ABSTRACT
There may be a mutually reinforcing relationship between hepatocellular carcinoma (HCC) and depression, but the mechanism is unknown. This study used bioinformatics to evaluate the relationship between HCC and depression at the genetic level. Genes associated with HCC and depression were obtained from pubmed2ensemble. Overlapping genes were annotated by gene ontology (GO) function and enriched by Kyoto Encyclopedia of Genes and Genomes (KEGG) signal pathway. The cluster-1 genes obtained by Cytoscape were analyzed by GEPIA for expression and overall survival in HCC and, finally, introduced target genes to DGIdb to get associated drugs. A total of 199 genes were found to be in common between HCC and depression. GO term enrichment analysis on DAVID found the top-6 biological processes to be mainly associated with cell death and apoptosis. The top-6 cellular component terms are extracellular. The top-6 of molecular function terms are mainly associated with receptor binding. The top-6 pathways enriched by KEGG are mainly related to inflammatory response. IGF1, VEGFA, and SERPINE1 had statistical differences in expression and 10-year survival rate. There are total 45 drugs that act on VEGFA and SERPINE1. Based on our findings, we hypothesize that the mechanism of the interaction between HCC and depression may be related to cell death or apoptosis. Further studies are needed to verify this hypothesis.
Introduction
Hepatocellular carcinoma is the third most common cause of cancer deaths worldwide [Citation1]. Due to various factors, including economic development and environmental factors, incidence is rapidly increasing across all age groups [Citation2]. Hepatitis B, hepatitis C, aflatoxin exposure, alcohol consumption, and smoking are HCC risk factors [Citation3]. Recent studies suggest that HCC is also associated with emotional and psychological factors. Numerous studies have reported high depression incidence in cancer patients. A Taiwanese prospective study involving 128 newly diagnosed HCC patients found that advanced Barcelona Clinic liver cancer stage, radiofrequency ablation therapy, or liver resection was most closely associated with worsening physical and psychological symptoms over time [Citation4]. Anxiety disorders and depression have also been associated with a significantly increased overall survival rate in HCC patients in a retrospective study involving 7304 patients treated for HCC from 1996 to 2010 [Citation5]. Thus, clinical professionals should pay attention to physical and psychological factors during treatment.
Depression is characterized by physical symptoms like fatigue, pain, or sleep disorder. Symptoms of depression may or may not occur [Citation6]. Numerous studies have associated depression with a variety of chronic diseases, including cancer, coronary heart disease, and diabetes. The relationship between depression and cancer risk is characterized by two aspects: on one hand, depression may increase risk of some cancers [Citation7,Citation8] and, on the other, depression and other mental disorders may accelerate deterioration in cancer patients. It is reported that somatic depressive symptoms are similar in depression cancer patients and depression patients without chronic somatic symptoms and that depression cancer patients show greater physical depression symptoms than depression patients without chronic somatic symptoms [Citation9]. The emergence of high levels of somatic symptoms should prompt clinicians to investigate potential complications in cancer patients. Due to the high prevalence of depressive symptoms and their impact on survival, psychological and drug intervention should be designed and implemented for patients diagnosed with liver cancer [Citation10]. Thus, further study of relationship between HCC and depression will greatly improve prevention and treatment of the disease.
Advances in information technology and interdisciplinary research have led to increased use of bioinformatics in clinical research. For example, with the rapid development of sequencing technology, the field of gene detection has been redefined based on the next-generation sequencing (NGS) and due to the complexity of resulting datasets, bioinformatics has become a necessary tool in clinical NGS testing [Citation11]. As for the treatment of cancer, more and more patients are being targeted by the detection of drug resistance genes. Thanks to the development of bioinformatics, researchers can design more small molecule targeted drugs to target gene therapy.
The potential mechanisms underlying HCC and depression are unknown. Here, we applied bioinformatics to identify common genes between HCC and depression. Genes associated with HCC and depression were identified on Pubmed2ensemble and those in common between the two subjected to GO and KEGG pathway analyses. PPI network analysis was used to identify core gene clusters, in order to provide more evidence for a relationship between HCC and depression at the genetic level.
