ABSTRACT
Fluoxetine is used to improve cognition, exercise ability, depression, and neurological functions in patients with cerebral ischemic stroke. Circular RNAs (circRNAs) play important regulatory roles in multiple diseases. However, studies regarding the fluoxetine-mediated circRNA–microRNA–messenger RNA (mRNA) axis have not been conducted. This study is aim to investigate the functions of fluoxetine and identification of fluoxetine-mediated circRNAs and mRNAs in cerebral ischemic stroke. The middle cerebral artery occlusion (MCAO) rat models were successfully established at fisrt, and then rats were intraperitoneally injected with 10-mg/kg fluoxetine hydrochloride for 14 d. Afterward, the cerebral infarction area was evaluated using triphenyltetrazolium chloride staining. High-throughput sequencing was adopted to screen the differential circRNAs and mRNAs. The candidate circRNAs, mRNAs, and potential microRNAs were verified using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). In addtion, microRNA and circRNA binding was verified using the dual-luciferase reporter assay. Results revealed that fluoxetine markedly diminished the cerebral infarction area in rats after MCAO. The circRNAs and mRNAs were differentially expressed, which includes 879 circRNAs and 815 mRNAs between sham and MCAO groups, respectively, and 958 circRNAs and 838 mRNAs between MCAO and fluoxetine groups, respectively. In which, circMap2k1 and Pidd1 expression was significantly increased in the MCAO group but suppressed after fluoxetine treatment. Moreover, circMap2k1 directly binds with miR-135b-5p. Taken together, we verified that fluoxetine could improve brain injury after cerebral ischemic stroke. Moreover, the circMap2k1/miR-135b-5p/Pidd1 axis is potentially involved in cerebral ischemic stroke.
Highlights
Fluoxetine improve brain injury after cerebral ischemic stroke.
The circMap2k1 directly binds with miR-135b-5p.
Fluoxetine regulates circMap2k1/miR-135b-5p/Pidd1 axis in cerebral ischemic stroke.
1. Introduction
Ischemic stroke is a type of clinical syndrome that causes irreversible damage and results in ischemic hypoxic necrosis of local tissues and corresponding neurological defects caused by a blood supply disorder in the brain [Citation1]. Ischemic stroke is characterized by the sudden and focal loss of nerve functions [Citation2]. The main clinical symptoms of ischemic stroke include sudden fainting, lateral appendage numbness accompanied by distortion of the commissure, aphasia, and hemiplegia, and these symptoms may be life-threatening [Citation3,Citation4]. Presently, the incidence, disability, and recurrence rates of ischemic stroke is increasing globally; it has become the third leading cause of death in the world [Citation5]. Studies have confirmed that ischemic stroke is a chronic non-communicable disease caused by interacting multiple environmental and genetic factors, and its pathogenesis is complex [Citation6,Citation7]. Currently, statins and aspirin are good secondary prevention drugs against ischemic stroke [Citation8,Citation9]. These drugs have a single target and can only act on a specific aspect in the pathogenesis of ischemic stroke. Moreover, they can only reduce the recurrence rate by approximately 15% [Citation10]. Therefore, an in-depth discussion of its risk factors and pathogenesis is necessary to diagnose, treat, and prevent ischemic stroke.
Fluoxetine, a selective 5-hydroxytriptamine (5-HT) reuptake inhibitor, is commonly marketed as fluoxetine hydrochloride [Citation11]. In the past, fluoxetine was clinically used as an antidepressant [Citation12]. A study showed that fluoxetine is mainly used to treat depression by inhibiting the reuptake of 5-HT by synaptic cells, thereby increasing extracellular 5-HT levels [Citation13]. Recent studies have also verified that fluoxetine can promote neuron regeneration and nerve function recovery [Citation14,Citation15]. Fluoxetine has also been reported to have various pharmacological effects, including antitumor, anti-inflammatory, antioxidant, and antiplatelet aggregation activities [Citation16,Citation17]. Currently, several studies have demonstrated the vital roles of fluoxetine in ischemic stroke, such as stroke recurrence [Citation18], neurological prognosis [Citation19], motor recovery [Citation20], and neurogenesis [Citation21]. However, the regulatory genes and mechanisms behind fluoxetine’s role in ischemic stroke have rarely been reported.
