ABSTRACT
Non-small cell lung cancer (NSCLC) is a common malignant tumor, with high morbidity and mortality. Circular RNA (circRNA) circ_0003028 was reported to be upregulated in NSCLC. This study is designed to explore the role and mechanism of circ_0003028 on NSCLC progression. In this work, circ_0003028, microRNA-1298-5p (miR-1298-5p), and glutamic oxaloacetic transaminase 2 (GOT2) level were detected by real-time quantitative polymerase chain reaction (RT-qPCR). The localization of circ_0003028 was analyzed by subcellular fractionation assay. Cell proliferation, colony number, cell cycle progression, apoptosis, migration, invasion, and angiogenesis were measured by Cell Counting Kit-8 (CCK-8), colony formation, flow cytometry, transwell, and tube formation assays. Protein levels of Beclin1, light chain 3 (LC3)-II/LC3-I, GOT2, proliferating cell nuclear antigen (PCNA) were examined by western blot assay. The binding relationship between miR-1298-5p and circ_0003028 or GOT2 was predicted by circular RNA Interactome or starbase and then verified by dual-luciferase reporter, RNA Immunoprecipitation (RIP), and RNA pull-down assays. The biological role of circ_0003028 on NSCLC tumor growth was examined by the xenograft tumor model in vivo. We reported that circ_0003028 and GOT2 were upregulated, and miR-1298-5p was decreased in NSCLC tissues and cells. Moreover, circ_0003028 knockdown curbed cell proliferative ability, migration, invasion, angiogenesis, and facilitate apoptosis and autophagy in NSCLC cells in vitro. Mechanical analysis discovered that circ_0003028 regulated GOT2 expression by sponging miR-1298-5p. Circ_0003028 silencing hindered the cell growth of NSCLC in vivo. Taken together, circ_0003028 knockdown could suppress NSCLC progression partly by regulating the miR-1298-5p/GOT2 axis, providing an underlying therapeutic target for NSCLC.
Introduction
As a most common malignant tumor, lung cancer has been considered a global problem with high morbidity and mortality [Citation1,Citation2], with non-small cell lung cancer (NSCLC) occupying about 80% of lung cancer cases [Citation3]. Although traditional treatments, including surgical resection and adjuvant chemotherapy, have made considerable progress, the prognosis of NSCLC patients remains dismal due to the recurrence and metastasis [Citation4]. This highlights an imperative need for exploring the mechanisms underlying NSCLC tumorigenesis to develop more effective therapies for NSCLC.
In recent years, some researchers have reported that approximately 90% of the human genome is likely to be actively transcribed into non-coding RNA [Citation5]. As a distinct group of non-coding RNAs, circular RNAs (circRNAs) are derived from back-splicing of a single pre-mRNA, with a covalently closed-loop structure [Citation6,Citation7]. Since circRNAs have remarkable stability and high abundance, emphasizing the importance of circRNAs in human diseases diagnosis and prognosis, including cancer [Citation8,Citation9]. In fact, a number of articles have documented that the abnormal expression of circRNAs has an inextricable correlation in the tumorigenesis and progression of NSCLC [Citation10]. For example, Li et al. displayed that the forced expression of hsa_circ_0046263 could aggravate the malignancy of the tumor by boosting carcinogenesis and metastasis in NSCLC cells [Citation11]. Analogously, Cheng et al. presented that the reduced expression of circSEC31A could repress cell migration, invasion, and glycolysis through binding to miR-376a in NSCLC cells [Citation12]. Notably, prior research described that circ_0003028, a novel circRNA originating from Fucosyltransferase 8 (FUT8), has been proven to be increased in liver cancer for the first time [Citation13]. Furthermore, the high expression of circ_0003028 was previously proved to have diagnostic value in NSCLC in GSE101586, GSEE101684, and GSE112214 datasets [Citation14], implying the vital role of circ_0003028 in NSCLC. However, the precise action and mechanism of circ_0003028 are far from being addressed in NSCLC.
To date, numerous studies discovered that circRNAs could serve as essential regulators in human cancer biology through sponging their target microRNAs (miRNAs) [Citation15,Citation16]. Here, we first found that circ_0003028 possessed some binding sites with miR-1298-5p. Furthermore, the tumor-suppressive role of miR-1298-5p has been reported in breast cancer, bladder cancer, and gastric cancer [Citation17–19]. Meanwhile, miR-1298-5p was proved to hinder cell proliferation, migration, and invasion in NSCLC cells. Therefore, we aimed to explore whether the regulatory role of circ_0003028 on NSCLC progression could be mediated by interacting with miR-1298-5p. Also, our findings could lay a new theoretical basis for developing targeted NSCLC clinical therapy.
Materials and methods
Clinical samples and cell culture
NSCLC tumor tissue samples (n = 53) and matched adjacent normal tissues (n = 53) were provided by NSCLC patients from Jianhu Hospital Affiliated to Nantong University in this research, which was approved by the Ethics Committee of Jianhu Hospital Affiliated to Nantong University. The written informed consent was signed by each participant.
In this study, all cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in a moist atmosphere with 5% CO2 at 37°C. Then, normal human bronchial epithelial cell-line BEAS-2B was cultured in BEBM kit (Lonza/Clonetics Corporation, Walkersville, MD, USA). Meanwhile, NSCLC cell lines (A-549, NCI-H1299, and NCI-H460) were grown in the Roswell Park Memorial Institute-1640 medium (RPMI; Gibico, Grand Island, NY, USA) with 10% fetal bovine serum (FBS; PAN Biotech, Aidenbach, Germany) and 1% penicillin/streptomycin (PAN Biotech).