Materials and methods
Acquisition of HCC and depression genes
Pubmed2ensemble data sources (http://pubmed2ensembl.ls.manchester.ac.uk/) are a customized and extended version of the Ensembl BioMart on genes. Input our target disease ‘hepatic carcinoma’ and ‘depression’ separately, choose ‘retrieve up to 100,000 document IDs, and select Homo sapiens genes (GRCh37) to get the gene set of related keywords.
Acquisition of overlapping genes between HCC and depression
Through the online tool: Venny2.1 (https://bioinfogp.cnb.csic.es/tools/venny/), genes related to HCC were put into List1, while related genes of depression were filled in List2. Then, we acquired the overlapping genes between them.
Functional and signal pathway enrichment analysis
Intersection gene obtained by Venny was introduced into DAVID 6.8 (https://david.ncifcrf.gov/), The Database for Annotation, Visualization and Integrated Discovery and analyzed using the parameters: OFFICIAL-GENE-SYMBOL, species: Homo sapiens. GOTERM_BP_FAT, GOTERM_CC_FAT, and GOTERM_MF_FAT were downloaded after GO and KEGG_PATHWAY analyses. BP, CC, and MF refer to biological process, cellular component, and molecular function, respectively.
Protein–protein interaction (PPI) analysis and gene module analysis
Intersection genes were analyzed on STRING (https://string-db.org) to create an interaction network using the settings: multiple proteins, species: Homo sapiens, minimum required interaction score: 0.900, hide disconnected nodes in the network, and download PPI network from TSV file. The TSV file was imported into the Cytoscape, and the MCODE plugin was used to analyze core functional groups of the PPI network. MCODE can be used to construct functional modules in large gene (protein) networks. Cluster-1, obtained after clustering, had 26 core genes.
Expression of cluster-1 gene in HCC and its effect on survival rate
The 26 genes were inputted into GEPIA (http://gepia.cancer-pku.cn/), which can analyze the expression of related genes in specific tumors and their effects on survival rate. First, we analyzed expression of the 26 genes in HCC and set the parameters: Log2FC cutoff: 1; p-value cutoff: 0.01. Then, we analyzed overall survival, group cutoff selected median, cutoff-high (%): 50; cutoff-low (%): 50. Expression of the genes was shown in boxplots and survival plots for HCC. Target genes for next analysis were brought together, which were both statistically significant between expression and survival rates.
Drug screening
Genes with statistically significant expression differences and genes with statistically significant survival rates were introduced into DGIdb (https://www.dgidb.org/search_interactions), which was used to search for drug–gene interactions. Entered the target genes, IGF1, VEGFA, and SERPINE1, preset Filters selected “Approved”.
Results
This study aimed to uncover the possible mechanism underlying the interaction between HCC and depression using bioinformatics. From our findings, we hypothesize that interaction between the two may exist in cell death/apoptosis. We also searched for drugs that can act on core genes and which may guide further research.
Common gene in HCC and depression
Through the gene2ensemble database, 2586 HCC-associated genes and 382 depression-associated genes were identified, with 199 genes common between HCC and depression.
Functional and signal pathway enrichment analysis
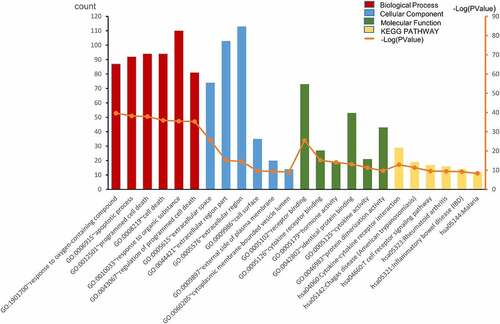
Based on GO enrichment analysis on DAVID, the top-6 biological processes were response to oxygen-containing compound, apoptotic process, programmed cell death, cell death, response to organic substance, and regulation of programmed cell death. The top-6 cellular component terms were extracellular space, extracellular region part, extracellular region, cell surface, external side of plasma membrane, and cytoplasmic membrane-bounded vesicle lumen. The top-6 molecular function terms were receptor binding, cytokine receptor binding, hormone activity, identical protein binding, cytokine activity, and protein dimerization activity. The top-6 KEGG pathways were cytokine–cytokine receptor interaction, Chagas disease (American trypanosomiasis), T cell receptor signaling pathway, rheumatoid arthritis, inflammatory bowel disease (IBD), and malaria ().
