year, about 530,000 CC cases are newly diagnosed, and approximately 275,000 of the CC patients are in danger of death, as reported in one survey [Citation1]. Although CC patients can receive effective treatments such as radiotherapy, chemotherapy, and surgical operation [Citation2], long-time survival ratio of CC patients remains unsatisfactory because of recurrence and metastasis [Citation3]. Studies have shown that various pathogenic factors, such as HPVs, early sexual life, early childbirth, and smoking, may lead to CC [Citation4]. However, the specific molecular mechanism of CC is still unclear. Thence, it is important to explore the underlying molecular mechanism in CC tumorigenesis.
Circular RNAs (circRNAs), a new category of non-coding RNAs firstly discovered in cells in the 1970s, are characterized by their lack of 3´ and 5´ ends and closed-loop structure [Citation5,Citation6]. Recently, there is increasing evidence supporting that circRNAs play vital roles in human diseases, including cancers [Citation7,Citation8]. A newly discovered circRNA, circRNA ZNF609 (circ-ZNF609), has been reported to accelerate the carcinogenesis of gastric cancer [Citation9], facilitate cell growth and metastasis in nasopharyngeal carcinoma [Citation10], and contribute to rhabdomyosarcoma progression [Citation11]. However, the potential regulating mechanism of circ-ZNF609 in CC was rarely studied.
MicroRNAs (miRNAs) constitute a large category of small non-coding RNAs with 21 to 23 nucleotides in length. Through interaction with 3’-UTRs of downstream targets, they regulate gene expression at the post-transcriptional level [Citation12]. Previous studies have proved that miRNAs play critical roles in cell proliferation, differentiation, invasion, and apoptosis [Citation13]. miR-197-3p expression is significantly lowered in various cancers, including gastric cancer [Citation14], hepatocellular carcinoma [Citation15], ovarian cancer [Citation16], and prostate cancer [Citation17], suggesting miR-197-3p is a presumptive tumor inhibitor. However, the potential regulating mechanism of miR-197-3p in CC is still unclear.
In this study, we intended to explore the biological role and molecular mechanism of circ-ZNF609 in CC progression and our finding could lay a new theoretical basis for developing targeted CC clinical therapy.
Materials and methods
Clinical samples
Thirty paired CC tissues and corresponding non-tumor tissues were acquired from patients hospitalized at the Third Affiliated Hospital of Soochow University. Then, the samples were processed and preserved in liquid nitrogen immediately till they were used. These specimens were diagnosed by two pathologists via blind review. None of the patients had received any local or systemic treatment. Written informed consent was collected from each patient enrolled. The Ethics Committee of the Third Affiliated Hospital of Soochow University permitted this study.
Cell culture and transfection
Normal human endocervical epithelial cell lines (End1/E6E7) as well as CC cell lines (HeLa, SiHa, HT-3, C-33A, and Ca-Ski) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM supplemented with 5% FBS (Gibco, USA) and 1% penicillin at 37°C with 5% CO2.
miR-197-3p mimics, NC mimics, miR-197-3p inhibitor, NC inhibitor, short hairpin RNA (shRNA) targeting circ-ZNF609 (sh-circ-ZNF609), shRNA negative control (sh-NC), pcDNA3.1-E2F6 overexpression vector (E2F6), and pcDNA3.1 empty vector (pcDNA) were bought from GenePharma (Shanghai, China). Lipofectamine 3000 (Invitrogen) was employed to perform cell transfection.
RT-qPCR assay
With TRIzol reagent (Invitrogen), total RNAs were extracted from CC tissues and cells following the manufacturer’s introductions. Concerning mRNA analysis, the complementary DNA (cDNA) was produced via the Reverse Transcription System Kit (Takara, Dalian, China). Concerning miRNA expression detection, cDNA was synthesized with Takara RNA PCR kit (Takara). Thereafter, SYBR Premix Ex Taq (Takara) was adopted to conduct RT-qPCR assay. By the 2−ΔΔCT method, relative expressions were calculated with GAPDH or U6 as the internal reference.