Real-time quantitative polymerase chain reaction (RT-qPCR)
According to the standard procedure of TRIzol reagent (Invitrogen, Paisley Scotland, UK), total RNAs were extracted from clinical specimens or cell lines, followed by quantization with a NanoDrop ND-1000 (NanoDrop Technologies, Waltham, Massachusetts, USA). After reverse transcription with PrimeScript RT Reagent Kit (TaKaRa, Tokyo, Japan) and TaqMan Advanced miRNA cDNA Synthesis Kit (Applied Biosystems, Foster City, CA, USA), the mixture of cDNA, SYBR (TaKaRa), and specific primer pairs (RiboBio, Guangzhou, China) was applied for RT-qPCR reaction on Thermal Cycler CFX6 System (Bio-Rad, Foster City, California, USA). With GAPDH for circ_0003028, linear FUT8, and Glutamic oxaloacetic transaminase 2 (GOT2) and U6 for miR-1298-5p as the corresponding internal reference, the relative RNA level was analyzed by the 2–ΔΔCt method, and the primers were shown as below:
circ_0003028: 5ʹ-GTCCAAGATTCTGGCAAAGC-3ʹ (sense), 5ʹ-TCAAAGAGATCCTCCTGGTGA-3ʹ (antisense);
linear FUT8: 5ʹ-ACTACTTTGTGTGCTGGGGC-3ʹ (sense), 5ʹ-GGACTCGGAAAATTCCCGGC-3ʹ (antisense);
miR-1298-5p: 5ʹ-TCATTCGGCTGTCCAGA-3ʹ (sense), 5ʹ-GAACATGTCTGCGTATCTC −3ʹ (antisense);
GOT2: 5ʹ-GAGCAGGGCATCAATGTCTG-3ʹ (sense), 5ʹ-GTTGGAATACAGGGGACGGA-3ʹ (antisense);
U6: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (sense), 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ (antisense);
GAPDH: 5ʹ-GGTCACCAGGGCTGCTTT-3ʹ (sense), 5ʹ-GGAAGATGGTGATGGGATT-3ʹ (antisense).
Subcellular fractionation assay and Ribonuclease R (RNase R) treatment
For subcellular fractionation assay, RNA was acquired from the nuclear and cytoplasmic fractions of A-549 and NCI-H1299 cells, referring to the instructions of the PARIS kit (Ambion, Austin, TX, USA). Subsequently, the assessment of circ_0003028, U6 (nucleus control), and 18S rRNA (cytoplasm control) was done by RT-qPCR assay. For RNase R digestion, the isolated RNA was incubated with RNase R (4 U/μg, Qiagen, Valencia, CA, USA) at 37°C for 60 min, followed by purification with an RNeasy MinElute Cleaning Kit (Qiagen). Then, the determination of circ_0003028 and linear FUT8 mRNA level was performed using RT-qPCR assay.
Cell transfection
For silencing circ_0003028, small interfering RNAs specific to circ_0003028 (si-circ_0003028#1: 5ʹ-GGCCGAATCTCTCCGCATGTA-3ʹ, si-circ_0003028#2: 5ʹ-CCGAATCTCTCCGCATGTAGA-3ʹ, si-circ_0003028#3: 5ʹ-TGGCCGAATCTCTCCGCATGT-3ʹ) and the relative si-NC (5ʹ-AAUUCUCCGAACGUGUCACGU-3ʹ) were purchased from RiboBio (Guangzhou, China). Then, 20 nM of aforementioned RNAs were transfected into A-549 and NCI-H1299 cells. Also, miR-1298-5p upregulation or downregulation was achieved using 20 nM of miR-1298-5p mimic (miR-1298-5p, RiboBio) or miR-1298-5p inhibitor (anti-miR-1298-5p), and their respective controls (NC or anti-NC). For GOT2 upregulation, the sequence of GOT2 was cloned into pcDNA empty vector (vector, Invitrogen, Carlsbad, CA, USA) to generate pcDNA-GOT2 (termed as GOT2), followed by transfection into A-549 and NCI-H1299 cells with 50 ng vector. In this study, all transfection was conducted using Lipofectamine 3000 (Invitrogen) for 48 h.
Cell counting kit (CCK-8) assay
After transfection for 48 h, cells were introduced into 96-well plates at the density of 2 × 103 cells/well. 24 h later, the medium of each well was added with 10 μL CCK-8 solution (Dojindo, Osaka, Japan), followed by incubation for another 4 h. At length, the optical density (OD) at 450 nm was observed using a microplate reader (Bio-Rad, Hercules, CA, USA) at different times (0, 24, 48, and 72 h).
Colony formation assay
After 48 h transfection, cells (500 cells/well) were uniformly dispersed in six-well plates, followed by incubation for 2 weeks. And then, 4% paraformaldehyde (Solarbio, Beijing, China) was used to fix the cells, which were next stained by 0.1% crystal violet (Sigma-Aldrich) for 30 min. Based on the operation manual of the microscope (Olympus, Toyko, Japan, magnification×40), the detection of visible colonies was performed in this assay.
Cell cycle and cell apoptosis assay
For cell cycle assay, after transfection for 48 h, cells were harvested and fixed in ice-cold ethanol, followed by incubation for 30 min. After treatment with RNAase (Sigma-Aldrich, St. Louis, MO, USA), the treated cells were stained with Propidium Iodide (PI, BD Biosciences, Franklin Lakes, NJ, USA) for 30 min. Finally, cell cycle distribution was analyzed according to the instruction guidelines of FACSan (Becton Dickinson, Franklin, NJ, USA) and Weasel 3.1 software (Becton Dickinson). For cell apoptosis assay, treated cells were re-suspended into 200 µL binding buffer, followed by staining with 5 µL Annexin (V-fluorescein isothiocyanate) V-FITC and 10 µL PI. After incubation for 15 min at 4°C, the assessment of apoptosis rate was conducted as per the supplier’s direction of FACSan (Becton Dickinson). Besides, the detection of caspase 3 activities in A-549 and NCI-H1299 cells was carried out, in accordance with the guidebook of caspase 3 activity assay kit (Solarbio) and a microplate reader at 405 nm.
Transwell assays
For migration assay, after transfection for 48 h, cells (5 × 104 cells/well) were re-suspended in serum-free medium, followed by an introduction into the upper chamber (BD Biosciences). Also, the bottom counterparts were filled with the medium with 10% FBS. Following incubation for 24 h, the cells attached to the bottom surface were stained with 0.1% crystal violet (Sigma-Aldrich) for 15 min after being fixing with methanol for 10 min. Finally, the measurement of migration was performed by a microscope (Olympus, magnification×100) and Image J v1.8 (NIH, Bethesda, MD, USA). For invasion assay, 1 × 105 cells were seeded into the Matrigel (BD Biosciences)-coated upper transwell chambers and other procedures were similar to the steps of cell migration assay.