Protein–protein interaction analysis and gene module analysis
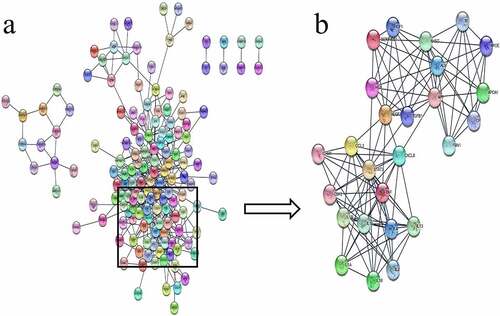
STRING analysis of the 199 common genes identified a PPI network comprising 148 points and 473 edges ()). In this network, the most prominent gene modules, cluster-1, was clustered using MCODE plugin on Cytoscape, with a total of 26 genes ()).
Expression of cluster-1 genes in HCC and its effect on survival rate
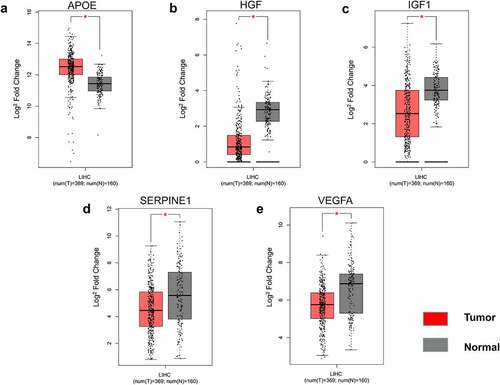
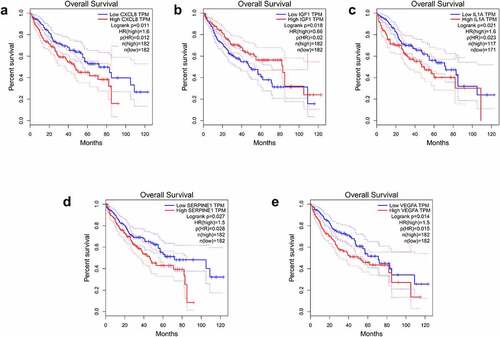
The 26 genes in cluster-1 were individually subjected to GEPIA analysis and disease was selected as liver hepatocellular carcinoma. This analysis identified IGF1, APOE, HGF, VEGFA, and SERPINE1 as differentially expressed in the HCC vs normal group (), with IGF1, HGF, VEGFA, and SERPINE1 poorly expressed and APOE highly expressed. The differences in 10-year survival rates were IGF1, IL1A, CXCL8, VEGFA, and SERPINE1 (). Among them, IGF1, VEGFA, and SERPINE1 had statistical differences in both expression and 10-year survival rate.
Drug selection
Through the DGIdb database, we screened 45 drugs for interaction with IGF1, VEGFA, and SERPINE1. No drugs with interaction with IGF1 were identified, while 26 drugs interact with VEGFA and 19 with SERPINE1 ().