Circular RNAs (circRNAs), a class of non-coding RNAs, are closed circular structures without 5ʹ end caps and 3ʹ end poly-A tails [Citation22]. CircRNAs are not easily degraded by the RNase exonuclease, and their structures are more stable than those of linear RNA [Citation23], allowing circRNAs to be stably expressed in various biological cells and tissues [Citation24]. Studies have also shown that circRNAs play important regulatory roles in multiple diseases, such as cardiovascular [Citation25,Citation26], neurological [Citation27,Citation28], tumoral diseases [Citation23,Citation29], and autoimmune diseases [Citation30]. Presently, numerous studies have reported that most circRNAs are relevant to cerebral ischemia–reperfusion injury [Citation31,Citation32]. MicroRNAs (miRNAs) are small non-coding RNAs with an intracellular length of approximately 22 bp that participate in many important biological processes, such as cell proliferation, differentiation, apoptosis, and metabolism [Citation33,Citation34]. Studies showed that miR-211 and miR-22 inhibited cerebral ischemia/reperfusion injury by decreasing cell apoptosis [Citation35,Citation36]. Moreover, circRNAs, an endogenous competitive RNA (ceRNA), play sponge adsorption roles on miRNAs and weaken the inhibitory effect of miRNAs on their target genes [Citation37,Citation38]. The circRNA–miRNA–messenger RNA (mRNA) axis has also been proven to significantly contribute to numerous biological processes, including cell differentiation, proliferation, apoptosis, metastasis, and angiogenesis [Citation39,Citation40]. In conclusion, circRNA–miRNA–mRNA axis may also participate in the development of fluoxetine-mediated ischemic stroke.
Therefore, this study is aim to explore the functions of fluoxetine and identification of fluoxetine-mediated circRNAs and mRNAs in cerebral ischemic stroke to find a new circRNA–miRNA–mRNA axis involved in the progression of fluoxetine treated ischemic stroke. The function of fluoxetine was evaluated by neurological function score and triphenyltetrazolium chloride (TTC) stainning. Then, the characterization of the expression profiles of fluoxetine-related circRNAs and mRNAs were investigated using RNA sequencing. Furthermore, the expression of candidate circRNAs, mRNAs, and potential binding miRNAs and the binding of circRNA and miRNA were verified. This study provides basic data for further research on the biological functions and mechanisms of circRNAs and transcriptome genes affecting the occurrence of cerebral ischemic stroke, and also provides a new insight into the molecular mechanism of fluoxetine-mediated ischemic stroke.
2. Materials and methods
2.1. Animal
Healthy male Sprague–Dawley (SD) rats weighing 200 ± 20 g were provided by the Guangdong Medical Laboratory Animal Center. The 21 SD rats were randomly divided into three groups, with seven rats in each group. The animals grew under natural conditions during the experiment, with the temperature set to 21°C–24°C and humidity at 50%–70%. This animal experiment was conducted according to the Animal Experiment Ethics Committee of Jinan University.
2.2. Establishment of the MCAO model
The MCAO rat models were established following the Zea-Longa method [Citation41]. All SD rats fasted for 8 h before the operation, after which the SD rats were anesthetized using an intraperitoneal injection of 2% sodium pentobarbital (2 mL/kg, cat. no. 57–33-0, Chemical book, China). SD rats were fixed on their back against the operating table, and the skin was cut along the middle of the neck. The right common carotid artery (CCA), external carotid artery (ECA), and internal carotid artery (ICA) were fully exposed under a microscope. The proximal end of the CCA and distal end of the ECA and ICA was ligated in turn, and the proximal end of the CCA was bifurcated with an artery clamp. Then, the nylon threads were inserted from the ECA through the ICA until it reached the middle cerebral artery (MCA). After the MCA was blocked for 2 h, the nylon threads were removed, and reperfusion was performed for 14 d. In the sham group, only the arteries were separated, and no nylon threads were inserted. In the fluoxetine treatment group, after pulling out the nylon threads for 30 min, the rats after MCAO were intraperitoneally injected with 10-mg/kg fluoxetine hydrochloride once daily for 14 d.