Tube formation assay
The angiogenic ability of HUVEC cells was detected by tube formation assay. In brief, growth factor-reduced Matrigel (BD Biosciences) was placed into 96-well plates. After that, HUVEC cells were suspended in the medium pre-conditioned with tumor cells, followed by addition to the plates pre-coated with Matrigel. After incubation for 18 h, the analysis of tube formation was conducted by a microscope and Image J v1.8 (NIH).
Western blot assay
After transfection for 48 h, lysate samples were prepared with RIPA lysis buffer (Beyotime, Shanghai, China), followed by separation with 10% SDS-PAGE and transferring to PVDF membranes (Millipore, Molsheim, France). Whereafter, the membranes were subjected to nonfat milk blockage, primary antibodies hybridization at 4°C overnight, and secondary antibody (ab6721, 1:10,000, Abcam, Cambridge, MA, USA) incubation for 2 h at room temperature. After interacting with the ECL reagent (Invitrogen), the blots were analyzed by Quantity One software (Bio-Rad). Meanwhile, the primary antibodies were presented as follows: Beclin1 (1:1000, #3738, Cell Signaling Technology, Beverly, MA, USA), light chain 3 (LC3)-II/LC3-I (1:1000, #12,741, Cell Signaling Technology), GOT2 (ab171739, 1:1000, Abcam), proliferating cell nuclear antigen (PCNA, 1:1000, #13,110, Cell Signaling Technology), and β-Actin (ab8227, 1:5000, Abcam).
Dual-luciferase reporter assay
Based on the analysis of the online software circular RNA Interactome (https://circinteractome.nia.nih.gov) or starbase (http://starbase.sysu.edu.cn), the binding relationship between miR-1298-5p and circ_0003028 or GOT2 3ʹ un-translated region (3ʹUTR) was predicted, as proved by a dual-luciferase reporter assay. Generally, the psiCHECK-2 vector (Promega, Fitchburg, WI, USA) was used for the structure of wild-type (wt) luciferase reporter vectors (circ_0003028 wt or GOT2 wt) via inserting the fragment of circ_0003028 or GOT2 harboring miR-1298-5p binding sequence. Likewise, the mutant-type (mut) vectors (circ_0003028 mut or GOT2 mut) were generated by mutating the binding sequences of miR-1298-5p. Whereafter, the constructed vectors were severally transfected into A-549 and NCI-H1299 cells along with miR-1298-5p or NC. At length, the luciferase activities were analyzed using a dual-luciferase reporter assay kit (Promega) after transfection for 48 h.
RNA immunoprecipitation (RIP)
In short, A-549 and NCI-H1299 cells at 80% confluency were disrupted using complete RIP lysis buffer (Millipore). Also, 50 μL magnetic beads of each system were resuspended with 100 μL RIP wash buffer, followed by an incubation with 5 μg antibody for 2 h. After that, the cell extract was incubated with the beads pre-coated with anti-Argonaute-2 (Ago2; Millipore) or anti-immunoglobulin G (IgG, Millipore) for 6 h at 4°C. After treatment with proteinase K (Invitrogen), the samples were subjected to RT-qPCR analysis.
RNA pull-down assay
Generally, A-549 and NCI-H1299 cells were transfected with 50 nM of biotinylated wild type or mutant miR-1298-5p (Bio-miR-1298-5p-WT or Bio-miR-1298-5p-MUT) or a negative control Bio-miR-NC, based on the Lipofectamine RNAiMax (Invitrogen). Subsequently, cells were cultured for 48 h and then harvested for lysis. M-280 streptavidin magnetic beads (Sigma-Aldrich) were added to the cellular lysates, followed by incubation at 4°C for 3 h. At length, the beads were washed in wash buffer, and the bound RNAs were purified and analyzed using RT-qPCR assay.
Tumor xenograft assay
Five-week-old male BALB/C nude mice (Vital River Laboratory, Beijing, China) were used for this animal experiment, which got the approval of the Animal Ethics Committee of the Jianhu Hospital Affiliated to Nantong University. The mice in the specific-pathogen-free environment were randomly divided into two groups, with six mice in each group. After that, 5 × 106 A-549 cells with Lenti-short hairpin (sh)-NC (Genechem, Shanghai, China) or Lenti-sh-circ_0003028 (Genechem) were injected subcutaneously into the flank of mice, respectively. Subsequently, the tumor size was assessed every 5 days with a caliper. Tumor volume was calculated according to the formula: volume = length × width 2 /2. Thirty days after cell injection, all mice were euthanized. The tumors were removed for weight and analysis. In addition, immunohistochemical staining was executed as described previously [Citation20]. The tissue sections were incubated with antibody specific for PCNA (1:5000, #13,110, Cell Signaling Technology) to evaluate proliferation, followed by analysis by at least two pathologists.
Statistical analysis
The data in this study were analyzed using GraphPad Prism7 (GraphPad Prism software, San Diego, CA, USA) and presented as the mean ± standard deviation (SD). Pearson correlation analysis was employed to assess the expression association of circ_0003028, miR-1298-5p, and GOT2. Significant differences were analyzed by Student’s t-test for two groups and one-way analysis of variance (ANOVA) with Tukey’s tests for multiple groups. A P-value of less than 0.05 was indicated statistically significant.
Results
Circ_0003028 expression was increased in NSCLC tissues and cells
First of all, to investigate the function of circ_0003028 in NSCLC, its expression pattern was detected by RT-qPCR assay. As exhibited in ), circ_0003028 level was upregulated in NSCLC tissue samples (n = 53) relative to adjacent normal tissue samples (n = 53). Moreover, the upregulation of circ_0003028 was also viewed in lung cancer cell lines (A-549, NCI-H1299, and NCI-H460) when compared with the normal human bronchial epithelial cell line (BEAS-2B) ()).