Cell proliferation
The transfected cells were inoculated onto 96-well plates (1x104 cells/well) and cultured in 100 μl DMEM at 37°C with 5% CO2 for the time indicated (24, 48, and 72 h). Then, 10 μL CCK-8 solution (Dojindo, Japan) was supplemented into each well and the cells were cultured at 37°C for another 2 h. Then, the optical density of each well was measured with a microplate reader at 450 nm (Bio-Rad, CA, USA).
Transwell assay
The cell migrative and invasive properties were analyzed with Transwell chambers possessing an 8-μm-pore polycarbonate membrane (Corning Costar, China) precoated without or with Matrigel (BD Biosciences). In brief, the transfected cells were transferred to the upper chamber containing serum-free DMEM. Then, 500 μL DMEM complete medium was added into the lower chamber. Then, the cells on the low surfaces were fixed with 4% formaldehyde and stained with 0.1% crystal violet following 24 hours’ incubation. At last, we randomly selected five visual fields to calculate the number of migrated and invaded cells with a microscope.
Bioinformatics analysis
To predict interactions between miR-197-3p and circ-ZNF609/E2F6, the StarBase database (http://starbase.sysu.edu.cn/) was employed to screen for possible binding sites.
Dual-luciferase reporter assay
Circ-ZNF609 or E2F6 3’UTR containing wild-type (WT) and mutant (MUT) miR-197-3p sequence was synthesized and cloned into pmirGLO vectors (Promega, Shanghai, China). Next, CC cells were transfected with 10 μg established luciferase reporter vectors combining with miR-197-3p mimics or NC mimics, respectively. Ultimately, the luciferase activity was observed with a dual-luciferase reporter assay kit (Promega).
Tumor xenografts
Male BALB/c nude mice (four-week-old) from Beijing Vital River Laboratory Animal Technology Company were provided and raised under pathogen-free conditions. CC cells stably expressing sh-circ-ZNF609 and sh-NC were injected subcutaneously into each nude mouse. Every 7 days, the volume of tumors was monitored. Twenty-eight days after injection, each nude mouse was sacrificed, and tumors were excised for weighting and further analysis. The Ethics Committee of the Third Affiliated Hospital of Soochow University.
Statistical analysis
Every experiment was performed independently three times. All the data were shown as mean value ± standard deviation (SD). GraphPad Prism 7 software was utilized for statistical analysis. Comparisons among multiple groups or between two groups were analyzed by one-way analysis of variance (ANOVA) or Student’s t-test, respectively. Pearson’s correlation analysis was adopted to perform a correlation study. p < 0.05 indicated a statistical significance.
Results
High Circ-ZNF609 expression in CC tissues and cell lines
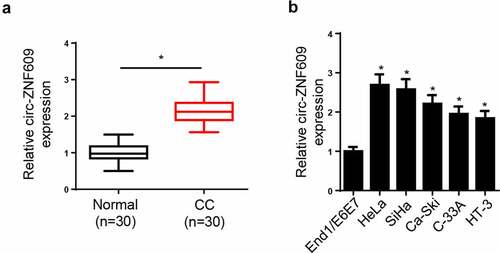
In order to explore the role of circ-ZNF609 in CC, circ-ZNF609 level was measured, and the results showed, relative to normal tissues and cell line, the circ-ZNF609 level was markedly increased in CC tissues and cell lines ( and B). Collectively, the results implied that circ-ZNF609 expression was dysregulated in CC, and circ-ZNF609 high expression might be correlative to CC progression.
