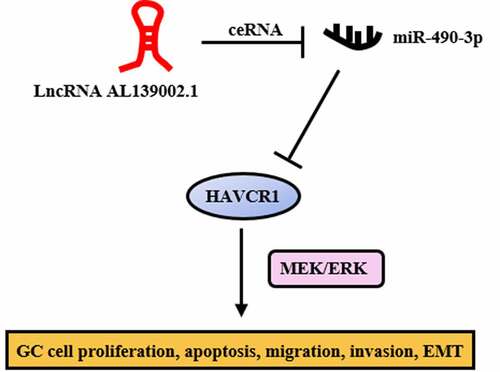
ABSTRACT
Mounting evidence suggests that lncRNA regulates many important diseases. However, the biological role of most lncRNAs in gastric cancer (GC) remain unclear. In this paper, we determined differential expression of lncRNAs and predicted ceRNA networks in the GC database by bioinformatics analysis and validated in GC cells. The effect of lncRNA AL139002.1 on GC cells biological function was assessed by flow cytometry, CCK-8, colony formation, wound healing assay, transwell, western blot, and qRT-PCR. And the relationship of lncRNA AL139002.1 or HAVCR1 with miR-490-3p was verified by luciferase reporter assay. The results showed that lncRNA AL139002.1 was highly expressed in GC cells and lncRNA AL139002.1 knockdown induced apoptosis, while suppressed cell proliferation, migration, invasion, and EMT. Functional examining indicated that lncRNA AL139002.1 regulated HAVCR1 expression by competitively binding miR-490-3p. In addition, lncRNA AL139002.1/miR-490-3p/HAVCR1 regulated EMT and metastasis through MEK/ERK signaling. In conclusion, lncRNA AL139002.1 was highly expressed in GC cells, and lncRNA AL139002.1/miR-490-3p/HAVCR1 functioned critically in GC by regulating MEK/ERK signaling. Our findings demonstrated that lncRNA AL139002.1 served as a potential therapeutic and anti-metastatic biotarget for GC.
Introduction
As a non-coding RNA, lncRNA has limited or no ability to code for proteins [Citation1]. In recent years, there has been increasing evidence that aberrant expression of lncRNA is associated with many diseases [Citation2,Citation3], and has also been shown to play a part in cancer metastasis [Citation4,Citation5] and drug resistance [Citation6,Citation7]. More than that, growing studies have demonstrated that lncRNA could compete for miRNA and regulate the expression of target genes by performing a competitive endogenous RNA (ceRNA). The upregulation of lncRNA HOXD-AS1 mediated by STAT3 promotes hepatocellular metastasis by regulating SOX4 as a ceRNA [Citation8]. MT1JP competitively binds miR-92a-3p to regulate FBXW7 in gastric cancer (GC) [Citation9]. LncRNA XLOC_006390 promotes cervical carcinogenesis and metastasis by sponging miR-331-3p and miR-338-3p [Citation10].
Currently, GC is the third leading cause of death related to cancer all over the world as a result of the lack of effective treatment and high morbidity [Citation11]. Most patients are diagnosed with advanced stages or have developed metastases for the reason that the lack of specific symptoms in the early stages [Citation12,Citation13]. Despite many significant medical advances regarding GC, its poor prognosis and survival rates remain a concern [Citation14]. Consequently, there is a crucial need to investigate the mechanisms underlying GC pathogenesis to improve diagnosis, prognosis and therapeutic targets.
It has been shown that multiple aberrantly expressed lncRNAs have been found in GC, which are closely related to the diagnosis and prognosis of GC. In other studies, lncRNA CCHE1 is contribute to proliferation and inhibits apoptosis in GC cells [Citation15]. ZEB1-AS1 exerts carcinogenesis by regulating GC cell migration, invasion and EMT processes [Citation16]. LncRNA ROR promotes drug resistance in GC [Citation17]. PTCSC3 [Citation18], VPS9D1-AS1 [Citation19] and RP11-397A15.4 [Citation20] could serve as a biomarker for the treatment and prediction of GC. Nevertheless, the role of lncRNA and its regulatory effect on GC is unclear. Therefore, this study identified lncRNA AL139002.1 and its downstream target genes through database analysis. We hypothesized that lncRNA AL139002.1 could target the downstream miRNA/mRNA to regulate the biological functions of GC cells. We studied the regulatory mechanism of them through experiments. Meanwhile, we deeply investigated their effects on GC progression, metastasis and other functions for enriching potential therapeutic targets for GC.
