ABSTRACT
In our previous studies, we discovered the congenital cold syndrome (CCS), which is characterized by ‘qi deficiency and qi stagnation, mixed cold and heat.’ And there is a type of syndrome with special incidence characteristic. However, the diagnosis of CCS still lacks an objective basis. In this study, we performed Tandem Mass Tag (TMT) based on quantitative proteomics technology to screen the significantly differentially expressed proteins (DEPs) in serum of patients with coronary heart disease (CHD) patients with CCS, patients with heart and kidney yang deficiency, and healthy people. A total of 22 DEPs (nine upregulated and 13 downregulated) were identified between patients with CCS and healthy subjects. Next, we performed GO and KEGG pathway enrichment analysis, we found the primary functions of DEPs of CCS were binding, catalytic activity, and molecular function regulator. These DEPs were mainly involved in important biological processes, such as cellular process, response to stimulus, localization, metabolic process, and biological regulation. The KEGG analysis revealed that the DEPs showed significant changes in fructose and mannose metabolism, Pentose phosphate pathway, and Arrhythmogenic right ventricular cardiomyopathy. After parallel reaction monitoring (PRM) verification, four upregulated target proteins (ALDOA, PCYOX1, Crisp3 and IGLV4-69) and three downregulated proteins (ALDOC, ADAMTSL-2 and C3) were accurately identified. These proteins were mainly related to immune response and glucose metabolism. These DEPs could be the marker proteins of coronary heart disease with CCS. This findings help to reveal the pathogenesis of CHD with CCS and provide potential therapeutic targets.
Introduction
There has been a gradual increase in the incidence of chronic diseases, such as cardiovascular diseases during the last few decades. Early diagnosis and treatment are known to improve the prognosis of chronic diseases [Citation1]. Health is a dynamic balance between human beings and the natural/social environments. Sub-health is the transition from yin and yang equilibrium to imbalance. Thus, the traditional Chinese medicine (TCM) uses comprehensive adjusting methods to eliminate abnormal and disordered pathological factors [Citation2]. The congenital cold syndrome (CCS, FuHanZ) has been recognized by TCM and is frequently found in people with sub–health as well as in the cases of hyperlipidemia, atherosclerosis, hypertension, coronary heart disease, stroke, diabetes, gastritis, hepatitis, cancer, gynecological diseases, and skin diseases [Citation3]. CCS is defined as cold limbs, especially hands and feet, despite living at a comfortable temperature (23-26°C) [Citation4]. Moreover, it has been observed that the family members of patients with CCS exhibit similar symptoms [Citation5]. The onset of CCS is a long-term process; early detection can help prevent the advancement of CCS [Citation6]. Nevertheless, the diagnosis of CCS is difficult and requires extensive clinical experiences [Citation3,Citation5]. Since there is a lack of objective signs for CCS diagnosis, objective phenotypes and biomarkers are needed.Current research shows that bioinformatics analysis has made a great contribution to the diagnosis and prognosis of various diseases. Udhaya Kumar et al. [Citation7] used the GEO database to identify 7 core genes closely related to familial hypercholesterolemia, indicating a higher risk of atherosclerosis. At the same time, they discovered potential targets for the treatment of diabetes from the obesity and co-morbid diseases database (OCDD) [Citation8]. In addition, Fu et al. [Citation9] analyzed the TCGA database to identify differentially expressed immune-related genes related to lung squamous cell carcinoma (LSCC), which can identify the disease progression and prognosis of patients with LSCC. The same is true of modern proteomics combined with bioinformatics research. It has made important contributions to the modernization of TCM [Citation10]. Previous studies have shown that TCM syndromes, such as kidney deficiency [Citation11], kidney-Yang deficiency [Citation12], spleen deficiency [Citation13], Yang deficiency [Citation14], blood stasis [Citation15], and pulmonary-qi deficiency [Citation16] are associated with significant omics changes. Isobaric relative and absolute quantitative labeling/tandem mass labeling (iTRAQ/TMT) is currently one of the most sensitive techniques used in proteomics, which improves the accuracy and relative quantification of protein identification [Citation12,Citation17]. Bo et al. [Citation18] used TMT quantitative technology to identify differentially expressed proteins (DEPs) in the lung tissue of rats exposed to silica. A study used 2D gel electrophoresis to analyze the changes in protein expression in myocardial ischemia caused by the qi deficiency and blood stasis syndrome [Citation19]. Similarly, the changes in serum protein levels in the phlegm-stasis syndrome were studied using 2D-GE [Citation20]. At present, TMT has not been used in CCS proteomics research.
In this study, TMT was used to detect and analyze DEPs in the serum of CCS patients, and Parallel reaction monitoring (PRM) further verified the results of TMT. To study the key proteins related to the occurrence of CCS and provide new targets for the occurrence, development, diagnosis, and prevention of CCS. We hope that this data set can provide a basis for further mining of CCS disease-specific biomarkers and implementation of early intervention.