Circ-ZNF609 knockdown suppresses malignant biological properties of CC cells
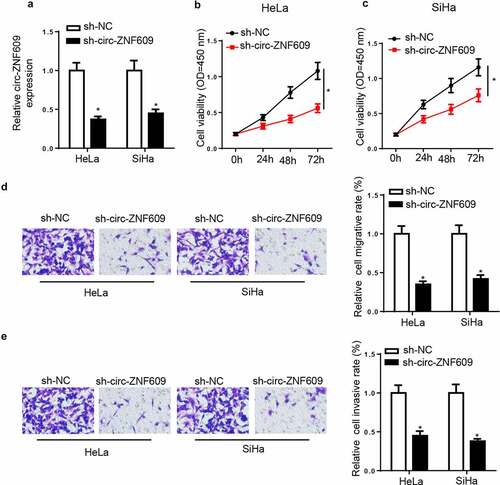
To assess the potential biological effects of circ-ZNF609 in CC progression, sh-circ-ZNF609 or sh-NC were used to transfect CC cells. Then, circ-ZNF609 expression showed an obvious downregulation in CC cells transfected with sh-circ-ZNF609 (). CCK-8 assay results showed circ-ZNF609 knockdown significantly suppressed cell proliferation in CC ( and C). Moreover, transwell experiments uncovered that CC cell migration and invasion were markedly reduced by circ-ZNF609 knockdown ( and E). The above data suggested that circ-ZNF609 knockdown suppressed the biological behaviors of CC cells.
Figure 2. Circ-ZNF609 knockdown suppressed malignant properties of CC cells. HeLa and SiHa cells were transfected with sh-circ-ZNF609 or sh-NC. (a) Circ-ZNF609 expression was detected via RT-qPCR. (b, c) Cell viability was measured by CCK-8 assay in HeLa and SiHa cells. (d, e) The migrated and invaded cells were analyzed with transwell assay. *p< 0.05
Circ-ZNF609 silencing suppressed tumor growth in vivo
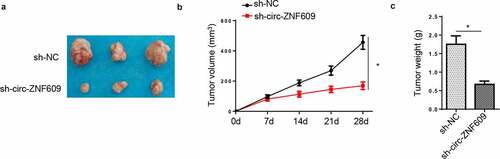
In this section, HeLa cells transfected with circ-ZNF609-silencing plasmids and sh-NC vectors were, respectively, injected into nude rats to assess the functions of circ-ZNF609 in CC tumorigenesis in vivo. Then, the tumors were excised from the above mice. As shown in , tumors harvested from the circ-ZNF609-silencing group were apparently smaller in size than the control group. Moreover, compared with the sh-NC group, tumor volume in the sh-circ-ZNF609 group was also smaller (). In addition, the weight of tumors excised from the sh-circ-ZNF609 group was lowered when compared with the control group (). All results together implied that circ-ZNF609 acted as a tumor-promoter gene in vivo.
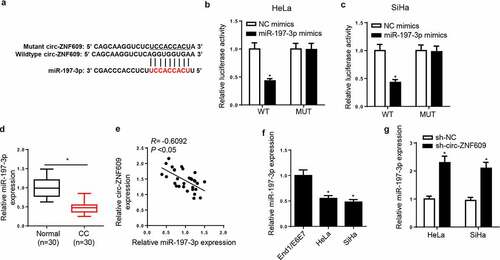
Circ-ZNF609 acts as a sponge for miR-197-3p in CC cells
Via StarBase v2.0 software, we predicted miR-197-3p as a direct target of circ-ZNF609 with a potential binding site (). To verify this prediction, we conducted a dual-luciferase reporter assay and found that miR-197-3p overexpression visibly reduced luciferase activity of circ-ZNF609-WT reporter in CC cells, whereas it had little effect on the circ-ZNF609-MUT reporter ( and C). Afterward, miR-197-3p expression in CC tumors and corresponding adjacent non-tumor tissues were detected by RT-qPCR assay, and a much lower miR-197-3p level was shown in CC tissues relative to adjacent non-tumor tissues (). In addition, miR-197-3p abundance was declined in CC cell lines (). In the meantime, a negative relationship between circ-ZNF609 and miR-197-3p expression was noticed in CC tissues (). Moreover, circ-ZNF609 inhibition was also identified to cause upregulation of miR-197-3p expression (). Based on the above results, we proved that circ-ZNF609 could interact with miR-197-3p and inversely modulate miR-197-3p level in CC cells.