Materials and methods
Bioinformatics
Gene expression quantification data and isoform expression quantification data for GC were obtained from TCGA database. In addition, the Data Transfer Tool (provided by GDC Apps) was used to download the level 3 mRNASeq gene expression data (HTSeq-Counts), miRNAseq data of samples, and clinical information of those patients (https://tcga-data.nci.nih.gov/). The sequenced data were derived from Illumina HiSeq RNASeq and Illumina HiSeq_miRNASeq platforms. The lncRNA sequences and mRNA sequences were included in the RNA sequences. Perl and R language were used to analyze the RNA data. Differentially expressed genes analysis using edgeR (|logFC|>3, padj < 0.05) was performed to identify differentially expressed lncRNAs (DElncRNAs), miRNAs (DEmiRNAs), and mRNAs (DEmRNAs). The lncRNA–miRNA relationship pairs were filtered in the miRcode database based on the differential lncRNAs obtained above, and the results were then compared with the differential miRNAs to obtain the lncRNA and miRNA relationship pairs. The predicted target genes from the database miRTarBase were then combined with the predicted target genes, and the predicted target genes were censored based on the differential mRNAs to obtain miRNA and mRNA relationship pairs. The resulting relationship pairs were used to construct the ceRNA network through R software. Using the survival time data for GC in TCGA, the ‘Survival’ package in R software (version 3.4.3) was used to analyze the specific lncRNAs and mRNAs associated with survival. Kaplan-Meier survival analysis was performed, and P < 0.05 was considered to indicate a statistically significant difference.
GC cell lines and culture
Six human gastric cancer cell lines (MKN-45, AGS, BGC823, SGC7901, MGC803, MKN-74) and nonmalignant gastric mucosal epithelial cell line (GES-1) (the Cell Bank of the Chinese Academy of Sciences, Shanghai) were employed in this experimental study. All cell lines were incubated in RPMI-1640 medium (Thermo Fisher Scientific, USA) with 5% CO2 humidified atmosphere at 37°C.
Cell transfection
In this experiment, siRNA, miR-490-3p mimic, lncRNA AL139002.1 overexpression vector and recombinant HAVCR1 pcDNA3.1(+) plasmid was synthesized by GenePharma (Shanghai, China). Transfection was completed by Lipofectamine 2000 (Invitrogen, USA) based on instructions.
Cell apoptosis
Cells were transfected for 24 h and then stained by FITC-Annexin V and Propidium iodide (PI). The apoptosis of the cells was analyzed by flow cytometry (BD Biosciences, USA).
Cell proliferation
Transfected cells were cultured into 96-well plates for 7 days at 37°C and 5% CO2 atmosphere. 10 μL of Cell Counting Kit-8 (CCK-8) solution (Dojindo, Tokyo, Japan) was added daily and the absorbance values at 450 nm were recorded to plot proliferation curves.
Colony formation
Cells were transfected for 24 h and then incubated in 6-cm dish with RPMI-1640 medium containing FBS (Gibco; USA) for 2 weeks. Samples were stained with 0.1% crystal violet (Sigma-Aldrich, USA) for 30 min after fixation with methanol. Visible colonies were imaged.
Cell migration
Wound healing assay was used in cell migration analysis. Treated cells were incubated in 6-well plates with serum-free medium and artificial scraped by pipette tip. Then wound closure was observed and imaged under the same field of view at 0 h and 24 h, respectively. The wound closure images were captured in the same field under magnification.
Cell invasion
Transwell chambers (8-μm, BD Biosciences, USA) with a Matrigel were used in this experiment. Cells were seeded into Matrigel-coated upper chambers with serum-free RPMI 1640 medium at 1 × 105 cells/well. The lower chamber served as a chemoattractant, which contained RPMI 1640 medium with 10% FBS. After 24 h, the cells on the upper chamber and basement membrane were wiped by cotton swabs. Cells were stained with 0.1% crystal violet for 20 min after fixation with methanol. Olympus microscope was applied for observation.
Western blot
Cells were solubilized by RIPA extraction reagent (Beyotime, China) containing protease inhibitor (Roche, Germany). Lysates were separated by 10% SDS-PAGE and then transferred to 0.22-μm polyvinylidene fluoride membranes (Millipore, USA) for specific antibody detection. Protein expressions were detected and quantified by ECL chromogenic substrate and Quantity One software (Bio Rad). HAVCR1, E-cadherin, vimentin, N-cadherin, p-MEK, MEK, p-ERK and ERK (Cell Signaling Technology, USA) were diluted 1000-fold. GAPDH was regarded as control.