2.3. Neurological function score
The Zea-Longa scoring criteria were used as follows: 0 – normal without neurological impairment; 1 – the left forepaw cannot be fully extended; 2 – turns to the left when walking; 3 – inclines to the left when walking; 4 – inability to walk spontaneously and exhibits loss of consciousness. The higher the score, the more severe the animal behavior disorder. Before the SD rats in each group were sacrificed, their neurological function was scored, and the MCAO model was considered successful if the rat scores were .2.
2.4. TTC staining
The SD rats in each group were fully anesthetized using an intraperitoneal injection of 2% sodium pentobarbital. After sacrifice, complete brain tissue was taken. After rinsing with cold saline, the brain tissue was frozen at −20°C for 10 min. Afterward, the brain tissue was sliced into 2-mm thick sections, and the sections were placed in 2% TTC solution (Sigma, St. Louis, MO, USA) at 37°C away from light for 20 min. After complete staining, the image information was collected using a digital camera.
2.5. RNA sequencing
The circRNA and mRNA sequencing were completed at Novogene Bioinformatics Technology Co. Ltd. (Beijing, China). In this study, total RNA was extracted from the infarcted tissue samples of the sham, MCAO model, and fluoxetine treatment groups using TRIzol (Invitrogen, CA, USA). NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA) was used to evaluate the integrity and concentration of the RNA samples. Ribosomal RNA (rRNA) was removed using Ribo-zero rRNA Removal Kit (Epicenter, Madison, WI, USA). The linear RNA was removed using RNase R (Sigma) for circRNA sequencing. Then, libraries were constructed according to the instruction of NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA). For mRNA sequencing, NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB) was used to establish libraries following the kit’s protocol. The circRNA and mRNA libraries were used to perform sequencing on the Illumina Hieq 4000 platform (Illumina, CA, USA).
2.6. Bioinformatic analysis
The R package was used to process quantile normalization and analyze the differentially expressed circRNAs and mRNAs. The circRNAs and mRNAs with log2f°ld change > 2 and P< 0.05 were considered as differentially expressed. Following guidelines set by previous studies [Citation42], the differential expression of the circRNAs and mRNAs among the samples were shown using heat and volcano map. Based on previous studies [Citation43], the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses for differential expression of mRNAs were performed using the KOBAS v.2.0 v (http://kobas.cbi.pku.edu.cn/).
2.7. Construction of circRNA–miRNA–mRNA regulation network
The differential circRNA parent gene involved in the NF-κB (nuclear factor kappa B) signaling pathway was selected. Then, six circRNAs were obtained using the following criteria: (1) circRNAs length between 450 and 2000 bp, (2) differential circRNAs with human homology, (3) parental genes not located on sex chromosomes, (4) upregulated in the MCAO group compared with those in the sham group; and downregulated in the fluoxetine treatment group compared with that in the MCAO group. According to the prediction in RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/), the circRNA–miRNA–mRNA regulation network was constructed using Cytoscape software.
2.8. Polymerase chain reaction (PCR) and quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assays
The total RNA was extracted from the infarcted tissue samples in each group, and then was subjected to reverse transcription using the First Strand cDNA Synthesis Kit (TaKaRa, Tokyo, Japan). Divergent and convergent primers were used for PCR amplification, and agarose gel electrophoresis was performed. Sanger sequencing was also conducted on the amplified products of divergent primers to verify the circular structure of circHipk3, circMap2k1, circSipa1l1, and circNav3. Furthermore, circHipk3, circMap2k1, circSipa1l1, circNav3, Cd40 (CD40 molecule), Pidd1 (P53-induced death domain protein 1), Il1b (interleukin 1 beta), LBP (lipopolysaccharide binding protein), rno-miR-18a-5p, rno-miR-135b-5p, rno-miR-135a-5p, and rno-miR-141-5p expressions were verified by qRT-PCR using SYBR Green qPCR Super Mix (Invitrogen, Carlsbad, CA, USA). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as the internal reference gene for circRNAs and mRNAs, and U6 was used as the internal reference gene of miRNAs. The sequences of primers used in this study are shown in .