Figure 1. Circ_0003028 was elevated in NSCLC tissues and cells. (a) RT-qPCR assay was applied to determine the expression level of circ_0003028 in 53 pairs of NSCLC tissue samples and adjacent normal tissues. (b) Circ_0003028 level was detected in normal human bronchial epithelial cell line (BEAS-2B) and lung cancer cell lines (A-549, NCI-H1299, and NCI-H460) by RT-qPCR assay. (c) The cellular localization of circ_0003028 in A-549 and NCI-H1299 cells was analyzed by Subcellular fractionation assay. (d) The levels of circ_0003028 and linear FUT8 mRNA were assessed in A-549 and NCI-H1299 cells treated with or without RNase R by RT-qPCR assay. *P < 0.05
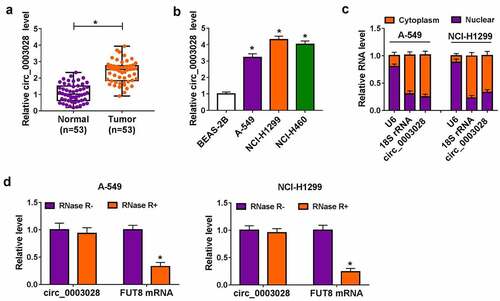
Meanwhile, we further verified that circ_0003028 was predominantly located in the cytoplasm of A-549 and NCI-H1299 cells, implying the possible post-transcriptional regulatory mechanism of circ_0003028 in NSCLC cells ()). Apart from that, to measure the stability of circ_0003028, A-549 and NCI-H1299 cells were treated with RNase R. As presented in ), the circular isoform was resistant to RNase R, while the linear isoform was apparently declined after RNase R treatment. Together, these data suggested that the involvement of circ_0003028 in NSCLC progression.
Downregulation of circ_0003028 suppressed proliferation, colony formation, cell cycle progression, migration, invasion, angiogenesis, and promoted apoptosis and autophagy of lung cancer cells in vitro
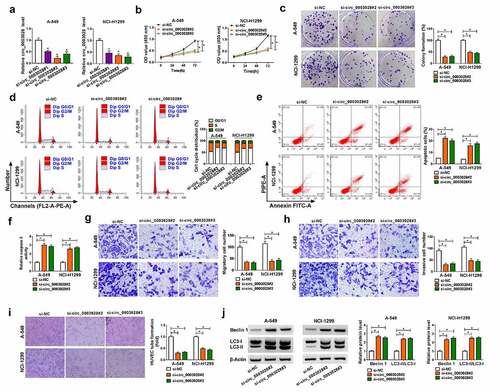
To assess the potential biological effects of circ_0003028 in NSCLC progression, we knocked down circ_0003028 expression in A-549 and NCI-H1299 cells. Also, the transfection efficiency of si-circ_0003028#1, si-circ_0003028#2, and si-circ_0003028#3 was assessed and presented in ). Particularly, the knockdown efficiency of si-circ_0003028#2 and si-circ_0003028#3 is more significant, so they were chosen for follow-up functional studies. CCK-8 and colony formation assay suggested that the deficiency of circ_0003028 repressed cell proliferation and clone number in A-549 and NCI-H1299 cells in comparison with their respective control groups (). Meanwhile, cell cycle results displayed that the cells in the si-circ_0003028#2 and si-circ_0003028#3 groups had an arrested cell cycle with a higher proportion of G0/G1-phase and a lower proportion of S-phase than the control groups ()). In addition, the knockdown of circ_0003028 could increase the apoptosis rate of A-549 and NCI-H1299 cells ()), as evidenced by enhanced caspase 3 activity ()). Synchronously, transwell assay exhibited that the silencing of circ_0003028 led to a substantial decline in migration and invasion in A-549 and NCI-H1299 cells versus their counterparts (). Consistently, the tube formation ability of HUVEC cells was also reduced due to the downregulation of circ_0003028 in A-549 and NCI-H1299 cells ()). Besides, autophagy has been reported to be related to apoptosis exists in NSCLC [Citation21], so we further identified the autophagy-associated proteins (Beclin1 and LC3-II/LC3-I) in A-549 and NCI-H1299 cells. Data showed that the protein levels of Beclin1 and LC3-II/LC3-I was improved on account of circ_0003028 knockdown in A-549 and NCI-H1299 cells ()), suggesting the promoting role of circ_0003028 downregulation on autophagy in NSCLC cells. Collectively, these results indicated that circ_0003028 deficiency could inhibit cell growth and metastasis of NSCLC cells in vitro.
Figure 2. The effects of circ_0003028 silencing on proliferation, colony formation, cell cycle progression, apoptosis, migration, invasion, and autophagy in NSCLC cells. (a) Relative circ_0003028 level was determined by RT-qPCR assay in A-549 and NCI-H1299 cells transfected with si-NC, si-circ_0003028#1, si-circ_0003028#2, and si-circ_0003028#3. (b-j) A-549 and NCI-H1299 cells were transfected with si-NC, si-circ_0003028#2, and si-circ_0003028#3. (b) CCK-8 assay was used to assess cell proliferation in transfected A-549 and NCI-H1299 cells. (c) Cell colony formation assay was performed to examine the clone number in transfected A-549 and NCI-H1299 cells. (d) Flow cytometry assay was carried out to analyze cell cycle distribution in transfected A-549 and NCI-H1299 cells. (e) Flow cytometry assay was conducted to test apoptosis rate in transfected A-549 and NCI-H1299 cells. (f) Caspase 3 activity analysis was detected by the special kit. (g and h) Transwell assay was applied to determine migration and invasion in transfected A-549 and NCI-H1299 cells. (i) Tube formation assay was used to detect the tube formation ability of HUVEC in transfected A-549 and NCI-H1299 cells. (j) Western blot assay was implemented to determine the protein levels of Beclin1 and LC3-II/LC3I in transfected A-549 and NCI-H1299 cells.*P < 0.05
Circ_0003028 could directly interact with miR-1298-5p
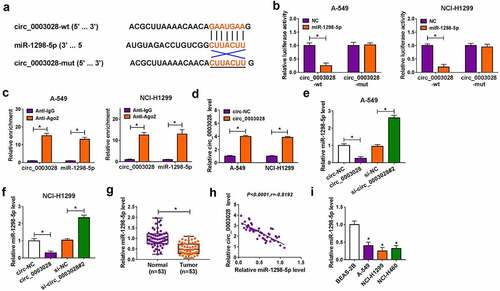
Further, we tried to explore the mechanism of action of circ_0003028 by seeking the putative circ_0003028-interacting miRNAs using circular RNA Interactome software. As a result, miR-1298-5p was found to have some binding sites with circ_0003028 ()), as verified by dual-luciferase reporter assay. Data exhibited that the forced expression of miR-1298-5p distinctly decreased the luciferase activity of circ_0003028-wt reporter, while had little effect on the luciferase activity of circ_0003028-mut reporter in A549 and NCI-H1299 cells ()). And then, in order to confirm the mutual effect of circ_0003028 and miR-1298-5p at the endogenous levels, RIP assay was carried out using antibody Ago2, which is an important component of the RNA-induced silencing complex (RISC). Data displayed that circ_0003028 and miR-1298-5p were significantly enriched in Ago2 pellets of A549 and NCI-H1299 cell extracts relative to the IgG control group ()). To further prove the sponge effect between circ_0003028 and miR-1298-5p, we carried out a biotin-coupled miRNA capture assay. Results displayed that the enrichment of circ_0003028 in the captured fraction was much higher in the Bio-miR-1298-5p-WT group than in the Bio-miR-1298-5p-MUT group and Bio-miR-NC group (Figure S1). Meanwhile, the overexpression efficiency of circ_0003028 was detected and exhibited in ). Also, we further determined that the authentic effect of circ_0003028 on miR-1298-5p in lung cancer cells. Results indicated that the expression of miR-1298-5p was decreased by circ_0003028 upregulation in A549 and NCI-H1299 cells, whereas miR-1298-5p level was increased due to the silencing of circ_0003028 (). In addition, we found that miR-1298-5p was downregulated in NSCLC tissues, and negatively associated with circ_0003028 (). Synchronously, our data also proved that the trend of miR-1298-5p expression in NSCLC cellular level was in line with that in tissues ()). In a word, circ_0003028 modulated the abundance of miR-1298-5p by binding to miR-1298-5p.