Materials and methods
Study design and patients
Patients with heart-kidney yang deficiency (HKYAD, XinShenY) and CCS were recruited from the outpatient department of the Affiliated Hospital of Changchun University of TCM between September 2018 and June 2019. The diagnoses were made by three associate–chief physicians (or above) of TCM. The healthy control group (JianKangR) constituted of hospital workers. The study was approved by the ethics committee of the Changchun University of TCM.
The inclusion criteria were 1) patients with HKYAD and CCS; 2) 35–72 years of age; 3) absence of risk factors for cardiovascular disease, diabetes, hypertension, hyperlipidemia, kidney disease, and digestive system disease, as well as biased symptoms, such as yin and yang, qi and blood loss in the healthy control group; 4) signed voluntary informed consent.
The exclusion criteria were 1) diagnosis of chronic diseases; 2) involved in another clinical trial within 1 month; 3) surgery within 4 weeks; 4) age < 35 y or > 72 y.
Diagnostic criteria of CHD
Referring to ‘2013 ESC guidelines on the management of stable coronary artery disease [Citation21]’, ‘2014 ACC/AHA/AATS/PCNA/SCAI/STS focused update of the Guideline for the Diagnosis Management of Patients with Stable ischemic Heart Disease [Citation22]’ formulated: 1) Coronary angiography diagnosed with one or more coronary arteries with a stenosis of more than 50% of the main branches of the coronary arteries. 2) Have typical symptoms of angina pectoris, with ischemic changes in ECG (or positive treadmill exercise test) during the attack. 3) History of previous myocardial infarction. 4) Coronary artery CT was positive. 5) Positive myocardial nuclide scan. The diagnosis can be made if 1 item is present.
Diagnostic criteria of CCS and HKYAD
The differential diagnosis of CCS was made based on the previous epidemiological studies [Citation3–6]. The major symptoms were cold hands and/or feet, fatigue, sighing, dry mouth, and upset. The secondary symptoms were short of breath, back pain, stomach pain or swelling, preferring warm food over cold food, poor appetite, insomnia, light purplish tongue or red tongue tip with dark red tongue body, thin and white coating on the tongue, and weak pulse. CCS could be diagnosed at approximately 14-year-old in women with dysmenorrhea, approximately 16-year-old men with lower abdominal pain or enuresis, or approximately 35-40-year-old in people of either sex with stomach pain or swelling, poor appetite, detesting cold food, acid regurgitation, and hiccups. The presence of all major symptoms was essential for diagnosis, with at least two items of disease history, and at least two items of secondary symptoms, along with tongue and pulse manifestations.
Diagnostic criteria of deficiency of heart and kidney yang
Referring to the ‘Guiding Principles of Clinical Research on New Chinese Medicines for Coronary Heart Disease’: chest pain, chest tightness, palpitations, shortness of breath, fatigue, pale complexion, cold limbs, loose stools, pale tongue, slippery fur, late or weak pulse.
Diagnosis: the main symptoms of chest pain and tightness could be diagnosed if there were two other symptoms and tongue and pulse support.
Serum collection and protein digestion
We collected the venous blood of three volunteers from each group, followed by centrifugation. Next, we used the Agilent Multiple Affinity Removal Column (Human-14) for multiple extractions to prepare serum after removing albumin, IgG, and other medium-abundance and high-abundance proteins. Then, we used the BCA quantitative method and SDS-PAGE to determine the quantity and quality of the protein in the sample. The samples were stored at −80°C for later use.
TMT labeling
The above qualified serum protein samples were enzymatically digested by Filter-aided proteome preparation (FASP) method [Citation19] and peptide quantification (OD280). Then, the obtained peptides were labeled with the corresponding TMT labeling reagents, and analyzed by high-precision mass spectrometry (Supplementary File 1).
Mass spectrometry data database analysis
We used Proteome Discoverer1.4 (Thermo, America) to perform qualitative and quantitative analysis of the proteomic contents in the serum sample based on the corresponding protein database as previously described [Citation18]. In 3 biological replicates, the average expression rate of two groups was defined as fold changes (FC). DEPs were accounted to be significant using p < 0.05 and fold changes > 1.1 (upregulated) or <0.90 (downregulated) as cutoff criteria.
Bioinformatics analysis
For DEPs, Gene Ontology (GO) analysis (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (https://www.kegg.jp/) and other specific bioinformatics analysis methods were used for further analysis as previously described [Citation23]. GO annotation was used to analyze the DEPs categorized by biological process (BP), molecular function (MF), and cellular component (CC). An in-depth understanding and mining of the specific functions of DEPs in patients with CCS to effectively and accurately find the diagnostic target protein.