RNA extraction and quantitative qRT-PCR
Total RNA in cells was separated through TRlzol reagent (Invitrogen, USA), and reverse transcribed by PrimeScript RT reagent Kit (TaKaRa, Japan). Expressions of lncRNA and mRNA were measured by ABI 7900HT RealTime PCR System (Applied Biosystem, USA) using SYBR Green assays (TaKaRa, Japan), GAPDH was regarded as control. MiRNA level was analyzed by TaqMan MicroRNA Assays (Applied Biosystems, USA), and U6 was regarded as control. Primer sequences used:
Luciferase reporter assay
LncRNA AL139002.1 or HAVCR1 3′-UTR fragment with the miR-490-3p putative binding site was amplified and cloned into the downstream of the luciferase gene within the pmirGlo vector (GenePharma, China). Mutant vectors for lncRNA AL139002.1 3′-UTR or HAVCR1 3′-UTR were constructed using lncRNA AL139002.1-mut or HAVCR1-mut. The cells were co-treated with miR-490-3p mimic and lncRNA AL139002.1-wt (or lncRNA AL139002.1-mut) or HAVCR1-wt (or HAVCR1-mut). After 72 h, luciferase activity of lysed cells was examined by the Dual-Luciferase Reporter Assay System (Promega Biotech, USA).
Statistical Analysis
GraphPad Prism version 6 for Windows was used to analyze the differences in different groups using Student’s t test or ANOVA followed by Tukey’s test. All data were represented as mean ± SD. Treatments were completed in triplicate. p < 0.05 was statistically significant. The correlation analysis between lncRNA AL139002.1 and HAVCR1 was performed by Pearson correlation analysis. Survival curves were evaluated and analyzed using KKaplan–Meiermethod and the log-rank test.
Results
1 Bioinformatics prediction of lncRNA AL139002.1 and targeting miRNA/mRNA and validation
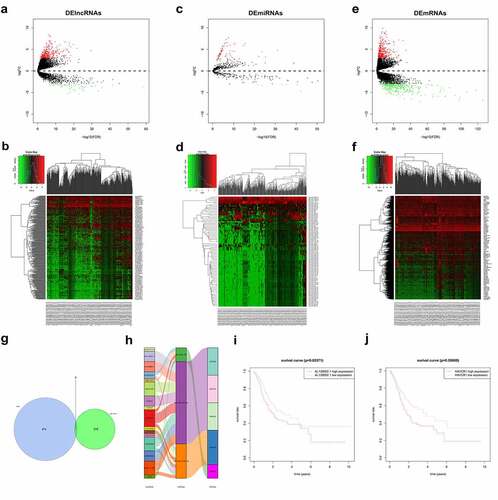
Differential analysis of downloaded GC data (TCGA) yielded 383 DElncRNAs ( and b), 76 DEmiRNAs ( and d) and 676 DEmRNAs ( and f). We screened DElncRNAs in the miRcode database (http://www.mircode.org/) for targeting miRNAs and compared them with DEmiRNAs, 16 lncRNAs and 3 miRNAs with targeted binding sites were obtained. We then used the miRTarBase database to predict the downstream regulatory target genes. By intersecting the predicted target genes with the DEmRNAs, 5 DEmRNAs with target-binding sites were finally obtained (). The ceRNA network was also mapped (). Among them, lncRNA AL139002.1 was highly expressed in the cancer group with significant difference and low survival rate, so lncRNA AL139002.1 was selected as the target of this study (). It was also found that lncRNA AL139002.1 was related to regulating miR-490-3p, and miR-490-3p expression was lower in the tumor group. MiR-490-3p targeted the regulation of 3 mRNAs, among which HAVCR1 was upregulated and had a low survival rate (). Therefore, we hypothesized that lncRNA AL139002.1 promoted GC development through the targeted regulation of HAVCR1 by sponging miR-490-3p.