Figure 3. Circ_0003028 directly bound with miR-1298-5p. (a) Schematic of a putative target sequence for miR-1298-5p in circ_0003028 and mutated miR-1298-5p-binding sites. (b) A dual-luciferase reporter assay was utilized to verify the binding relationship in A-549 and NCI-H1299 cells. (c) miR-1298-5p endogenously associated with circ_0003028 was determined in A-549 and NCI-H1299 cell extracts by RIP assay. (d) Circ_0003028 level was detected in circ-NC or circ_0003028-transfected A-549 and NCI-H1299 cells by RT-qPCR assay. (e and f) miR-1298-5p level was measured by RT-qPCR assay in A-549 and NCI-H1299 cells transfected with circ-NC, circ_0003028, si-NC, and si-circ_0003028. (g) miR-1298-5p level was examined by RT-qPCR assay in NSCLC tissue samples and adjacent normal tissues. (h) The expression association between circ_0003028 and miR-1298-5p in NSCLC tumor tissues was assessed by Pearson correlation analysis. (i) miR-1298-5p level was determined in BEAS-2B, A-549, NCI-H1299, and NCI-H460 cells by RT-qPCR assay. *P < 0.05
Knockdown of circ_0003028 inhibited NSCLC progression by interacting with miR-1298-5p in vitro
To further tunnel whether miR-1298-5p participated in the promotion of NSCLC progression induced by circ_0003028, we performed rescue assays in A549 and NCI-H1299 cells. Meanwhile, the knockdown efficiency of anti-miR-1298-5p was first assessed and exhibited in ). Subsequently, RT-qPCR assay indicated that the co-transfection of miR-1298-5p inhibitor could counteract the circ_0003028 silencing-triggered enhancement in miR-1298-5p level in A549 and NCI-H1299 cells ()). Functional analysis suggested that the cell proliferative ability impaired by circ_0003028 deletion was notably abrogated by miR-1298-5p downregulation in A549 and NCI-H1299 cells (). Meanwhile, the deficiency of circ_0003028 could induce cell cycle arrest in A549 and NCI-H1299 cells, which was obviously mitigated by miR-1298-5p reduction ()). Apart from that, the promotion role of apoptosis rate by circ_0003028 silencing was markedly overturned by the re-introduction of anti-miR-1298-5p in A549 and NCI-H1299 cells ()), as evidenced by lower caspase 3 activity ()). Also, transwell assay suggested that si-circ_0003028 resulted in a significant decline in migration and invasion, while miR-1298-5p downregulation could evidently reverse these effects in A549 and NCI-H1299 cells (). Besides, our data presented that the re-transfection of miR-1298-5p inhibitor could abolish the repression effect of circ_0003028 knockdown on the tube formation of HUVEC cells ()). In terms of autophagy, the pro-autophagy proteins Beclin1 and LC3-II/LC3-I expression were improved by circ_0003028 silencing and then was partly abrogated by miR-1298-5p knockdown in A549 and NCI-H1299 cells ()). These results reveal that circ_0003028 downregulation could suppress proliferation, migration, invasion, angiogenesis, and accelerate apoptosis and autophagy of NSCLC by targeting miR-1298-5p in vitro.