Table 1. The primer sequences in this study
2.9. Bioinformatics prediction and dual-luciferase reporter assay
We used TargetScan (http://www.targetscan.org/vert_72/) to predict the binding site of rno-miR-135b-5p and circMap2k1. To verify the binding of circMap2k1 to rno-miR-135b-5p, wild-type (WT) or mutant (MUT) circMap2k1 fragments were constructed and inserted into pmirGLO vector (Promega, Madison, WI, USA). We used the Lipofectamine 3000 reagent (Thermo Fisher Scientific) to transfect WT (or MUT or pmirGLO) vectors and miR-135b-5p mimics (or mimics negative control (mimics NC)) into 293 T cells, according to the manufacturer’s instruction. After 24 h, cells were collected, and dual-luciferase reporter (Promega) was used to measure the luciferase activity.
2.10. Statistical analysis
All experiments were repeated at least three times, and the data are shown as mean ± standard deviation. Quantitative data in this study were evaluated using the SPSS v.23.0 (Chicago, IL, USA). Furthermore, differences between the two were analyzed using Student’s t-test. Analysis of variance was used to compare the difference in three or more groups, followed by Dunnett post-hoc test. P < 0.05 was used to denote statistical significance.
3. Results
The circRNA–miRNA–mRNA axis significantly contributes to numerous biological processes. However, studies regarding the fluoxetine-mediated circRNA–miRNA–mRNA axis in ischemic stroke have not been conducted. This study investigated whether there is a circRNA–miRNA–mRNA axis regulated by fluoxetine in ischemic stroke. We established MCAO in rats followed by fluoxetine treatment. Then, we further elucidated the expression profiles of fluoxetine-related circRNAs and mRNAs in ischemic stroke using high-throughput sequencing. Furthermore, the circRNA–miRNA–mRNA network was preliminarily constructed using bioinformatics analysis. The candidate circRNAs, mRNAs, and potential binding miRNAs were verified using qRT-PCR. Also, the binding of circRNA and miRNA was verified using dual-luciferase reporter assay. This study provides a new insight into the molecular mechanism of fluoxetine-mediated ischemic stroke.
3.1. Fluoxetine markedly caused weight gained and reduced the cerebral infarction area in rats after MCAO
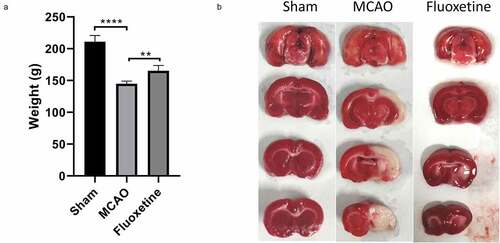
To estimate the effects of fluoxetine on the rat weight and cerebral infarction after MCAO, the MCAO model was successfully established in rats followed by fluoxetine treatment. We discovered that the weight was significantly reduced in rats after MCAO compared with those of the sham group. In contrast, the weight was markedly increased after fluoxetine treatment in rats after MCAO (P< 0.001, )). Also, TTC staining showed that the cerebral infarction area was markedly elevated in the MCAO model group relative to that in the sham group. Alternatively, this significant elevation in rats after MCAO could be markedly weakened by fluoxetine treatment ()).
Figure 1. Fluoxetine treatment caused increased weight and reduced cerebral infarction area in rats after MCAO (middle cerebral artery occlusion). (a) The weights of rats in each group were monitored and measured after the experiment. (b) The cerebral infarction area was determined using triphenyltetrazolium chloride staining on the three groups of rats. Sham: the sham group (n = 7), MCAO: the MCAO model group (n = 7), Fluoxetine: the fluoxetine treatment group (n = 7); ** P< 0.01, **** P< 0.0001
3.2. Fluoxetine observably improved nerve function in MCAO rats
Next, we determined the influence of fluoxetine on the nerve function of rats after MCAO. Based on the Zea-Longa neurological function scores, we discovered that in the sham group, there was no observable neurological deficit (0 score: n= 7); whereas, in the MCAO model group, there was a neurological deficit (while walking, the rats circled or tilted to the left, 2 score: n= 4, 3 score: n= 3). Relative to the MCAO model group, the fluoxetine treatment group notably improved the neurological deficit scores (1 score: n= 3, 2 score: n= 4). These results showed that fluoxetine observably improved nerve function in MCAO rats.