Figure 1. Bioinformatics prediction of lncRNA AL139002.1 and targeting miRNA/mRNA and validation. (a-f) Volcano plot and Heatmap of DElncRNAs, DEmiRNAs, and DEmRNAs in the TCGA database. (g) TCGA and miRcode database consistently predicted five mRNAs interacted with lncRNA. (h) LncRNA-miRNA-mRNA regulatory network. (i, j) Kaplan-Meier survival analysis showed the relationship between lncRNA AL139002.1 or HAVCR1 expression with the prognosis of GC patients
2 LncRNA AL139002.1 and HAVCR1 expression increased, miR-490-3p expression decreased in GC cell lines
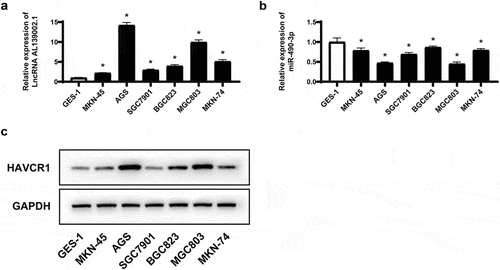
We first determined lncRNA AL139002.1 and miR-490-3p levels in different GC cell lines. and c showed that lncRNA AL139002.1 and HAVCR1 were highly expressed and miR-490-3p was lowly expressed in GC cell lines compared to GES-1. In combination with the above experimental results, AGS and MGC-803 cell lines were selected for further research. The above results approved that lncRNA AL139002.1 and HAVCR1 expression increased, and miR-490-3p expression decreased in GC cell lines.
Figure 2. LNCRNA AL139002.1 and HAVCR1 expression increased, miR-490-3p expression decreased in GC cell lines. qRT-RCR was applied to detect the relative expression of lncRNA AL139002.1 (a) and miR-490-3p (b) in GC cell lines (MKN-45, AGS, BGC823, SGC7901, MGC803, MKN74) and (GES-1). (c) Western blot of HAVCR1 proteins in GC cell lines (MKN-45, AGS, BGC823, SGC7901, MGC803, MKN74) and (GES-1)
3 Silencing of lncRNA AL139002.1 regulated proliferation, apoptosis, migration, invasion and EMT of GC cells
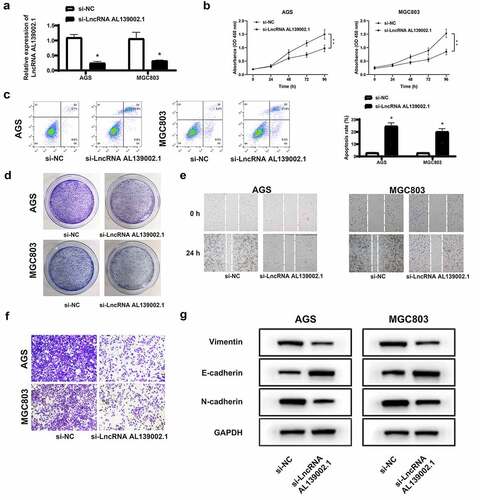
To explore the role of lncRNA AL139002.1 in GC cells, we knocked down lncRNA AL139002.1 by siRNA. The silencing efficiency of lncRNA AL139002.1 in AGS, MGC-803 cells was verified in . The regulating effect of lncRNA AL139002.1 on GC cell proliferation, apoptosis and cloning ability was first investigated. In CCK-8 assay, the proliferation of cells treated with lncRNA AL139002.1 siRNA was markedly reduced compared to the negative control (si-NC) (). In flow cytometry, the apoptosis of AGS, MGC-803 transfected with lncRNA AL139002.1 siRNA was significantly elevated (). The same trend as CCK-8 assay was observed in clone formation experiments (). Compared with the si-NC group, the clone formation of lncRNA AL139002.1 siRNA group was signally decreased in AGS and MGC-803 cells. Migration analysis showed that the wound distance of LncRNA AL139002.1 siRNA group was wider than that of the si-NC group in AGS and MGC-803 cells, and transwell analysis showed that lncRNA AL139002.1 siRNA transfection notably suppressed invasion of GC cells ( and f). Since lncRNA AL139002.1 influenced cell migration and invasion, epithelial-mesenchymal transformation (EMT) markers protein expressions were measured. LncRNA AL139002.1 siRNA transfection increased E-cadherin level and decreased vimentin and N-cadherin levels (). The above results indicated that silencing of lncRNA AL139002.1 regulated proliferation, apoptosis, migration, invasion, and EMT processes of GC cells.