PPI network construction
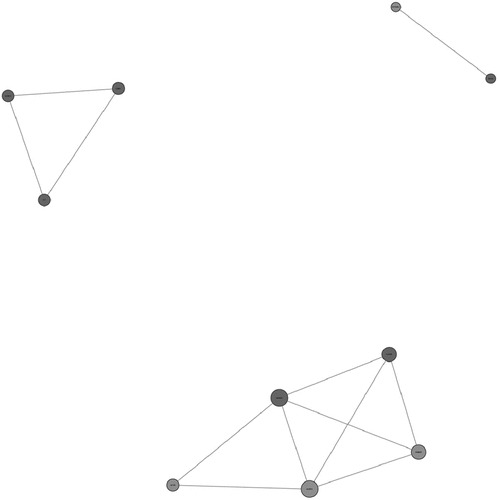
The online database STRING (v11.0, http://www.string-db.org/)was used to visualize the PPIs between the statistically significant DEG-encoded proteins in the resultant dataset [Citation24]. The dataset contained more than 10,000 DEGs. To avoid an inaccurate PPI network, we used a cutoff ≥ 0.9 (highconfidence interaction score) to obtain the significant PPIs. We used Cytoscape software v3.7.1 (http://www.cytoscape.org/) to visualize the PPI network obtained from the STRING database [Citation25]. Based on the log fold change values, the PPI network was plotted for both the upregulated and downregulated DEGs. The interrelation analysis of the identified genes was performed by using the GeneMANIA online tool [Citation26].
PRM verified differential protein
The target proteins were verified by PRM according to the study of Bo et al. [Citation18] Based on the results of TMT, the LC-MS/MS system was used to quantitatively compare the target protein in the DEPs. The mass spectrometry-based PRM technique was used to target and quantify the relevant proteins. The three sets of samples were detected thrice with PRM, and finally, the software Skyline 3.7.0 was used to analyze the original PRM files and quantify the target protein and target peptide.
Statistical analysis
We used the SPSS 20.0 for windows statistical software package (Chicago, IL). The measurement data are expressed as mean ± standard deviation (SD). Under the condition that the test data conformed to the normal distribution and the homogeneity of variance, the independent sample t-test was used to compare the averages of the two sets of samples. If the normal distribution or the homogeneity of variance was not satisfied, a nonparametric test was used. The results are expressed as a P-value and P < 0.05 was considered statistically significant.
Results
Baseline characteristics of the participants
The average age of the three patients in the CCS group was 59.6 ± 5.7 years and included one male and two female subjects. The average age of the three patients in the HKYAD group was 55.2 ± 6.3 years and included two males and one female subject. The average age of the three healthy volunteers was 50.23 ± 4.7 years and included one male and two female subjects. There were no differences in age, gender, blood pressure, heart rate, BMI, total cholesterol, triglycerides, aspartate transaminase, alanine transaminase, red blood cells, and white blood cells, before experiment (all P > 0.05) ().
Table 1. Characteristics of the participants
DEPs screening
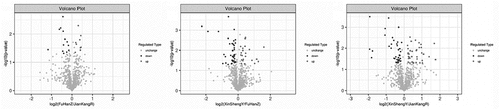
We identified 1058 de–redundant proteins and 21,945 peptides (Supplementary File 2-3) in this study. And we screened the DEPs between the groups based on the expression fold change of 1.1 times or more (upregulation > 1.1-fold or downregulation < 0.90-fold) and P-value < 0.05. showed the histogram of the results of protein quantification and differential analysis (Supplementary File 4). We found the presence of some common or overlapping protein between them in CCS, healthy people and HKYAD. Compared with the patients in the CCS group and HKYAD group, there were more differential proteins, indicating that the two were different in physique or the tendency of future diseases. The fold difference in protein expression between the two groups of samples (fold change > 1.1 times and P value < 0.05) and the P-value obtained by the t-test were used to draw a volcano graph ().
Figure 1. DEPs in the CCS
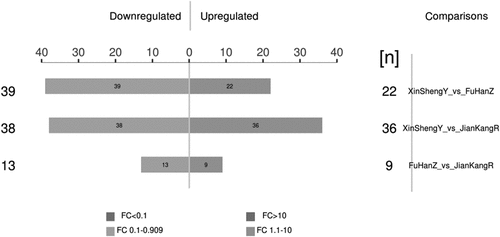
Figure 2. Vocano plot of the DEPs
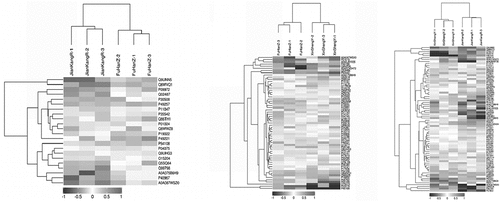
Protein cluster analysis
The hierarchical clustering algorithm (Hierarchical Cluster) was used to cluster the DEPs of the comparison group, and the data is displayed in the form of heat map (). The DEPs obtained by standard screening with a fold change > 1.1 times and P-value < 0.05 (Student t-test or One-way ANOVA) could effectively separate the comparison groups indicating that the DEPs screened in this study were reasonable.