Figure 4. Downregulation of miR-1298-5p abolished the effects of circ_0003028 knockdown on proliferation, colony formation, cell cycle progression, apoptosis, migration, invasion, and autophagy in NSCLC cells. (a) miR-1298-5p level was detected in anti-NC or anti-miR-1298-5p-transfected A-549 and NCI-H1299 cells by RT-qPCR assay. (b-k) A-549 and NCI-H1299 cells were transfected with si-NC, si-circ_0003028#2, si-circ_0003028#2+ anti-NC, and si-circ_0003028#2+ anti-miR-1298-5p. (b) miR-1298-5p level was determined by RT-qPCR assay in transfected A-549 and NCI-H1299 cells. (c) Proliferation was assessed by CCK-8 assay in transfected A-549 and NCI-H1299 cells. (d) Colony number was calculated by colony formation assay in transfected A-549 and NCI-H1299 cells. (e and f) Cell cycle distribution and apoptosis rate were analyzed by flow cytometry assay in transfected A-549 and NCI-H1299 cells. (g) Caspase 3 activity was detected by special kit in transfected A-549 and NCI-H1299 cells. (h and i) Migration and invasion were measured by transwell assay in transfected A-549 and NCI-H1299 cells. (j) HUVEC tube formation ability was examined by tube formation assay in transfected A-549 and NCI-H1299 cells. (k) Protein levels of Beclin1 and LC3-II/LC3I were tested by western blot assay in transfected A-549 and NCI-H1299 cells. *P < 0.05
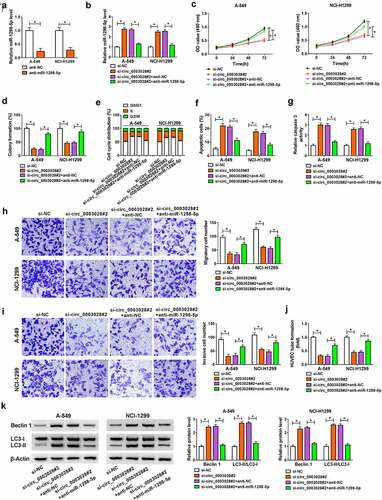
GOT2 acted as a direct target of miR-1298-5p
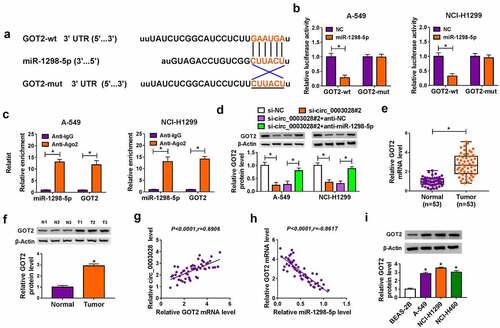
To further explore the molecular mechanism of miR-1298-5p in NSCLC, starbase software was applied to search the underlying candidate target mRNAs of miR-1298-5p. As exhibited in ), miR-1298-5p comprised the binding sites in GOT2 3ʹUTR. The next dual-luciferase reporter assay confirmed the prediction. Data suggested that the luciferase activity in A549 and NCI-H1299 cells transfected with GOT2-wt and miR-1298-5p was notably reduced compared with that in cells transfected with GOT2-wt and NC, while there was no evident impact in the cells with GOT2-mut ()). Meanwhile, the Ago2 RIP analysis was also performed to verify their binding. As presented in ), when compared with the negative control IgG group, miR-1298-5p and GOT2 were specially enriched on the same Ago2-based complex. Apart from that, the results of RNA pull-down assay suggested that GOT2 was pulled down when using Bio-miR-1298-5p-WT rather than Bio-miR-1298-5p-MUT and Bio-miR-NC in in A549 and NCI-H1299 cells (Figure S1). In addition, western blot results displayed that the knockdown of circ_0003028 could block the protein level of GOT2, while the co-transfection of anti-miR-1298-5p counteract the effect in A549 and NCI-H1299 cells ()), implying that circ_0003028 could serve as a sponge of miR-1298-5p to regulate GOT2 expression. Besides, the upregulation of GOT2 was noticed in NSCLC tissues relative to the normal tissues (). As expected, GOT2 expression was positively related to circ_0003028, and inversely associated with miR-1298-5p in NSCLC tissues (). We also verified that the expression level of GOT2 was increased in NSCLC cell lines in comparison with the BEAS-2B cells ()). Overall, these data suggested that GOT2 was a direct target of miR-1298-5p in NSCLC cells.
Figure 5. GOT2 was a direct target of miR-1298-5p. (a) The potential binding sites between GOT2 and miR-1298-5p were predicted using starbase software. (b) The binding relationship was proved by a dual-luciferase reporter assay. (c) RIP assay was performed to verify the interaction between GOT2 and miR-1298-5p. (d) GOT2 protein level was measured by western blot assay in A-549 and NCI-H1299 cells transfected with si-NC, si-circ_0003028#2, si-circ_0003028#2+ anti-NC, and si-circ_0003028#2+ anti-miR-1298-5p. (e and f) GOT2 expression was examined by RT-qPCR and western blot assay in NSCLC tissues. (g and h) Pearson correlation analysis was used to analyze the expression correlation of GOT2 with miR-1298-5p and circ_0003028 in NSCLC tissues. (i) GOT2 protein level was determined by western blot assay in BEAS-2B, A-549, NCI-H1299, and NCI-H460 cells. *P < 0.05
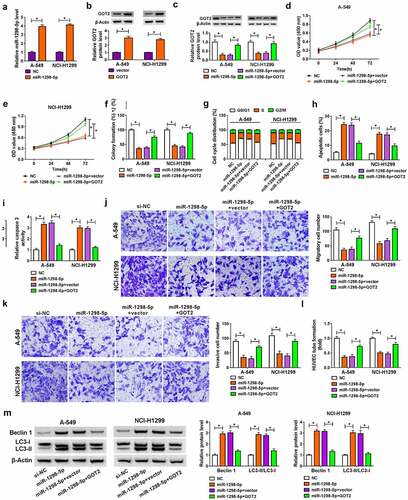
miR-1298-5p upregulation could block NSCLC progression by targeting GOT2 in vitro
Then, we further investigated whether the regulatory role of miR-1298-5p on NSCLC development could be mediated by regulating GOT2. The transfection efficiency of miR-1298-5p mimic and pcDNA-GOT2 was measured and exhibited in . After that, western blot results displayed that the upregulation of miR-1298-5p decreased GOT2 protein level in A-549 and NCI-H1299 cells, which was counteracted after pcDNA-GOT2 introduction ()). Functionally, the forced expression of GOT2 attenuated the inhibitory action of miR-1298-5p on cell proliferative ability in A-549 and NCI-H1299 cells (–). Also, decreased cell cycle progression caused by miR-1298-5p mimic was weakened by GOT2 upregulation in A-549 and NCI-H1299 cells ()). Moreover, the overexpression of GOT2 relieved the facilitation effect of miR-1298-5p upregulation on apoptosis rate in A-549 and NCI-H1299 cells ()), as reflected by reduced caspase 3 activity ()). Apart from that, the re-introduction of pcDNA-GOT2 could effectively reverse miR-1298-5p mimic-triggered decline in migration and invasion in A-549 and NCI-H1299 cells (). Likewise, GOT2 elevation could ameliorate the negative action of miR-1298-5p on the tube formation ability of HUVEC cells ()). In addition, western blot results indicated that the upregulation of miR-1298-5p resulted in an overt improvement in Beclin1 and LC3-II/LC3I level, which was reversed by pcDNA-GOT2 re-introduction in A-549 and NCI-H1299 cells ()). All of these data implied that GOT2 upregulation could partly abolish the suppressive action of miR-1298-5p on NSCLC development.