Figure 3. Silencing of lncRNA AL139002.1 regulated proliferation, apoptosis, migration, invasion and EMT of GC cells. LncRNA AL139002.1 was silenced in AGS and MGC-803 cells. (a) qRT-RCR was applied to detect the relative expression of lncRNA AL139002.1 in cells. (b, c) Proliferation and apoptosis assays of cells through CCK-8 assay and flow cytometry. (d) The colony formation of cells results. (e) Migration assay of cells by wound healing analysis (40×). (f) Invasion assay of cells by transwell assay. (g) Western blot of E-cadherin, vimentin and N-cadherin proteins in cells
4 LncRNA AL139002.1 regulated HAVCR1 by sponging miR‑490‑3p
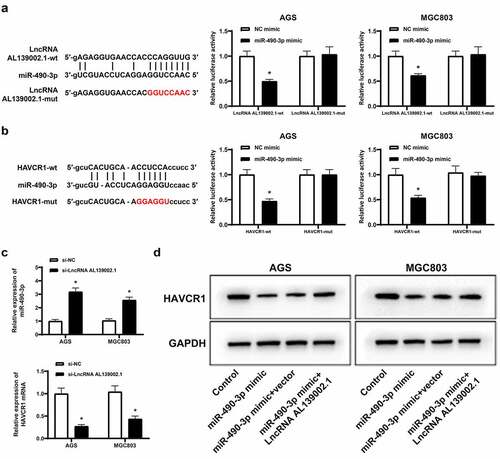
Based on the results of the bioinformatics analysis, we first validated the ceRNA network by luciferase reporter assay. The luciferase activity of lncRNA AL139002.1-wt was effectively reduced by overexpressing miR-490-3p in GC cells (), and also remarkably decreased HAVCR1-wt luciferase activity, but not HAVCR1-mut (). We then detected the levels of miR-490-3p and HAVCR1. The results displayed an increase in miR-490-3p levels and a decrease in RNA levels of HAVCR1 (). In addition, miR-490-3p overexpression suppressed HAVCR1 expression, while overexpression of lncRNA AL139002.1 abolished inhibiting effect of miR-490-3p mimic on HAVCR1 expression (). These findings uncovered that lncRNA AL139002.1 regulated HAVCR1 by sponging miR‑490‑3p.
Figure 4. LncRNA AL139002.1 regulated HAVCR1 by sponging miR‑490‑3p. Relative luciferase activities of lncRNA AL139002.1-wt and lncRNA AL139002.1-mut (a) or HAVCR1-wt and HAVCR1-mut (b) reporter plasmid in AGS and MGC-803 cells co-treated with miR-490-3p mimic. (c) LncRNA AL139002.1 was silenced in AGS and MGC-803 cells. qRT-RCR was conducted to detect the relative expressions of miR-490-3p and HAVCR1. (d) Western blot of HAVCR1 protein in AGS and MGC-803 cells treated with LncRNA AL139002.1 and miR‑490‑3p mimic
5 LncRNA AL139002.1 regulated the proliferation, apoptosis, migration, invasion and EMT of GC cells by miR‑490‑3p/HAVCR1
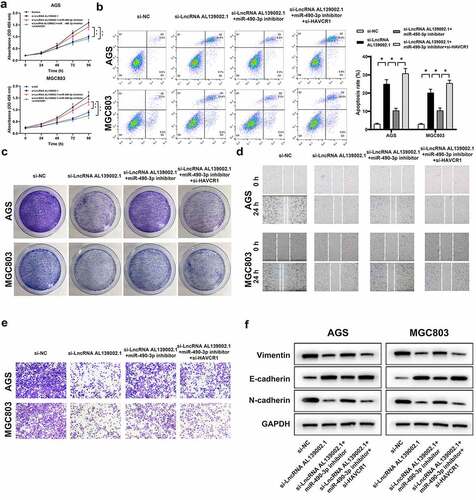
Next, we verified that miR-490-3p, HAVCR1 were involved in the effects of lncRNA AL139002.1 knockdown on GC cells. Rescue experiments showed that the miR-490-3p inhibitor alleviated the anticancer effect of lncRNA AL139002.1 siRNA. After transfection of si-HAVCR1 on this basis, restoration of cell proliferation, migration and invasion were inhibited, and suppression of apoptosis was recovered. Interestingly, vimentin and N-cadherin expression was downregulated by lncRNA AL139002.1 siRNA transfection, and upregulation of E-cadherin expression was inhibited by miR-490-3p inhibitor. In contrast, transfection with si-HAVCR1 decreased vimentin and N-cadherin expression, and increased E-cadherin expression ().