Figure 3. Heatmap of the DEPs
Bioinformatics analysis
We used the Blast2Go (https://www.blast2go.com/) software to annotate GO functions for all proteins identified in this study, followed by Fisher’s exact test to perform GO function enrichment analysis on DEPs ( A-F). The studied proteins were blasted against the online Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://geneontology.org/) to retrieve their KEGG orthology identifications and then mapped to pathways in KEGG. Finally, the DEPs were analyzed by Fisher’s exact test method ( A-F).
Figure 4. Gene Ontology (GO) term enrichment analysis of DEPs
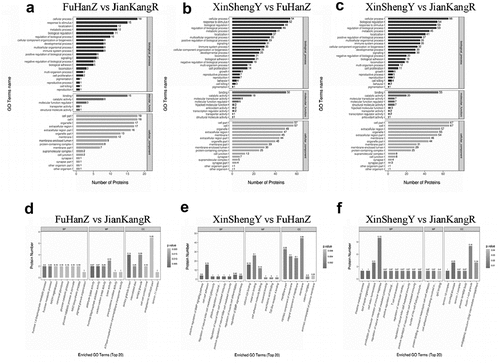
Figure 5. Signaling pathway enrichment analysis of DEPs
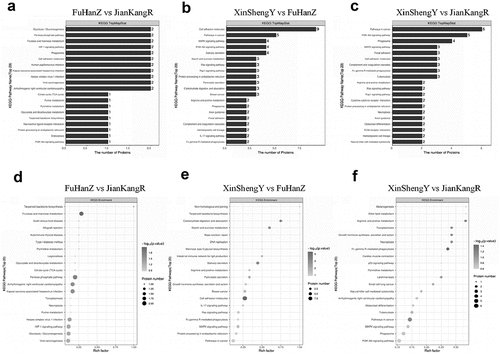
After GO function and KEGG pathway enrichment analysis, we found that the functions of DEPs of CCS were mainly binding, catalytic activity, and molecular function regulation. These DEPs were mainly involved in cellular process, response to stimulus, localization, metabolic process, biological regulation and other important biological processes. Through pathway enrichment analysis, we found that the differential proteins in the CCS group had significant changes in important pathways, such as fructose and mannose metabolism, pentose phosphate pathway, and arrhythmogenic right ventricular cardiomyopathy.
Construction of the PPI Network
To evaluate the PPIs between the DEGs, we used the STRING tool to identify the PPI networks for both the up- and downregulated genes (). A combined score of ≥0.9 for the nodes was considered to indicate a significant PPI interaction.
Validate protein based on PRM technology
We used the targeted PRM technology to detect 22 candidate proteins. The PRM quantitative analysis was performed on eight target peptides of the seven target proteins identified. There were four upregulated proteins (fructose-bisphosphate aldolase A (ALDOA), prenylcysteine oxidase 1 (PCYOX1), cysteine–rich secretory protein 3 (Crisp3), and immunoglobulin lambda variable 4–69 (IGLV4-69)) and three downregulated proteins (fructose-bisphosphate aldolase C (ALDOC), ADAMTS-like protein 2 (ADAMTSL–2), and complement C3 (C3)) in CCS. Supplementary File 5 shows the Skyline analysis results of the target peptide. It contains information, such as peptide chromatographic peaks, original peak areas, and comparison histograms of original peak areas. Supplementary File 6-7 show the quantitative information of the target peptide in the sample. The isotope-relabeled peptides were used to normalize the quantitative information, and then the target peptides and target proteins were quantitatively analyzed. The analysis results showed that there were certain differences in the expression levels of the seven target proteins under six different conditions (Supplementary File 7).
We found that the overall trend of the quantitative results of TMT and PRM was consistent. Therefore, the DEPs verified by screening might be the marker protein of coronary heart disease and CCS.
Discussion
Previous study used 1H-NMR for the comparative analysis between CCS patients with CHD and non-CCS patients [Citation1] and found that there were significant differences regarding energy, lipid, and glucose metabolism between those two groups. Additionally, transcriptomics was used to identify the specific alterations in CCS patients at the gene level, and it was found there were 116 differentially expressed genes between CCS patients and healthy controls [Citation27]. Another study by Wang et al. [Citation28] showed that there were 15 differentially expressed genes between CCS and controls, all involving energy metabolism. A recent study used proteomics to show that the DEPs between CCS patients and controls, were mostly associated with the complement pathway [Citation29]. This study was firstly report on the use of TMT quantitative proteomics technology combined with 2DLC-MS/MS to study CCS as well as the initial screening of the relevant DEPs of CCS. The targeted PRM mode was used to analyze 22 types of proteins. The peptide sequence of the candidate protein was targeted for monitoring. In addition, GO analysis suggested that these DEPs were mainly involved in important biological processes, such as cellular processes, stimulus responses, positioning, metabolic processes, and biological regulation. Through KEGG pathway enrichment analysis, we found that the DEPs in the CCS group were enriched in fructose and mannose metabolism, pentose phosphate pathway, and arrhythmogenic right ventricular cardiomyopathy.