Table 1. Drugs acting on VEGFA and SERPINE1
Discussion
In recent decades, hepatocellular carcinoma (HCC) incidence has increased in many countries. HCC is the main histological type of liver cancer and accounts for most liver cancer deaths [Citation12]. Depression is a highly common mental disorder, and many patients do not respond adequately to existing treatments [Citation13]. Chronic or early life stress is one of the key risk factors for depression. Epidemiological investigation found that the prevalence of depression in cancer patients is higher than that in the general population [Citation14]. Cancer incidence is higher in patients with depression, and depression is linked to the increase in cancer-specific mortality, which is consistent with findings that depression and anxiety were significantly associated with increased risk of cancer (adjusted RR: 1.13, 95% CI: 1.06–1.19), cancer-specific mortality (1.21, 1.16–1.26), and all-cause mortality in cancer patients (1.24, 1.13–1.35) [Citation15]. The estimated absolute risk increases (ARIs) associated with depression and anxiety were 34.3 events/100,000 person years (15.8–50.2) for cancer incidence and 28.2 events/100,000 person years (21.5–34.9) for cancer-specific mortality. However, so far, we have seen more clinical observational and epidemiological survey data. The genetic mechanism of the interaction between HCC and depression has not been determined. Here, we used bioinformatics to identify genes that are co-expressed in HCC and depression and to identify the biological process, cellular component, molecular function, and signaling pathways involved. Using DGIdb database, we identified drugs that may be used in both.
Our data show that HCC and depression share 199 genes. GO enrichment analysis shows that the top-6 biological processes are response to oxygen-containing compound, apoptotic process, programmed cell death, cell death, response to organic substance, and regulation of programmed cell death. Four of these are related to cell death/apoptosis. We hypothesize that before depression is apparent, the biological mechanism underlying the interaction between HCC and depression may be associated with cell death or apoptosis. Most anticancer therapies trigger apoptosis [Citation16,Citation17], and with molecular technology advances, anticancer drugs have targeted apoptosis with increasingly higher specificity. Exposition of pathophysiology of diseases from the gene level may better provide strategies for the development of new cancer treatments [Citation18]. Li et al. proved experimentally that deletion or inactivation of genetic targeting of Survivin, an inhibitor of apoptosis, sensitized HCC cells to TNFα-induced cell death and significantly suppressed human and mouse HCC cells [Citation19]. With regard to depression, we know that brain-derived neurotrophic factor (BDNF) is a growth factor, which influences neuronal survival, growth, and neuroplasticity. The tumor suppressor gene, Pdcd4 is an endogenous inhibitor of BDNF translation. BDNF expression changes are associated with severe depressive disorder. Animal studies show that blocking Pdcd4 in depression promotes BDNF expression [Citation20], indicating that Pdcd4 may be a potential anti-depression target.
Four of the top-6 pathways are related to inflammation. At present, the mechanism of hepatocellular carcinogenesis due to viral hepatitis is clinically recognized [Citation21]. Other studies show that T cells play a role in the immune escape mechanism of liver cancer [Citation22]. For depression, mounting evidence shows a link between depression and inflammatory processes. Clinical trials have shown that the antidepressant effect of anti-inflammatory drugs can be used as an additional treatment or a single treatment, but important challenges remain [Citation23–25].
Cluster-1 genes were inputted into GEPIA database to analyze their expression and 10-year survival rate in HCC. We concluded that IGF1, VEGFA, and SERPINE1 had statistical significance in the expression and 10-year survival rate between hepatocellular carcinoma group and normal group. Although the expression of VEGFA and SERPINE1 is low in HCC compared with normal group, survival rate in low expression group is higher than that in high expression group. This result is based on the statistics of the high and low expression groups of patients with liver cancer, which has nothing to do with normal people, so it is not contradictory. As for depression, histological analysis by Kondo et al. [Citation26] showed that a serotonin type 3 receptor (5HT3R) and IGF1 are expressed in the same neurons in the subgranular region of the dentate gyrus of the hippocampus. 5HT3R regulates extracellular IGF1 levels in the hippocampus, a novel 5HT3R-IGF1 mechanism that differs from fluoxetine-induced responses. This provides a novel therapeutic target for depression, especially in depression patients with selective resistance serotonin reuptake inhibitors. However, genetic association analysis of polymorphisms in 10 genes belonging to the IGF-I system (IGF1, IGF1R, IGFBP1 to IGFBP7, and IGFBPL1) in the largest genetic databases for major depression (Psychiatric Genomics Consortium) revealed nominal associations with susceptibility to depression and treatment response, although results were not remain significant after multiple testing correction [Citation27]. Cui [Citation28] measured the VEGFA mRNA and protein in hippocampus tissues and sera from a rat model of depression and found that they were downregulated in hippocampi and blood of the depressed rats. Our results also showed low VEGFA expression in the tumor group, suggesting a new avenue for the pathology and treatment of HCC complicated by depression. Yi Y’s [Citation29] study shows that SERPINE1 is upregulated in ovarian cancer patients with severe depression and worsens survival, but the mechanism is unknown. In patients with liver cancer complicated by depression, IGF1/VEGFA/SERPINE1 may play an important role in the pathogenesis and development of the disease, but the specific mechanism has not been clarified.