Figure 6. Overexpression of GOT2 abrogated miR-1298-5p-mediated NSCLC progression in vitro. (a) miR-1298-5p level was examined in NC or miR-1298-5p-transfected A-549 and NCI-H1299 cells by RT-qPCR assay. (b) GOT2 protein level was assessed in NC or miR-1298-5p-transfected A-549 and NCI-H1299 cells by western blot assay. (c-m) A-549 and NCI-H1299 cells were transfected with NC, miR-1298-5p, miR-1298-5p+vector, and miR-1298-5p+ GOT2. (c) GOT2 protein level was measured in transfected A-549 and NCI-H1299 cells by western blot assay. (d and e) The detection of cell proliferation was performed by CCK-8 assay. (f) The calculation of colony number was conducted by colony formation assay. (g and h) The analysis of cell cycle distribution and apoptosis rate was carried out by flow cytometry assay. (i) The determination of caspase 3 activity was executed by special kit. (j and k) The assessment of migration and invasion was implemented by transwell assay. (l) The tube formation of HUVEC cells was detected by tube formation assay in transfected A-549 and NCI-H1299 cells. (m) The detection of Beclin1 and LC3-II/LC3I protein levels was conducted by western blot assay in transfected A-549 and NCI-H1299 cells. *P < 0.05
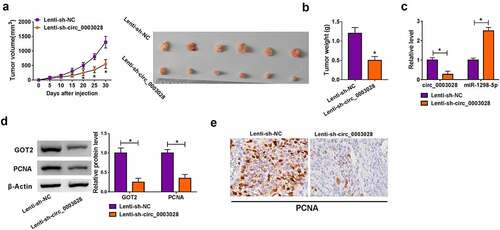
Circ_0003028 deficiency repressed cell growth of NSCLC in vivo
As mentioned above, we inferred that circ_0003028 knockdown might hinder tumor growth of NSCLC in vivo. To verify the speculation, we established mice xenograft models of NSCLC. Data exhibited that tumor volume and weight dropped in response to circ_0003028 downregulation (), indicating that the deficiency of circ_0003028 dampened cell growth of NSCLC in vivo. Furthermore, RT-qPCR and western blot analysis exhibited that circ_0003028, GOT2, and PCNA (a pro-proliferation marker) expression was prominently lower in tumor tissues from the Lenti-sh-circ_0003028 group than the Lenti-sh-NC group, whereas the expression level of miR-1298-5p presented the opposite trend in this xenograft (). Also, Immunohistochemical staining verified that the expression level of PCNA was impeded by circ_0003028 downregulation in the xenograft ()). Thus, it is concluded that circ_0003028 silencing could restrain cell growth of NSCLC in vivo.
Figure 7. Circ_0003028 deficiency repressed NSCLC cell growth in vivo. A-549 cells introduced with Lenti-sh-NC or Lenti-sh-circ_0003028 were inoculated subcutaneously into the nude mice. (a and b) Tumor volume and tumor weight were detected in the xenografts. (c) Circ_0003028 and miR-1298-5p expression were examined by RT-qPCR assay in this xenograft. (d) Protein levels of GOT2 and PCNA were assessed by western blot assay in this xenograft. (e) The expression of PCNA was determined by Immunohistochemical staining of sections from the NSCLC xenograft model in nude mice. *P < 0.05
Discussion
Up to now, plenty of circRNAs have drawn great attention on account of the important roles in various cancers [Citation22,Citation23], containing lung cancer [Citation24]. Unlike linear mRNA, the high stability and abundance of circRNA have been highlighted as potential promising biomarkers [Citation25]. Accumulating evidence is pointing toward the dysregulation of circRNAs can widely participate in the modulation of the occurrence and progression in diverse cancer, including NSCLC [Citation26]. In this paper, we identified the upregulation of circ_0003028 in NSCLC, in agreement with a former study [Citation14]. Meanwhile, circ_0003028 was found to be unusually stable and resistant to RNase R, implying the underlying biomarker for NSCLC prognosis. Functionally, the deficiency of circ_0003028 retard proliferation, migration, invasion, angiogenesis, and expedite apoptosis of NSCLC cells in vitro. Autophagy, a highly conserved cellular process of self-digestion, has been reported to be related to tumor development [Citation27]. It has been confirmed that autophagy could take part in cell death in NSCLC [Citation21,Citation28]. In this article, our results suggested that circ_0003028 knockdown could induce autophagy of NSCLC cells in vitro. Consistently, the inhibitory action of circ_0003028 silencing on tumor growth of NSCLC was validated in nude mice. That was to say, circ_0003028 exerts an oncogenic activity of NSCLC in vitro and in vivo.
As widely believed, circRNAs could perform as miRNAs sponges that reduce the binding of miRNAs to their target genes [Citation29]. Apart from that, the current study confirmed the mainly cytoplasmic localization of circ_0003028 in NSCLC, providing a possibility for the interaction with miRNAs. Of note, we first proved that circ_0003028 bound with miR-1298-5p and inhibited its expression. In addition, miR-1298 has been reported to block cell growth and boost the apoptosis of lung cancer cells [Citation30]. In our research, the low expression of miR-1298-5p in NSCLC tissues and cells was demonstrated, consistent with the earlier study [Citation31]. Furtherly analysis suggested that the downregulation of miR-1298-5p could partly abolish the repression role of circ_0003028 knockdown on NSCLC progression. Additionally, circ_0003028 was reported to partake in the regulation of cell growth and metastasis in liver cancer and NSCLC by sponging multiple miRNAs [Citation13,Citation32]. These results inducted that circ_0003028 involves in complex regulatory networks. Likewise, using the bioinformatics software, we found that GOT2 might be the latent target gene of miR-1298-5p. According to previous studies, GOT2 serves as a key transfer enzyme participating in the amino acid metabolism of tumor cells [Citation33]. Moreover, GOT2 has been pointed out to accelerate proliferation, migration, invasion, and metabolic regulation of NSCLC cells [Citation34]. In this research, GOT2 expression was increased in NSCLC, in line with the prior work [Citation35]. Synchronously, our data discovered that the overexpression of GOT2 could partially reverse miR-1298-5p-mediated repression on NSCLC development. Additionally, GOT2 expression could be positively regulated by circ_0003028/miR-1298-5p in NSCLC, further supporting the circ_0003028/miR-1298-5p/GOT2 axis in NSCLC. Besides, it has been reported that GOT2 can be involved in the transformation of glutamine [Citation36,Citation37], which is a transitional demand for the growth of tumor cells [Citation38]. In future research, we will focus on the regulatory effect of circ_0003028/miR-1298-5p/GOT2 axis on glutamine metabolism in NSCLC. Compared with previous NSCLC studies [Citation39], the novelty of this paper is that it not only enriches the circRNA-miRNA-mRNA network based on competitive endogenous RNA (ceRNA), but also explores the functional role of circRNA-based experiments in nude mice. These findings provide an important preclinical basis for NSCLC treatment.