Furthermore, the mass spectrometry results and PRM verification accurately identified seven target proteins. There were four upregulated proteins in CCS (ALDOA, PCYOX1, Crisp3 and IGLV4-69) and three downregulated proteins (ALDOC, ADAMTSL-2 and C3). The analysis of differential protein expression between CCS and healthy people and between HKYAD and healthy people, we identified some crossover protein. Among them, IGLV4-69 was found to be upregulated and ALDOC was downregulated, indicating that there were certain commonalities in the occurrence and development of the disease between CCS and HKYAD. ALDOA and PCYOX1 were downregulated in the analysis of HKYAD and CCS group, indicating that these two were different in physique or disease tendency.
Aldolase (ALD) is widely found in various tissues. Both ALDOA and ALDOC are involved in glycolysis [Citation30]. ALD has a strong binding affinity with various proteins, including actin, α-tubulin, light-chain dynein, Violet syndrome protein, phospholipase D (PLD2), glucose transporter GLUT4, inositol triphosphate, V-ATPase, and ARNO (guanine nucleotide exchange factor of ARF 6). These associations are probably related to cell structure, and functions involving cell endocytosis, parasite invasion, cytoskeletal rearrangement, cell movement, membrane protein transport and recycling, signal transduction and tissue separation [Citation31,Citation32]. Some studies have shown that the increased expression of ALD 1A is a key factor leading to mitochondrial dysfunction and acute myocarditis [Citation33]. This study revealed that the DEPs in the CCS group included ALDOA and ALDOC, which might act as important targets for CCS.
PCYOX1 is a low-density lipoprotein pro-oxidase [Citation34]. The oxidized low-density lipoprotein (oxLDL) is known to be the most characteristic autoantigen in atherosclerosis. The pro-inflammatory properties of oxLDL are known to be closely related to oxidative modification involving enzymes that lead to the release of pro-inflammatory phospholipids and lipid peroxides. Also, oxLDL is formed in the body and induces a humoral immune response [Citation35]. Enhancing the immune system’s response to oxLDL can slow down the progression of atherosclerosis [Citation36]. The increased activity of PCYOX1 might cause an increase in the levels of hydrogen peroxide, which might help spread the oxidation of LDLs, making it a potential pharmacological target and new biomarker for cardiovascular disease.
Crisp-3 is a glycoprotein belonging to the family of Crisps. Elevated expression of Crisp-3 mRNA was originally found in human salivary glands, pancreas, and prostate [Citation37]. Bjartell et al. [Citation38] also detected Crisp-3 protein in human tissue fluids, such as saliva, sweat, blood and seminal plasma. Pathak et al. [Citation39] reported that the increase in CRISP-3 levels was closely related to the occurrence of prostate tumors. This is the first study to identify Crisp-3 as a DEPs of coronary heart disease and CCS. Human Crisp-3 exists in exocrine fluid and the secretory granules of neutrophils and is believed to play a role in innate immunity. GO function analysis found that it was mainly involved in defense response and innate immune response and other biological processes. Thus, we hypothesized that Crisp-3 probably participated in the occurrence of arteriosclerosis through immune response.
Immunoglobulin (Ig) is a glycoprotein. Most natural Ig monomers (molecular weight approximately 150 kDa) are composed of two light (L) chains and two heavy (H) chains. Both the light chain and the heavy chain contain a variable region (V, at the N-terminus) as well as a constant region (C, at the C–terminus) [Citation40]. Ig light chains are divided into κ and λ chains. In humans, approximately 40% of Ig light chains in serum are λ type, indicating that these chains play an important role in antibody response [Citation41]. Clinically, autoimmune diseases, such as systemic lupus erythematosus (SLE) often occur with AS. There are differences in V lambda gene expression between SLE patients and normal people [Citation42]. The differential protein Ig λ variable 4-69 was found in the CCS group and was mainly involved in the biological processes of adaptive immune response, immune response, and immunoglobulin production. It is known to exist in the extracellular space and plasma membrane of the cell. Its effect on arteriosclerosis might also be achieved by participating in the immune response.