Finally, through the DGIdb database, 45 drugs were identified for VEGFA and SERPINE1. Because no drugs were identified for IGF1, its therapeutic value will be investigated in subsequent studies. The selected drugs can be validated via in vitro studies to provide reference values for clinical practice.
Limitations
This study only explained the mechanism between hepatocellular carcinoma and depression using bioinformatics, and no experimental tests were done on the proposed hypothesis.
Conclusion
In summary, we found the possible mechanism linking HCC and depression. By enriching the biological functions of related genes, we propose that the biological mechanism of the interaction between HCC and depression may be associated with cell death or apoptosis. These bioinformatics findings need experimental validation.
Highlights
• Put forward the hypothesis that the mechanisms underlying the interaction between HCC and depression may be related to cell death or apoptosis.
• First explored relationship between HCC and depression was through data mining.
• Pointed pharmacological targets.
Ethics approval
Not applicable. This article does not involve human and animal experiments. This article extracts relevant information based on public databases for text mining and data analysis, which is not within the scope of ethical review.
Author contribution
(1) Conception and design: Danhua Li, Yingchun Zhou
(2) Provision of study materials or patients: Tiantian Han
(3) Collection and assembly of data: Tiantian Han, Yingchun Zhou
(4) Data analysis and interpretation: Tiantian Han
(5) Manuscript writing: All authors.
(6) Final approval of manuscript: All authors.
Supplemental Material
Download ()Data availability statement
In this article, we used four databases and a web page mapping tool, as listed below:
pubmed2ensembl (http://pubmed2ensembl.ls.manchester.ac.uk).
VENNY (https://bioinfogp.cnb.csic.es/tools/venny/).
DAVID (https://david.ncifcrf.gov/).
STRING (http://string-db.orgis).
GEPIA (http://gepia.cancer-pku.cn/).
DGIdb (http://www.dgidb.org/search).
Abbreviations
HCC: hepatocellular carcinoma; GO: gene ontology; KEGG:Kyoto Encyclopedia of Genes and Genomes; NGS: next generation of sequencing; PPI: protein–protein interaction
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
References
- Galle P. Extended abstract: management of liver cancer. Dig Dis. 2016;34(4):438–439.
- Chen W, Dong W, Wang J, et al. Elevated expressions of survivin and endoglin in patients with hepatic carcinoma. Cancer Biother Radiopharm. 2019;34(1):7–12.
- Fan J, Wang J, Jiang Y, et al. Attributable causes of liver cancer mortality and incidence in China. Asian Pac J Cancer Prev. 2013;14(12):7251–7256.
- Chen J, Huang S, Li I, et al. Prognostic association of demographic and clinical factors with the change rates of symptoms and depression among patients with hepatocellular carcinoma. Support Care Cancer. 2019;27(12):4665–4674.
- Lee K, Lin J, Shi H. Anxiety and depression are associated with long-term outcomes of hepatocellular carcinoma: a nationwide study of a cohort from Taiwan. World J Biol Psychiatry. 2018;19(6):431–439.
- Rakel R. Depression. Prim Care. 1999;26(2):211–224.
- Jia Y, Li F, Liu Y, et al. Depression and cancer risk: a systematic review and meta-analysis. Public Health. 2017;149:138–148.