3.3. The changes of circRNAs expression profiles in rat brain tissue
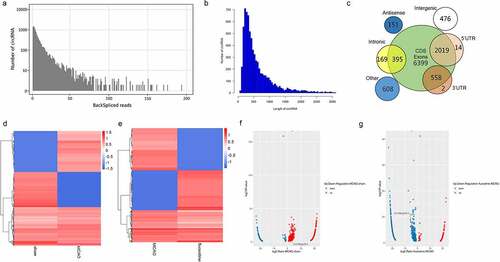
To further examine fluoxetine-related circRNAs in the brain tissue of rats after MCAO, RNA sequencing was used, followed by bioinformatics analysis. All circRNAs numbers with the same back-splicing reads are shown in ). The length distribution of circRNAs was mainly within 1000 bp ()), and the genomic origin of circRNAs is from coding domain sequence and exons ()). Furthermore, a heat and volcano map was used to reveal the differentially expressed circRNAs (|Fold Change| > 1.0) between the sham and MCAO groups and between the MCAO and fluoxetine groups ()). There were 879 differentially expressed circRNAs between the sham and MCAO groups, of which 592 were upregulated and 287 were downregulated ()). Compared with the MCAO group, 958 circRNAs (|Fold Change| > 1.0) were differentially expressed in the fluoxetine treatment group, of which 353 were upregulated and 605 were downregulated (, g)). These results suggest that fluoxetine changes the circRNAs expression in rats after MCAO.
Figure 2. The expression profiles of fluoxetine-mediated circRNAs in the rats’ brain tissue after MCAO (middle cerebral artery occlusion). (a) The back-splicing reads in the sequences of sham, MCAO model, and fluoxetine treatment groups. (b) The length distribution of circRNAs. (c) The genomic origin of circRNAs. The differentially expressed circRNAs were exhibited in heat and volcano maps between the sham and MCAO groups (d and f). The differentially expressed circRNAs were exhibited in heat and volcano maps between the MCAO and fluoxetine treatment groups (e and g)
3.4. The changes in mRNAs expression profiles in rat brain tissue
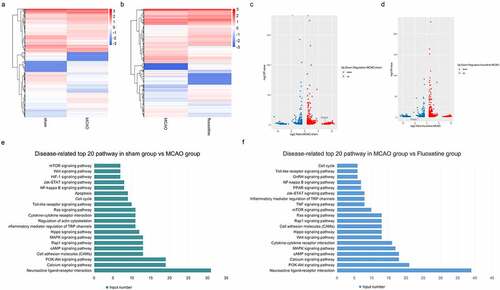
Also, we screened the changes of mRNAs in brain tissue in the sham, MCAO, and fluoxetine groups. As shown in ), compared with the sham group, there were 815 differentially expressed mRNAs (|Fold Change| > 1.0) in the MCAO group, of which 483 were upregulated and 332 were downregulated. Compared with the MCAO group, there were 838 differentially expressed mRNAs (|Fold Change| > 1.0) in the fluoxetine treatment group, of which 440 were upregulated and 398 were downregulated ()). Next, KEGG analysis was used to determine the top 20 enrichment pathways between the sham and MCAO groups, and between the MCAO and fluoxetine groups ()). The shared signaling pathways of differentially expressed mRNAs between the sham and MCAO groups and between the MCAO and fluoxetine groups were neuroactive ligand–receptor interaction, PI3K-Akt signaling pathway, calcium signaling pathway, CAMs, cAMP, and MAPK (mitogen-activated protein kinase) signaling pathway ()). The above results revealed that fluoxetine could change the mRNAs expression in rats after MCAO by regulating different pathways.