The ADAMTS superfamily includes 19 secreted metalloproteases and seven ADAMTS-like proteins, each of which is the product of different genes. They are extracellular matrix proteins with a wide range of activities and functions. ADAMTSL2, is a secreted protein that resembles the ancillary domains of the ADAMTS proteases, but with distinct structural features [Citation43]. The cDNA cloning of ADAMTSL2 and the complete presumptive primary structure of ADAMTSL2 was first reported by the Kazusa DNA Institute as the KIAA0605 gene during a large-scale study to identify genes expressed in human brain [Citation44]. Adamtsl2 is known to be primarily expressed in musculoskeletal tissues, such as tendons and muscles [Citation45]. In the past, only a few studies have investigated the role of ADAMTSL2 in cardiovascular diseases. However, in recent years, there have been many studies on ADAMTS family members, which are closely related to diseases. This has led to the identification of some important pathophysiological processes, such as collagen processing (ADAMTS 2, 3, 14), matrix proteoglycan degradation (ADAMTS 4, 5), inhibition of the formation of blood vessels (ADAMTS1, 8), maintenance of a stable coagulation environment (ADAMTS13), etc. [Citation46,Citation47].
Here, we found that C3 was downregulated in CCS compared with the healthy people, and C3 participated in multiple metabolic pathways, among which complement and coagulation cascades were closely related to the occurrence of coronary heart disease. The complement system is known to participate in specific and nonspecific immune responses. Inflammation is closely associated with heart coronary diseases, and immune response is a vital initiation factor for inflammation. Therefore, the activation of the complement system and the expression of complement factors are related to the genesis and development of coronary disease [Citation28]. Additionally, increasing number of studies have indicated that complement factors are involved in the initiation, development, and the whole process of atherosclerosis [Citation48]. The mild form of atherosclerosis found in patients with systemic lupus erythematosus (SLE) was also found to be associated with altered expression of C3 and C4 [Citation49]. A study found that the vascular tonus could be adjusted by interactions between C3 and C4 to collagens, which was related to the pathogenesis of atherosclerosis [Citation50]. In this study, the level of C3 in Fuhan Syndrome was relatively down-regulated. The possible reason is that patients with stable angina pectoris in CHD may be in the early stage or stable disease.
Most of the DEPs found in this study were related to immune response, which could regulate the occurrence and development of atherosclerosis. Innate immunity is the natural immune defense function formed by the body in the process of germline development and evolution, i.e., the nonspecific defense function that is already present after birth, also known as nonspecific immunity. This is similar to the clinical manifestations of patients with CCS. We found that at the beginning of ‘male and female conjugation,’ the congenital yang qi inherited from the blood of the father’s sperm and mother’s blood was insufficient, and the yin qi was excessive, causing the yin and cold to fall inward. At the beginning of the embryo, the yin and yang of the human body were in a biased state, constructing the nature of congenital cold syndrome. It also determined the law of future disease occurrence and development. Therefore, we speculated that the DEPs exhibited by patients with CCS could explain their unique clinical manifestations.
Additionally, this study had several limitations. The sample size was small and from a single center. Also, only one omics was used and thus, additional studies are necessary to identify the mechanisms involved in CCS. Additional studies are also necessary to test the identified proteins and determine their diagnostic value.
Conclusion
In conclusion, we identified the DEPs related to immune response and glucose metabolism in serum of CCS patients, suggesting that these processes were probably involved in the occurrence and development of congenital cold. Therefore, the DEPs verified by screening might act as the marker proteins of CHD and CCS, and provide a reference for the screening of protein markers of CHD and CCS.
Highlights
Four upregulated target proteins (ALDOA, PCYOX1, Crisp3 and IGLV4-69) and three downregulated proteins (ALDOC, ADAMTSL-2 and C3) were accurately identified in CCS patients.
The primary functions of DEPs of CCS were binding, catalytic activity, and molecular function regulator.
The DEPs showed significant changes in fructose and mannose metabolism, Pentose phosphate pathway, and Arrhythmogenic right ventricular cardiomyopathy.
Abbreviations
CCS congenital cold syndrome
CHD coronary heart disease
DEPs differentially expressed proteins
TMT Tandem Mass Tag
TCM traditional Chinese medicine
HKYAD Patients with heart-kidney yang deficiency
GO Gene Ontology
KEGG Kyoto Encyclopedia of Genes and Genomes
PRM parallel reaction monitoring
ALDOA fructose-bisphosphate aldolase A
PCYOX1 prenylcysteine oxidase 1
Crisp3 cysteine-rich secretory protein 3
IGLV4-69 immunoglobulin lambda variable 4-69
ALDOC fructose-bisphosphate aldolase C
ADAMTSL-2 ADAMTS-like protein 2
C3 complement 3.
Acknowledgements
Thanks to the National Natural Science Foundation of China for funding this project. Thanks to Shanghai applied protein Technology Co., Ltd. for its technical support for this project.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Yan YX, Liu YQ, Li M, et al. Development and evaluation of a questionnaire for measuring suboptimal health status in urban Chinese. J Epidemiol. 2009;19(6):333–341.