Figure 4. Circ-ZNF609 acted as a sponge for miR-197-3p in CC cells. (a) The binding sites between circ-ZNF609 and miR-197-3p were listed. (b, c) Luciferase activity was analyzed in HeLa and SiHa cells co-transfected with WT-circ-ZNF609+ NC mimic, WT-circ-ZNF609+ miR-197-3p mimic, MUT-circ-ZNF609+ NC mimic, and MUT-circ-ZNF609+ miR-197-3p mimic. (d, e) MiR-197-3p expression was measured via RT-qPCR in CC tissues and cell lines. (f) The correlation between circ-ZNF609 and miR-197-3p was analyzed by Pearson’s correlation analysis. (g) MiR-197-3p expression was assessed via RT-qPCR in HeLa and SiHa cells after transfection with sh-circ-ZNF609 or sh-NC. *p< 0.05
Circ-ZNF609 knockdown suppresses CC progression through binding miR-197-3p
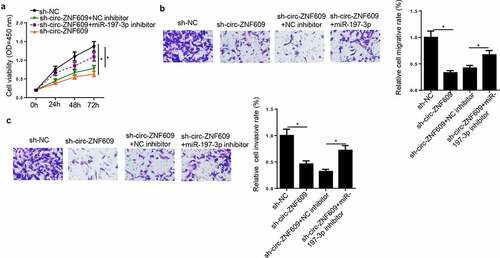
To investigate whether miR-197-3p participated in the promotion of CC progression induced by circ-ZNF609, sh-NC, sh-circ-ZNF609, sh-circ-ZNF609+ NC inhibitor, and sh-circ-ZNF609+ miR-197-3p inhibitor were transfected into HeLa cells, respectively. CCK-8 and transwell assays exhibited that circ-ZNF609 depletion caused suppression on cell viability, migration and invasion (), which could be upended via miR-197-3p reduction. Hence, it was indicated that circ-ZNF609 inhibition downregulated CC progression by binding miR-197-3p.
Figure 5. Circ-ZNF609 knockdown suppressed biological behaviors of CC Cells through binding miR-197-3p. HeLa cells were transfected with sh-NC, sh-circ-ZNF609, sh-circ-ZNF609+ miR-197-3p inhibitor, and sh-circ-ZNF609+ NC inhibitor. (a, b) Cell proliferation was analyzed by CCK-8 assay. (c, d) Transwell assay was adopted to detect cell migration and invasion abilities. *p< 0.05
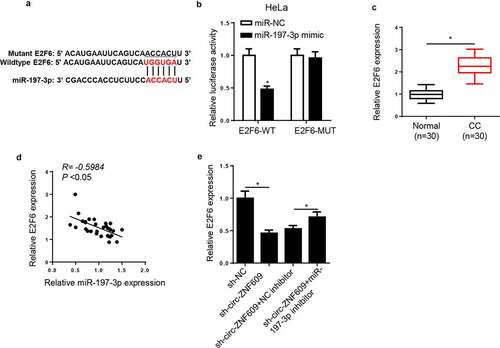
miR-197-3p directly targets E2F6 in CC cells
Next, we screened downstream effectors of miR-197-3p. In line with the prediction provided by StarBase2.0 program, E2F6 may be a downstream target of miR-197-3p in CC cells (). The dual-luciferase reporter assay showed that miR-197-3p high expression obviously decreased the luciferase activity of E2F6-WT reporter, whereas there was almost no change in the luciferase activity of E2F6-MUT reporter in HeLa cells (), indicating that there is an interaction between miR-197-3p and E2F6. Then, E2F6 expression was detected. Compared with the E2F6 level in normal tissues, a significant increase in E2F6 level was showed in CC tissues (). There was an adverse correlation between miR-197-3p and E2F6 expression in CC tissues (). Additionally, the inhibition of miR-197-3p neutralized the suppressing effect of circ-ZNF609 blocking on E2F6 expression (). In sum, circ-ZNF609 positively regulated E2F6 abundance by sponging miR-197-3p.