Figure 3. The expression profiles of fluoxetine-mediated mRNAs and functional analysis in the rats’ brain tissue after MCAO (middle cerebral artery occlusion). (a) and (c) is the heat and volcano maps of differentially expressed mRNAs between the sham and MCAO group. (b) and (d) is the heat and volcano maps of differentially expressed mRNAs between the MCAO and fluoxetine treatment groups. (e) KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis was used to show the top 20 signaling pathways between the sham and MCAO model groups. (f) KEGG analysis was used to show the top 20 signaling pathways between the MCAO and fluoxetine treatment groups
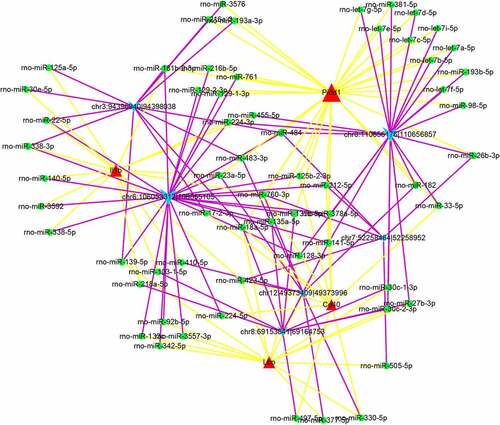
3.5. The ceRNA regulation network of six circRNAs
The differential circRNAs parent gene involved in the NF-κB signaling pathway was selected. Then, six circRNAs were obtained using the following criteria: (1) circRNA length between 450 and 2000 bp, (2) differential circRNAs with human homology, (3) parental genes not located on sex chromosomes, (4) upregulated in the MCAO group compared with those in the sham group, and downregulated in the fluoxetine treatment group compared with that in the MCAO group. According to the prediction in RegRNA2.0, the circRNA–miRNA–mRNA regulation network was constructed by Cytoscape software ().
3.6. Identification of screened circRNAs and mRNAs
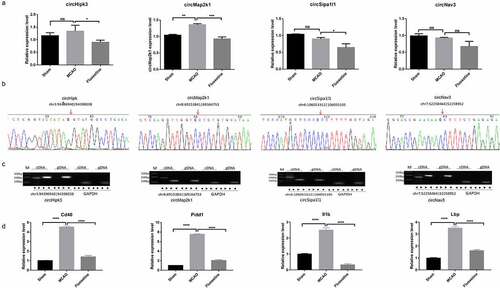
We searched the references of the six selected circRNAs and their parent genes; four circRNAs related to growth and nerve were verified. The results showed that circMap2k1 was significantly upregulated in the MCAO group compared with those in the sham group, which was consistent with the sequencing results. circHipk3, circSipa1l1, and circNav3 expression had no difference between the sham and MCAO groups, which was inconsistent with the sequencing results. circHipk3, circMap2k1, and circSipa1l1 were downregulated in the fluoxetine treatment group compared with those in the MCAO group, which was consistent with the sequencing results ()). Then, Sanger sequencing and agarose gel electrophoresis were used to verify the circular structure of four circRNAs, and the results showed that the four circRNAs had a back-splicing structure ()). Altogether, circMap2k1 is completely consistent with the sequencing results. In addition, qRT-PCR was used to detect the expression of four mRNAs, Cd40, Pidd1, Il1b, and LBP, in the circRNA–miRNA–mRNA network. The results showed that the expression trends of the four mRNAs were consistent with the sequencing results, in which the expression of Pidd1 had the largest change between the sham and MCAO groups, and between the MCAO and fluoxetine groups ()). So circMap2k1 and Pidd1 were selected for further research.