- Barrera I, Spiegel D. Review of psychotherapeutic interventions on depression in cancer patients and their impact on disease progression. Int Rev Psychiatry. 2014;26(1):31–43.
- Nikendei C, Terhoeven V, Ehrenthal J, et al. Depression profile in cancer patients and patients without a chronic somatic disease. Psychooncology. 2018;27(1):83–90.
- Steel J, Geller D, Gamblin T, et al. Depression, immunity, and survival in patients with hepatobiliary carcinoma. J Clin Oncol. 2007;25(17):2397–2405.
- Oliver G, Hart S, Klee E. Bioinformatics for clinical next generation sequencing. Clin Chem. 2015;61(1):124–135.
- McGlynn K, Petrick J, El-Serag H. Epidemiology of hepatocellular carcinoma. Hepatology. 2021 Jan;73 Suppl 11:4–13
- Cruz-Pereira J, Rea K, Nolan Y, et al. Trinity: dysregulated stress, immunity, and the microbiome. Annu Rev Psychol. 2020;71:49–78.
- Szelei A, Döme P. Cancer and depression: a concise review. Orv Hetil. 2020;161(22):908–916.
- Wang Y, Li J, Shi J, et al. Depression and anxiety in relation to cancer incidence and mortality: a systematic review and meta-analysis of cohort studies. Mol Psychiatry. 2020;25(7):1487–1499.
- Carneiro B, El-Deiry W. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17(7):395–417.
- Pentimalli F, Grelli S, Di Daniele N, et al. Cell death pathologies: targeting death pathways and the immune system for cancer therapy. Genes Immun. 2019;20(7):539–554.
- Ke B, Tian M, Li J, et al. Targeting programmed cell death using small-molecule compounds to improve potential cancer therapy. Med Res Rev. 2016;36(6):983–1035.
- Li D, Fu J, Du M, et al. Hepatocellular carcinoma repression by TNFα-mediated synergistic lethal effect of mitosis defect-induced senescence and cell death sensitization. Hepatology. 2016;64(4):1105–1120.
- Li Y, Jia Y, Wang D, et al. Programmed cell death 4 as an endogenous suppressor of BDNF translation is involved in stress-induced depression. Mol Psychiatry. 2020. DOI:10.1038/s41380-020-0692-x.
- Idilman R, De Maria N, Colantoni A, et al. Pathogenesis of hepatitis B and C-induced hepatocellular carcinoma. J Viral Hepat. 1998;5(5):285–299.
- Wang W, Wang Z, Qin Y, et al. Th17, synchronically increased with T and B, promoted by tumour cells via cell-contact in primary hepatic carcinoma. Clin Exp Immunol. 2018;192(2):181–192.
- Kohler O, Krogh J, Mors O, et al. Inflammation in depression and the potential for anti-inflammatory treatment. Curr Neuropharmacol. 2016;14(7):732–742.
- Majd M, Saunders E, Engeland C. Inflammation and the dimensions of depression: a review. Front Neuroendocrinol. 2020;56:100800.
- Halaris A. Inflammation and depression but where does the inflammation come from? Curr Opin Psychiatry. 2019;32(5):422–428.
- Kondo M, Koyama Y, Nakamura Y, et al. A novel 5HT3 receptor-IGF1 mechanism distinct from SSRI-induced antidepressant effects. Mol Psychiatry. 2018;23(4):833–842.
- Kopczak A, Stalla G, Uhr M, et al. IGF-I in major depression and antidepressant treatment response. Eur Neuropsychopharmacol. 2015;25(6):864–872.
- Cui J, Gong C, Cao B, et al. MicroRNA-27a participates in the pathological process of depression in rats by regulating VEGFA. Exp Ther Med. 2018;15(5):4349–4355.
- Yi Y, Liu Y, Wu K, et al. The core genes involved in the promotion of depression in patients with ovarian cancer. Oncol Lett. 2019;18(6):5995–6007.