- Liao Y, Lin Y, Zhang C, et al. Intervention effect of baduanjin exercise on the fatigue state in people with fatigue-predominant subhealth: a cohort study. J Altern Complement Med. 2015;21(9):554–562.
- L J. Theories about Congenital Fu-Han. J Chin Med. 2014;29:41–43.
- Yang L, Wang M, Wu W, et al. Transcriptome analysis of cold syndrome using microarray. Am J Chin Med. 2007;35(4):609–620.
- Cui YGJ, Huang Y. Clinical observation for coronary heart disease of congenital Fu-Han: 126 case. Chin Med. 2010;51:516–519.
- Huang YGJ, Deng Y. The theory of Congenital Fu-Han and its significance in clinical practice. J Pract Trad Chin Int Med. 2007;21:3–7.
- Udhaya Kumar S, Thirumal Kumar D, Bithia R, et al. Analysis of differentially expressed genes and molecular pathways in familial hypercholesterolemia involved in atherosclerosis: a systematic and bioinformatics approach. Front Genet. 2020;11:734.
- Udhaya Kumar S, Bithia R, Thirumal Kumar D, et al. Involvement of essential signaling cascades and analysis of gene networks in diabesity. Genes (Basel). 2020;11. 10.3390/genes11111256
- Fu D, Zhang B, Yang L, et al. Development of an immune-related risk signature for predicting prognosis in lung squamous cell carcinoma. Front Genet. 2020;11:978.
- Wang P, Chen Z. Traditional Chinese medicine ZHENG and Omics convergence: a systems approach to post-genomics medicine in a global world. Omics. 2013;17(9):451–459.
- Shen ZY, Huang JH, Chen WH. Comparison of gene expression profiles between rats of Shen deficiency syndrome and Shen-yang deficiency syndrome differentiated according to therapeutic efficacy of drugs used. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2007;27(2):135–137.
- Ni HMWY, He YM. Differential expression genes in teenagers with kidney-yang asthenia constitution by genechip technique. Shanghai J Trad Chin Med. 2004;38:3–5.
- Wang SCX, Zou SJ, Gang WJ. Gene expression changes in the cerebral cortex of reserpine-induced mice model of spleen deficiency syndrome. Chinese Archives of Traditional Chinese Medicine. 2003;21:1512–1515. .
- Ding SP, Fang ZQ, L U W-L, et al. Specially up-regulated and highly expressed genes in thymus of the H_(22) tumor-bearing mice with pathogenic toxin-exuberance syndrome. 2010;30(04):353–357.
- Ma XJ, Yin HJ, Chen KJ. Differential gene expression profiles in coronary heart disease patients of blood stasis syndrome in traditional Chinese medicine and clinical role of target gene. Chin J Integr Med. 2009;15(2):101–106.
- Li ZG, Tong JB, Peng B. [Preliminary study on differential expressed genes in T lymphocyte of chronic obstructive pulmonary disease patients with Fei-qi deficiency syndrome. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2006;26(12):1082–1085.
- Yang YHLZ. Study on brain gene chip of mouse model with deficiency of kidney-yang syndrome. China J Trad Chin Med Pharm. 2008;23:396–401.
- Bo C, Geng X, Zhang J, et al. Comparative proteomic analysis of silica-induced pulmonary fibrosis in rats based on tandem mass tag (TMT) quantitation technology. PLoS One. 2020;15(10):e0241310.
- Wang Y, Chuo WJ, Li C, et al. Energy metabolism disorder and myocardial injury in chronic myocardial ischemia with Qi deficiency and blood stasis syndrome based on 2-DE proteomics. Chin J Integr Med. 2013;19(8):616–620.
- Liu JL, Song JN, Yan L. Differential plasma protein profiles in patients with hyperlipidemia & atherosclerosis of different patterns of phlegm-stasis syndrome. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2010;30(5):482–487.
- Montalescot G, Sechtem U, Achenbach S, et al. ESC guidelines on the management of stable coronary artery disease: the Task Force on the management of stable coronary artery disease of the European Society of Cardiology. Eur Heart J. 2013;34:2949–3003.
- Fihn SD, Blankenship JC, Alexander KP, et al. ACC/AHA/AATS/PCNA/SCAI/STS focused update of the guideline for the diagnosis and management of patients with stable ischemic heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines, and the American Association for Thoracic Surgery, Preventive Cardiovascular Nurses Association, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. Circulation. 2014;130(19):1749–1767.
- Li M, Wu M, Qin Y, et al. Differentially expressed serum proteins in children with or without asthma as determined using isobaric tags for relative and absolute quantitation proteomics. PeerJ. 2020;8:e9971.
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–52.
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504.
- Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46:W60–w64.