Figure 6. E2F6 was a target of miR-197-3p in CC cells. (a) The binding sites between E2F6 and miR-197-3p were predicted. (b) Luciferase activity was analyzed in HeLa cells co-transfected with E2F6-WT+NC mimic, E2F6-WT+miR-197-3p mimic, E2F6-MUT+NC mimic, and E2F6-MUT+miR-197-3p mimic. (c) E2F6 expression was measured in CC tissues and paired normal tissues via RT-qPCR. (d) The correlation between E2F6 and miR-197-3p was analyzed by Pearson’s correlation analysis. (e) Via RT-qPCR, E2F6 expression was measured in HeLa cells transfected with sh-NC, sh-circ-ZNF609, sh-circ-ZNF609+ NC inhibitor, and sh-circ-ZNF609+ miR-197-3p inhibitor. *p< 0.05
Circ-ZNF609 knockdown inhibits biological behaviors in CC cells through modulating E2F6
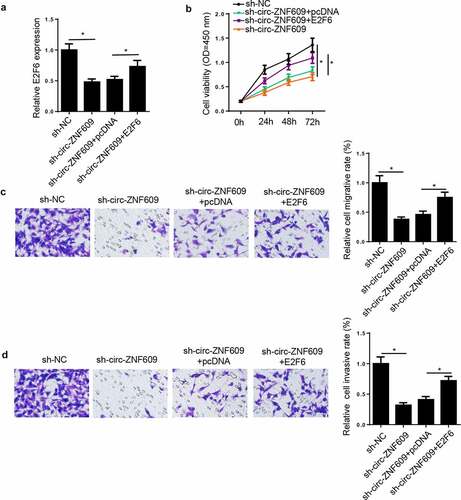
In view of the result that circ-ZNF609 modulated E2F6 expression via miR-197-3p, we presumed that E2F6 might be in relation to the regulation of CC cells induced by circ-ZNF609. To prove this presumption, HeLa cells were transfected with sh-NC, sh-circ-ZNF609, sh-circ-ZNF609+ pcDNA, and sh-circ-ZNF609+ E2F6. Then, we discovered E2F6 expression was inhibited by circ-ZNF609 downregulation and restored by E2F6 upregulation in HeLa cells (). Subsequently, rescue assay indicated that E2F6 overexpression upended the inhibition on HeLa cell proliferation, migration, and invasion () induced by circ-ZNF609 deletion. Therefore, we illustrated that circ-ZNF609 silencing inhibited CC progression via modulating E2F6.
Figure 7. Circ-ZNF609 knockdown inhibited CC progression through modulating E2F6. HeLa cells were transfected with sh-NC, sh-circ-ZNF609, sh-circ-ZNF609+ pcDNA, and sh-circ-ZNF609+ E2F6. (a) E2F6 expression was measured via RT-qPCR. (b) Cell proliferation was analyzed by CCK-8 assay. (c, d) Transwell assay was adopted to detect cell migration and invasion abilities. *p< 0.05
Discussion
Increasing studies showed that circRNAs might participate in multiple biological processes of tumor cells and regulate human cancer progression, including CC development [Citation18]. For instance, circ_0067934 accelerates CC progression by regulating the miR-545/EIF3C pathway [Citation19]. circ_0023404 promotes CC metastasis as a sponge for miR-5047 [Citation20]. Hsa_circ_0000515 upregulates ELK1 through interaction with miR-326, thus contributing to CC progression [Citation21]. All these studies suggested the crucial regulatory function of circRNAs in CC progression.