Figure 5. LncRNA AL139002.1 regulated the proliferation, apoptosis, migration, invasion and EMT of GC cells by miR‑490‑3p/HAVCR1. Silencing of lncRNA AL139002.1, miR‑490‑3p inhibitor and silencing of HAVCR1 were transfected into AGS and MGC-803 cells. (a, b) Proliferation and apoptosis assays of cells by CCK-8 assay and flow cytometry. (c) The results of the colony formation of cells. (d) Migration assay of cells by wound healing analysis (40×). (e) Invasion assay of cells by transwell assay. (f) Western blot of E-cadherin, vimentin and N-cadherin proteins in AGS and MGC-803 cells
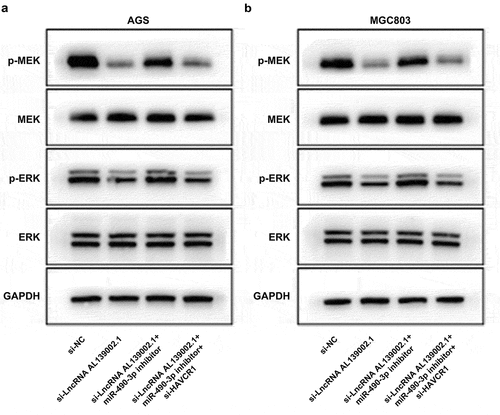
6 LncRNA AL139002.1/miR‑490‑3p/HAVCR1 regulated GC through MEK/ERK signaling
Afterward, the potential mechanism of the role of lncRNA AL139002.1/miR-490-3p/HAVCR1 on GC cells was then investigated. MEK/ERK signaling has been reported to be activated by carcinogenic gene, and could regulate various cellular processes including migratory and invasion [Citation21]. It has been shown that HAVCR1 regulates the proliferation of GAC cells through the MEK/ERK signaling [Citation22]. Therefore, we hypothesized that lncRNA AL139002.1 sponged miR-490-3p to regulate HAVCR1, and thus regulating MEK/ERK signaling to affect GC cells biological process. Western blot results displayed that miR-490-3p inhibitor was capable to significantly up-regulate the level of p-MEK and p-ERK inhibited by si-lncRNA AL139002.1. After transfection with si-HAVCR1, p-MEK and p-ERK expressions were significantly down-regulated ( and b). These findings supported the idea that lncRNA AL139002.1/miR‑490‑3p/HAVCR1 modulated GC cells through MEK/ERK signaling.
Discussion
In this paper, we predicted differential expression of lncRNA AL139002.1 and ceRNA network by bioinformatics analysis. We hypothesized that lncRNA AL139002.1 could regulate the biological functions of GC cells by targeting the downstream miRNA/mRNA. And we verified the hypothesis through flow cytometry, CCK-8, colony formation, wound healing assay, transwell, western blot, qRT-PCR. The results showed that lncRNA AL139002.1 regulated HAVCR1 expression by competitively binding miR-490-3p, and lncRNA AL139002.1 regulated cell proliferation, migration, invasion, and EMT.
LncRNAs serve crucial roles in multiple biological processes of diverse diseases, including cancer [Citation23]. This study identified a GC-associated lncRNA AL139002.1 by bioinformatics, which was upregulated in GC cell lines. Furthermore, lncRNA AL139002.1 deficiency induced apoptosis and inhibited cell proliferation, migration, invasion and EMT. Our results indicated that lncRNA AL139002.1 had oncogenic effects in GC and was a potential prognostic indicator.
LncRNAs regulate GC development by competitively sponging miRNAs. For example, Linc00483 activates MAPK to regulate GC cells development by sponging miR-30a-3p [Citation24]. LncRNA HOTAIR modulates HER2 expression, proliferation and migration of GC cells by competing miR-331-3p [Citation25]. This article identified miR-490-3p as a downstream gene of lncRNA AL139002.1 by bioinformatics and cell experiment analysis. MiR-490-3p had been verified to play the inhibitory effect in several diseases. MiR-490-3p restrains autophagy by targeting ATG7, thereby suppressing cell proliferation and enhancing apoptosis in hepatocellular carcinoma cells [Citation26]. MiR-490-3p inhibits oncogene VDAC1 expression to exert tumor suppressive effects in colorectal cancer [Citation27]. MiR-490-3p targets downregulation of HMGA2 to inhibit esophageal squamous cell carcinoma development [Citation28]. Downregulation of miR-490-3p promotes cell development by targeting RAB14, predicting poor prognosis in colorectal cancer [Citation29]. In this paper, miR-490-3p was notably down-regulated in GC cancer cells. Consistent with this result, the downregulation of miR-490-3p in GC induced by helicobacter pylori is strongly related to poor clinical outcome [Citation30]. MiR-490-3p inhibits GC development [Citation31]. Epigenetic knockdown of miR-490-3p activates SMARCD1, thereby promoting the helicobacter pylori-induced GC [Citation32]. Echoing previous studies, we found that lncRNA AL139002.1 competitively sponged miR-490-3p and inhibited GC.