Figure 5. The identification of the candidate circRNAs and mRNAs. (a) qRT-PCR (quantitative reverse transcriptase polymerase chain reaction) detected the expression of circHipk3, circMap2k1, circSipa1l1, and circNav3 in the sham, MCAO model, and fluoxetine treatment groups. (b) and (c) Sanger sequencing and agarose gel electrophoresis was used to verify the circular structure of circHipk3, circMap2k1, circSipa1l1, and circNav3. (d) qRT-PCR detected the four candidate mRNAs, Cd40 (CD40 molecule), Pidd1 (P53-induced death domain protein 1), Il1b (interleukin 1 beta), and LBP (lipopolysaccharide binding protein), that had competitive RNA effect with circRNAs circHipk3, circMap2k1, circSipa1l1, and circNav3. * P< 0.05, ** P< 0.01, ** P< 0.001, **** P< 0.0001
3.7. Fluoxetine might exert a protective effect on ischemic stroke through the circMap2k1/miR-135b-5p/Pidd1 axis
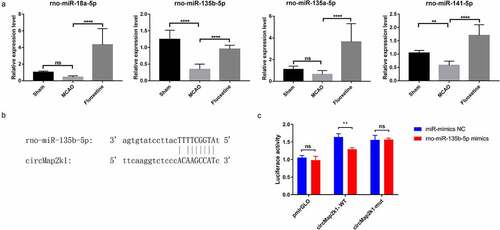
Firstly, we performed qRT-PCR verification on miRNAs that have binding sites with circMap2k1 and Pidd1. The results showed that in the MCAO group, rno-miR-135b-5p and rno-miR-141-5p were downregulated compared with those in the sham group. In the fluoxetine treatment group, rno-miR-18a-5p, rno-miR-135b-5p, rno-miR-135a-5p, and rno-miR-141-5p were significantly upregulated compared with those in the MCAO groups ()). As rno-miR-135b-5p had the most changes between the sham and MCAO groups and between the MCAO and fluoxetine groups, we selected this miRNA for further analysis. To investigate the targeting relationship between circMap2k1 and miR-135b-5p, TargetScan confirmed the binding sites of circMap2k1 and miR-135b-5p ()). Dual-luciferase reporter assay showed that the luciferase activity of WT circMap2k1 was significantly decreased after cotransfection with miR-135b-5p mimics ()). The above results indicate that fluoxetine participates in the protective effect of ischemic stroke through the circMap2k1/miR-135b-5p/Pidd1 axis.
Figure 6. The effect of fluoxetine on circMap2k1/miR-135b-5p/Pidd1 axis. (a) Quantitative reverse transcriptase polymerase chain reaction detected miRNAs rno-miR-18a-5p, rno-miR-135b-5p, rno-miR-135a-5p, and rno-miR-141-5p that has binding sites with circMap2k1 and Pidd1 (P53-induced death domain protein 1). (b) TargetScan predicted the binding sites of rno-miR-135b-5p and circMap2k1. (c) Dual-luciferase reporter assay verified the binding of rno-miR-135b-5p and circMap2k1. ** P< 0.01, **** P< 0.0001
4. Discussion
Cerebral ischemic stroke is a series of neurological defects caused by cerebral ischemia and hypoxic necrosis due to cerebral blood flow disorder in the brain [Citation44]. Fluoxetine, with its characteristics of high selectivity and specificity, can significantly improve the serotonin levels in the presynaptic space of patients, has a significant antidepressant effect, and can greatly reduce the symptoms in stroke patients [Citation45]. Previous studies have also shown that fluoxetine can effectively stimulate the motor nerves of patients, causing the spinal cord nerves of patients to be excited, which in turn can promote the recovery of patients’ nerve function [Citation14,Citation46]. Moreover, recent studies have also revealed that fluoxetine can restore neurological function and avoid the anti-choline effect in patients, thereby significantly enhancing its therapeutic effect [Citation47,Citation48]. Our study first verified that fluoxetine had significant reduction effects on the body weight of rats and on the cerebral infarction area in rats after MCAO.