Circ-ZNF609 was identified to play a vital part in cancer progression. To cite an instance, circ-ZNF609 facilitates hepatocellular carcinoma tumorigenesis by sponging miR-342-3p and increasing PAP2C level [Citation22]. Silenced circ-ZNF609 represses cancerogenesis in nasopharyngeal carcinoma by the miR-188/ELF2 pathway [Citation23]. Circ-ZNF609 expedites renal carcinoma progression via the miR-138-5p/FOXP4 axis as a ceRNA [Citation24]. The above studies suggested the important role of circ-ZNF609 in CC development. Consistent with all these studies, the present research also demonstrated that circ-ZNF609 could accelerate CC progression. In this research, the circ-ZNF609 level was found to be increased in CC, suggesting the important role of circ-ZNF609 in CC tumourigenicity. Then, various assays were conducted, and we found circ-ZNF609 knockdown inhibited CC cell viability, migration, and invasion.
A growing body of studies have proposed that the interaction within circRNA-miRNA-mRNA networks involving ceRNAs exists extensively [Citation25–27]. In such networks, circRNA may compete for binding miRNA, thus modulating the expression of the targeted mRNA. To cite an instance, circ-ATP8A2 acts as a ceRNA to facilitate cell proliferation and invasion in cervical cancer via miR-433/EGFR axis [Citation28]. Circ-FOXM1 targets PPDPF and MACC1 to aggravate the development of non-small cell lung cancer by competitively binding to miR-1304-5p [Citation29]. Circ_0102049 contributes to osteosarcoma cell proliferation and metastasis by the miR-1304-5p/MDM2 axis as a ceRNA [Citation30]. In the present study, it was predicted via the bioinformatics databases that circ-ZNF609 might interact with miR-197-3p. This prediction was confirmed with the dual-luciferase reporter assay. Besides, expression analysis demonstrated circ-ZNF609 knockdown promoted miR-197-3p expression, implying the regulatory relation between circ-ZNF609 and miR-197-3p. Afterward, functional assays were performed, and circ-ZNF609 could repress CC progression via binding miR-197-3p in CC cells.
E2F family is a group of vital regulators in cell progression [Citation31]. As an isoform of the E2F family, E2F6 has been identified as a critical effector in regulating cancer progression [Citation32]. In the present study, abnormal E2F6 expression was also observed in CC. Therefore, E2F6 was speculated and identified as a downstream target of miR-197-3p. Moreover, E2F6 presented a negative correlation with miR-197-3p. Co-transfection analysis confirmed that circ-ZNF609 regulated E2F6 expression in CC cells as a ceRNA for miR-197-3p. In addition, E2F6 overexpression could upend the inhibiting effects on CC progression induced by circ-ZNF609 knockdown.
Conclusion
The present study demonstrated circ-ZNF609 increased in CC tissues and cell lines, and high circ-ZNF609 expression facilitated malignant biological properties of CC cells. Also, we identified the circ-ZNF609/miR-197-3p/E2F6 regulating network in CC development. These findings offer a novel therapeutic target for CC treatment.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Guo H, Yang S, Li S, et al. LncRNA SNHG20 promotes cell proliferation and invasion via miR-140-5p-ADAM10 axis in cervical cancer. Biomed Pharmacother. 2018;102:749–757.
- Han D, Wang J, Cheng G. LncRNA NEAT1 enhances the radio-resistance of cervical cancer via miR-193b-3p/CCND1 axis. Oncotarget. 2018;9(2):2395–2409.
- Gao F, Feng J, Yao H, et al. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):776–782.
- Chen J, Fu Z, Ji C, et al. Systematic gene microarray analysis of the lncRNA expression profiles in human uterine cervix carcinoma. Biomed Pharmacother. 2015;72:83–90.
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388.
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–40.
- Bian L, Zhi X, Ma L, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505(2):346–352.
- Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellula carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):98.
- Liu Z, Pan HM, Xin L, et al. Circ-ZNF609 promotes carcinogenesis of gastric cancer cells by inhibiting miRNA-145-5p expression. Eur Rev Med Pharmacol Sci. 2019;23(21):9411–9417.