As a target of miRNA, mRNA is also a vital part of the ceRNA network. HAVCR1 was predicted as the target gene of miR-490-3p by database and experimental verification. Furthermore, miR-490-3p inhibited HAVCR1 expression. HAVCR1, also known as hepatitis A virus cellular receptor 1 [Citation33], is associated with disease susceptibility [Citation34]. HAVCR1 is significantly upregulated in human colorectal cancer [Citation34]. Targeted silencing of HAVCR1 inhibits clear cell renal cell carcinoma cells growth in vitro and in vivo [Citation35]. In addition, HAVCR1 has been found to modulate the MEK/ERK pathway in GAC and influence tumor progression and patient outcomes [Citation22]. The ERK signaling cascade response is a central MAPK pathway, and MEK acts as a key protein upstream of ERK in a variety of cellular processes [Citation36]. YAP has been reported to promote GC cell survival and migration/invasion via the ERK pathway [Citation37]. Recombinant Newcastle disease virus (rL-RVG) inhibits GC migration by regulating α7-nAChR-MEK/ERK-EMT [Citation38]. Consistent with previous studies, HAVCR1 was highly expressed in GC cells, and HAVCR1 knockdown induced apoptosis and inhibited proliferation, migration, invasion, and EMT. Furthermore, the detection of MEK/ERK pathway-associated proteins revealed that lncRNA AL139002.1 sponged miR-490-3p to regulate HAVCR1, thereby modulating ERK/MEK signaling to regulate GC cell biological process.
Conclusion
In conclusion, this article identified and validated a new GC-related lncRNA AL139002.1. LncRNA AL139002.1 regulated HAVCR1 by targeting miR-490-3p, and exerted carcinogenesis through MEK/ERK signaling in proliferation, apoptosis, migration, and invasion of GC cells. This study enriched the ceRNA network during GC pathogenesis and provided the basis for effective treatment and prognosis of GC.
Author contributions
Yurong Chen: Data curation, Writing-Original draft preparation, Visualization, Investigation, and Validation.
Renchao Zhang: Conceptualization, Methodology, Software, Supervision, Writing, Reviewing, and Editing.
Availability of data and material
The datasets used and/or analyzed during the current study are available from
the corresponding author on reasonable request.
Ethics approval
Not applicable.
Highlights
Prediction of DElncRNA AL139002.1 and ceRNA network related to GC.
AL139002.1 regulated proliferation, migration, invasion and EMT of GC cells.
AL139002.1 regulated HAVCR1 by sponging miR‑490‑3p.
AL139002.1 regulated the biological function of GC cells by miR‑490‑3p/HAVCR1.
AL139002.1/miR‑490‑3p/HAVCR1 regulated GC through MEK/ERK signaling.
Acknowledgements
None.
Disclosure statement
The authors declare there are no competing interests.
Additional information
Funding
References
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181.
- Garcia-Venzor A, Mandujano-Tinoco EA, Lizarraga F, et al. Microenvironment-regulated lncRNA-HAL is able to promote stemness in breast cancer cells. Biochim Biophys Acta Mol Cell Res. 2019;1866(12):118523..
- Zhu H, Zhao H, Zhang L, et al. Dandelion root extract suppressed gastric cancer cells proliferation and migration through targeting lncRNA-CCAT1. Biomed Pharmacother. 2017;93:1010–1017.
- Li J, Meng H, Bai Y, et al. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23(5):205–217.
- Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50(12):1705–1715..
- Ren J, Ding L, Zhang D, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932–3948..
- Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16(1):9..
- Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136..
- Zhang G, Li S, Lu J, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17(1):87..
- Luan X, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol. 2018;29(6):e95.
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
- Van Cutsem E, Ducreux M. Colorectal and gastric cancer in 2015: the development of new agents and molecular classifications. Nat Rev Clin Oncol. 2016;13(2):69–70.
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet. 2016;388(10060):2654–2664.
- Wang XN, Liang H. Some problems in the surgical treatment of gastric cancer. Chin J Cancer. 2010;29(4):369–373.