Conclusion
Taken together, these results suggested that circ_0003028 could act as a sponge of miR-1298-5p to regulate GOT2 expression, thereby promoting NSCLC progression. Our findings provided an important preclinical basis for NSCLC treatment.
Research highlights
Circ_0003028 was upregulated in NSCLC tissues and cells.
Circ_0003028 boosted NSCLC cell progression
Circ_0003028/miR-1298-5p/GOT2 axis in NSCLC.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated or analysed during this study are included in this published article (and its supplementary information files).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Siegel RL, Miller KD. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
- Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103:463–473.
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454.
- Sekihara K, Hishida T, Yoshida J, et al. Long-term survival outcome after postoperative recurrence of non-small-cell lung cancer: who is ‘cured’ from postoperative recurrence? Eur J Cardiothorac Surg. 2017;52:522–528.
- Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816.
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442.
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565.
- Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127.
- Sun Q, Li X, Xu M, et al. Differential expression and bioinformatics analysis of circRNA in non-small cell lung cancer. Front Genet. 2020;11:586814.
- Li G, Zhao C, Zhang H, et al. Hsa_circ_0046263 drives the carcinogenesis and metastasis of non-small cell lung cancer through the promotion of NOVA2 by absorbing Mir-940 as a molecular sponge. Cancer Manag Res. 2020;12:12779–12790.
- Cheng F, Yu J, Zhang X, et al. CircSEC31A promotes the malignant progression of non-small cell lung cancer through regulating SEC31A expression via sponging miR-376a. Cancer Manag Res. 2020;12:11527–11539.
- Ren S, Xin Z. Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle. 2017;16:2204–2211.
- Li L, Sun D, Li X, et al. Identification of key circRNAs in non-small cell lung cancer. Am J Med Sci. 2021;361:98–105.
- Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18.
- Chen W, Lu Q, Li S, et al. microRNA-1298 inhibits the malignant behaviors of breast cancer cells via targeting ADAM9. Biosci Rep. 2020;40:BSR20201215.
- Li G, Sun L, Mu Z, et al. MicroRNA-1298-5p inhibits cell proliferation and the invasiveness of bladder cancer cells via down-regulation of connexin 43. Biochem Cell Biol. 2020;98:227–237.
- Qiu ZK, Liu N, Zhao SF, et al. MiR-1298 expression correlates with prognosis and inhibits cell proliferation and invasion of gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22:1672–1679.
- Jiang G, Wu AD, Huang C, et al. Isorhapontigenin (ISO) inhibits invasive bladder cancer formation in vivo and human bladder cancer invasion in vitro by targeting STAT1/FOXO1 Axis. Cancer Prev Res (Phila). 2016;9:567–580.
- Liu G, Pei F, Yang F, et al. Role of autophagy and apoptosis in non-small-cell lung cancer. Int J Mol Sci. 2017;18:367.
- Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7.
- Feng J, Chen K, Dong X, et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer. 2019;18:35.
- Braicu C, Zimta AA. The function of non-coding RNAs in lung cancer tumorigenesis. Cancers (Basel). 2019;11:605.
- Ojha R, Nandani R, Chatterjee N, et al. Emerging role of circular RNAs as potential biomarkers for the diagnosis of human diseases. Adv Exp Med Biol. 2018;1087:141–157.
- Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145:2875–2889.
- Onorati AV, Dyczynski M, Ojha R, et al. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318.
- Zhang Z, Zhang M, Liu H, et al. AZD9291 promotes autophagy and inhibits PI3K/Akt pathway in NSCLC cancer cells. J Cell Biochem. 2019;120:756–767.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388.
- Zhou Y, Dang J, Chang KY, et al. miR-1298 inhibits mutant KRAS-driven tumor growth by repressing FAK and LAMB3. Cancer Res. 2016;76:5777–5787.
- Du Z, Wu J, Wang J, et al. MicroRNA-1298 is downregulated in non-small cell lung cancer and suppresses tumor progression in tumor cells. Diagn Pathol. 2019;14:132.
- Zhu H, Lu Q, Lu Q, et al. Matrine regulates proliferation, apoptosis, cell cycle, migration, and invasion of non-small cell lung cancer cells through the circFUT8/miR-944/YES1 axis. Cancer Manag Res. 2021;13:3429–3442.
- Hsu PP, Sabatini DM. Cancer cell metabolism: warburg and beyond. Cell. 2008;134:703–707.
- Jin M, Shi C, Hua Q, et al. High circ-SEC31A expression predicts unfavorable prognoses in non-small cell lung cancer by regulating the miR-520a-5p/GOT-2 axis. Aging (Albany NY). 2020;12:10381–10397.
- Gao L, He RQ, Wu HY, et al. Expression signature and role of miR-30d-5p in non-small cell lung cancer: a comprehensive study based on in silico analysis of public databases and in vitro experiments. Cell Physiol Biochem. 2018;50:1964–1987.
- Yang S, Hwang S, Kim M, et al. Mitochondrial glutamine metabolism via GOT2 supports pancreatic cancer growth through senescence inhibition. Cell Death Dis. 2018;9:55.
- Feist M, Schwarzfischer P, Heinrich P, et al. Cooperative STAT/NF-κB signaling regulates lymphoma metabolic reprogramming and aberrant GOT2 expression. Nat Commun. 2018;9:1514.
- DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350.
- Zhang CC, Li Y, Feng XZ, et al. Circular RNA circ_0001287 inhibits the proliferation, metastasis, and radiosensitivity of non-small cell lung cancer cells by sponging microRNA miR-21 and up-regulating phosphatase and tensin homolog expression. Bioengineered. 2021;12:414–425.