- Zhu L, Liu Y, Yang Y, et al. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci. 2019;23(7):2817–2826.
- Rossi F, Legnini I, Megiorni F, et al. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene. 2019;38(20):3843–3854.
- Meng X, Li Z, Zhou S, et al. miR-194 suppresses high glucose-induced non-small cell lung cancer cell progression by targeting NFAT5. Thorac Cancer. 2019;10(5):1051–1059.
- Shao S, Wang C, Wang S, et al. LncRNA STXBP5-AS1 suppressed cervical cancer progression via targeting miR-96-5p/PTEN axis. Biomed Pharmacother. 2019;117(p):109082.
- Niu W, Guo LY, Zhang JY, et al. E2F1-induced upregulation of lncRNA HCG18 stimulates proliferation and migration in gastric cancer by binding to miR-197-3p. Eur Rev Med Pharmacol Sci. 2020;24(19):9949–9956.
- Ni J-S, Zheng H, Huang Z-P, et al. MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol Lett. 2019;17(2):2317–2327.
- Xie W, Shui C, Fang X, et al. miR-197-3p reduces epithelial–mesenchymal transition by targeting ABCA7 in ovarian cancer cells. 3 Biotech. 2020;10(8):375.
- Huang Q, Ma B, Su Y, et al. miR-197-3p Represses the Proliferation of Prostate Cancer by Regulating the VDAC1/AKT/β-catenin Signaling Axis. Int J Biol Sci. 2020;16(8):1417–1426.
- Chaichian S, Shafabakhsh R, Mirhashemi SM, et al. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2019;23(7):718–724.
- Hu C, Wang Y, Li A, et al. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J Cell Physiol. 2019;234(6):9225–9232.
- Guo J, Chen M, Ai G, et al. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed Pharmacother. 2019;115:108957.
- Tang Q, Chen Z, Zhao L, et al. Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging (Albany NY). 2019;11(22):9982–9999. .
- Liao X, Zhan W, Tian B, et al. Circular RNA ZNF609 promoted hepatocellular carcinoma progression by upregulating PAP2C expression via sponging miR-342-3p. Onco Targets Ther. 2020;13:7773–7783.
- Li M, Li Y, Yu M. CircRNA ZNF609 knockdown suppresses cell growth via modulating miR-188/ELF2 axis in nasopharyngeal carcinoma. Onco Targets Ther. 2020;13(p):2399–2409.
- Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2019;234(7):10646–10654.
- Sang M, Wu M, Meng L, et al. Identification of epithelial-mesenchymal transition-related circRNA-miRNA-mRNA ceRNA regulatory network in breast cancer. Pathol Res Pract. 2020;216(9):153088.
- Guan Y-J, Ma J-Y, Song W. Identification of circRNA–miRNA–mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int. 2019;19(1):183.
- Liu Q, Zhang W, Wu Z, et al. Construction of a circular RNA-microRNA-messengerRNA regulatory network in stomach adenocarcinoma. J Cell Biochem. 2020;121(2):1317–1331.
- Ding L, Zhang H. Circ-ATP8A2 promotes cell proliferation and invasion as a ceRNA to target EGFR by sponging miR-433 in cervical cancer. Gene. 2019;705:103–108.
- Liu G, Shi H, Deng L, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;513(1):207–212.
- Jin Y, Li L, Zhu T, et al. Circular RNA circ_0102049 promotes cell progression as ceRNA to target MDM2 via sponging miR-1304-5p in osteosarcoma. Pathol Res Pract. 2019;215(12):152688.
- Hiebert SW, Chellappan SP, Horowitz JM, et al. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 1992;6(2):177–185.
- Li Y, Jiang L, Lv S, et al. E2F6-mediated lncRNA CASC2 down-regulation predicts poor prognosis and promotes progression in gastric carcinoma. Life Sci. 2019;232:116649.