- Xu G, Zhang Y, Li N, et al. LncRNA CCHE1 in the proliferation and apoptosis of gastric cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(9):2631–2637.
- Li Y, Wen X, Wang L, et al. LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric cancer. Surg Oncol. 2017;26(4):527–534..
- Wang S, Chen W, Yu H, et al. lncRNA ROR Promotes Gastric Cancer Drug Resistance. Cancer Control. 2020;27(1):1073274820904694..
- Hu M, Han Y, Zhang Y, et al. lncRNA TINCR sponges miR-214-5p to upregulate ROCK1 in hepatocellular carcinoma. BMC Med Genet. 2020;21(1):2.
- Chen M, Wu X, Ma W, et al. Decreased expression of lncRNA VPS9D1-AS1 in gastric cancer and its clinical significance. Cancer Biomark. 2017;21(1):23–28..
- Chen H, Xu Z, Liu X, et al. Increased Expression of Lncrna RP11-397A15.4 in Gastric Cancer and Its Clinical Significance. Ann Clin Lab Sci. 2018;48(6):707–711.
- Liu W, Zhang Z, Zhang Y, et al. HMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biol Ther. 2015;16(4):511–517..
- Xue J, Li Y, Yi J, et al. HAVCR1 Affects the MEK/ERK Pathway in Gastric Adenocarcinomas and Influences Tumor Progression and Patient Outcome. Gastroenterol Res Pract. 2019;2019:6746970.
- Sun TT, He J, Liang Q, et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6(7):784–801..
- Li D, Yang M, Liao A, et al. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J Cell Mol Med. 2018; 22(8):3875–3886..
- Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13(1):92..
- Ou Y, He J, Liu Y. MiR-490-3p inhibits autophagy via targeting ATG7 in hepatocellular carcinoma. IUBMB Life. 2018;70(6):468–478.
- Liu X, He B, Xu T, et al. MiR-490-3p Functions As a Tumor Suppressor by Inhibiting Oncogene VDAC1 Expression in Colorectal Cancer. J Cancer. 2018;9(7):1218–1230..
- Kang NN, Ge SL, Zhang RQ, et al. MiR-490-3p inhibited the proliferation and metastasis of esophageal squamous cell carcinoma by targeting HMGA2. Eur Rev Med Pharmacol Sci. 2018;22(23):8298–8305.
- Wang B, Yin M, Cheng C, et al. Decreased expression of miR4903p in colorectal cancer predicts poor prognosis and promotes cell proliferation and invasion by targeting RAB14. Int J Oncol. 2018;53(3):1247–1256..
- Qu M, Li L, Zheng WC. Reduced miR-490-3p expression is associated with poor prognosis of Helicobacter pylori induced gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21(15):3384–3388.
- Luo M, Liang C. LncRNA LINC00483 promotes gastric cancer development through regulating MAPK1 expression by sponging miR-490-3p. Biol Res. 2020;53(1):14.
- Shen J, Xiao Z, Wu WK, et al. Epigenetic silencing of miR-490-3p reactivates the chromatin remodeler SMARCD1 to promote Helicobacter pylori-induced gastric carcinogenesis. Cancer Res. 2015;75(4):754–765..
- Cuadros T, Trilla E, Sarro E, et al. HAVCR/KIM-1 activates the IL-6/STAT-3 pathway in clear cell renal cell carcinoma and determines tumor progression and patient outcome. Cancer Res. 2014;74(5):1416–1428..
- Wang Y, Martin TA, Jiang WG. HAVcR-1 expression in human colorectal cancer and its effects on colorectal cancer cells in vitro. Anticancer Res. 2013;33(1):207–214.
- Xu J, Sun L, Sun W, et al. Targeted silencing of Kim-1 inhibits the growth of clear cell renal cell carcinoma cell line 786-0 in vitro and in vivo. Oncol Res. 2018;26(7):997–1003.
- Deschenes-Simard X, Kottakis F, Meloche S, et al. ERKs in cancer: friends or foes? Cancer Res. 2014;74(2):412–419.
- Liu H, Mei D, Xu P, et al. YAP promotes gastric cancer cell survival and migration/invasion via the ERK/endoplasmic reticulum stress pathway. Oncol Lett. 2019;18(6):6752–6758.
- Bu X, Zhang A, Chen Z, et al. Migration of gastric cancer is suppressed by recombinant Newcastle disease virus (rL-RVG) via regulating alpha7-nicotinic acetylcholine receptors/ERK- EMT. BMC Cancer. 2019;19(1):976..