- Z. W. Transcriptomic studies to Congenital Fu-Han. Changchun: PhD Thesis of Changchun Chinese Medicine University. 2013
- L W. Complement system and cardiac I-R injury. Int Med Anethiology. 2001;22:340–368.
- Lh. J. Serum proteomics study of people with “congenital fusiform cold syndrome” caused by the etiology of “congenital fuyu cold. Changchun University of Traditional Chinese Medicine; 2015.
- Walther EU, Dichgans M, Maricich SM, et al. Genomic sequences of aldolase C (Zebrin II) direct lacZ expression exclusively in non-neuronal cells of transgenic mice. Proc Natl Acad Sci U S A. 1998;95(5):2615–2620.
- Merkulova M, Hurtado-Lorenzo A, Hosokawa H, et al. Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution. Am J Physiol Cell Physiol. 2011;300(6):C1442–55.
- Rangarajan ES, Park H, Fortin E, et al. Mechanism of aldolase control of sorting nexin 9 function in endocytosis. J Biol Chem. 2010;285(16):11983–11990.
- Choi S, Chung JH, Nam MH, et al. Elevated aldolase 1A, retrogene 1 expression induces cardiac apoptosis in rat experimental autoimmune myocarditis model. Can J Physiol Pharmacol. 2020;98(6):373–382.
- Herrera-Marcos LV, Lou-Bonafonte JM, Martinez-Gracia MV, et al. Prenylcysteine oxidase 1, a pro-oxidant enzyme of low density lipoproteins. Front Biosci (Landmark Ed). 2018;23(3):1020–1037.
- Palinski W, Rosenfeld ME, Ylä-Herttuala S, et al. Low density lipoprotein undergoes oxidative modification in vivo. Proc Natl Acad Sci U S A. 1989;86(4):1372–1376.
- Freigang S, Hörkkö S, Miller E, et al. Immunization of LDL receptor-deficient mice with homologous malondialdehyde-modified and native LDL reduces progression of atherosclerosis by mechanisms other than induction of high titers of antibodies to oxidative neoepitopes. Arterioscler Thromb Vasc Biol. 1998;18(12):1972–1982.
- Krätzschmar J, Haendler B, Eberspaecher U, et al. The human cysteine-rich secretory protein (CRISP) family. Primary structure and tissue distribution of CRISP-1, CRISP-2 and CRISP-3. Eur J Biochem. 1996;236(3):827–836.
- Bjartell AS, Al-Ahmadie H, Serio AM, et al. Association of cysteine-rich secretory protein 3 and beta-microseminoprotein with outcome after radical prostatectomy. Clin Cancer Res. 2007;13(14):4130–4138.
- Pathak BR, Breed AA, Nakhawa VH, et al. Growth inhibition mediated by PSP94 or CRISP-3 is prostate cancer cell line specific. Asian J Androl. 2010;12(5):677–689.
- Schroeder HW Jr., Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010;125(2):S41–52.
- Lefranc MP. L G: The immunoglobulin facts book. suffolk: Academic Press; 2001.
- Trevisan GL, Tamia-Ferreira MC, Junta CM, et al. Immunoglobulin V-lambda transcription profiling of systemic lupus erythematosus patients reveals biased usage of genes located near the Jlambda-Clambda segments. Scand J Immunol. 2004;59(4):395–399.
- Koo BH, Le Goff C, Jungers KA, et al. ADAMTS-like 2 (ADAMTSL2) is a secreted glycoprotein that is widely expressed during mouse embryogenesis and is regulated during skeletal myogenesis. Matrix Biol. 2007;26(6):431–441.
- Nagase T, Ishikawa K, Miyajima N, et al. Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5(1):31–39.
- Hubmacher D, Taye N, Balic Z, et al. Limb- and tendon-specific Adamtsl2 deletion identifies a role for ADAMTSL2 in tendon growth in a mouse model for geleophysic dysplasia. Matrix Biol. 2019;82:38–53.
- Tang BL. ADAMTS: a novel family of extracellular matrix proteases. Int J Biochem Cell Biol. 2001;33(1):33–44.
- Porter S, Clark IM, Kevorkian L, et al. The ADAMTS metalloproteinases. Biochem J. 2005;386(1):15–27.
- Carter AM. Complement activation: an emerging player in the pathogenesis of cardiovascular disease. Scientifica (Cairo). 2012;2012:402783.
- Parra S, Vives G, Ferré R, et al. Complement system and small HDL particles are associated with subclinical atherosclerosis in SLE patients. Atherosclerosis. 2012;225(1):224–230.
- Shields KJ, Stolz D, Watkins SC, et al. Complement proteins C3 and C4 bind to collagen and elastin in the vascular wall: a potential role in vascular stiffness and atherosclerosis. Clin Transl Sci. 2011;4(3):146–152.