Currently, many circRNAs have also been confirmed to contribute significantly to cerebral ischemic stroke. For instance, the knockdown of circHIPK2 could induce functional recovery of neural stem cells after ischemic stroke [Citation49]. Among the circRNAs, circHECTD1 was crucial to the inflammation, risk, severity, and recurrence of ischemic stroke. It was also involved in regulating astrocyte activation through autophagy in ischemic stroke [Citation50,Citation51]. Alternatively, circDLGAP4 alleviates ischemic stroke by miR-143 [Citation52], while circTLK1 could accelerate neuronal injury and neurological deficits by regulating miR-335-3p/TCDD inducible poly(ADP-Ribose) polymerase in ischemic stroke [Citation53]. Our study used high-throughput sequencing to screen fluoxetine-related circRNAs with differential expression in rats after MCAO and found that circMap2k1 may participate in this progress. Moreover, our data showed that the fluoxetine-related mRNAs were mainly concentrated in neuroactive ligand–receptor interaction, PI3K-Akt signaling pathway, calcium signaling pathway, CAMs, cAMP, and MAPK signaling pathway. These results provide a clue for the potential mechanism of fluoxetine treatment in MCAO.
In line with the annotation and differential expression of fluoxetine-related circRNAs and mRNAs that we screened, we also found that fluoxetine can downregulate circRNA Map2k1 and transcriptional gene Pidd1. After bioinformatics analysis, We further discovered that there were binding sites between circMap2k1 and miR-135b-5p, which were verified using dual-luciferase reporter assay. Previous studies have also demonstrated that miR-135b-5p is a vital miRNA in regulating apoptosis [Citation54]. miR-135b-5p overexpression can significantly alleviate hippocampus neuron damage and oxidative stress [Citation55], and miR-135b-5p can also be a diagnostic marker for mild craniocerebral injury during exercise [Citation56]. Moreover, studies have also confirmed that Pidd, a critical switch between NF-κB- and radiation-induced apoptosis, can regulate microglial activation induced by radiation [Citation57]. In contrast, Pidd silencing can inhibit NF-κB activation and reduce inflammation, as inflammation can activate the microglia [Citation58]. Therefore, we showed that fluoxetine-mediated circMap2k1/miR-135b-5p/Pidd1 axis has significant effects on cerebral ischemic stroke.
However, although our study provided a potential mechanism; more studies are needed to confirm that the circMap2k1/miR-135b-5p/Pidd1 axis was involved in fluoxetine-mediated cerebral ischemic stroke in vivo and in vitro.
5. Conclusion
We proved that fluoxetine has observable alleviating effects on cerebral ischemic stroke; moreover, we showed a mass of candidate circRNAs and mRNAs related to fluoxetine in cerebral ischemic stroke. We also demonstrated a potential signal axis (circMap2k1/miR-135b-5p/Pidd1) in cerebral ischemic stroke. This study provides a potential clue for using fluoxetine on the clinical treatment of cerebral ischemic stroke. It provides basic data for further research on the biological functions and mechanisms of circRNAs and transcriptome genes affecting the occurrence of cerebral ischemic stroke.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the author(s).
Ethics approval
This animal experiment was carried out in accordance with the Animal Experiment Ethics Committee of Jinan University.
Abbreviations
circRNA: circular RNA mRNA: messenger RNA MCAO: middle cerebral artery occlusion qRT-PCR: quantitative reverse transcriptase polymerase chain reaction PCR: polymerase chain reaction; miRNA: microRNAs 5-HT: 5-hydroxytriptamine ceRNA: competitive RNA SD: sprague–dawley CCA: common carotid artery ECA: external carotid artery ICA: internal carotid artery MCA: middle cerebral artery TTC: triphenyltetrazolium chloride rRNA: ribosomal RNA KEGG: Kyoto Encyclopedia of Genes and Genomes NF-κB: nuclear factor kappa B Cd40: CD40 molecule Pidd1: P53-induced death domain protein 1 Il1b: interleukin 1 beta LBP: lipopolysaccharide binding protein GAPDH: glyceraldehyde-3-phosphate dehydrogenase WT: wild-type MUT: mutant type mimics NC: mimics negative control MAPK: mitogen-activated protein kinase
Data availability statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Randolph SA. Ischemic stroke. Workplace Health Saf. 2016;64(9):444.
- Wu S, Yuan R, Wang Y, et al. Early prediction of malignant brain edema after ischemic stroke. Stroke. 2018;49(12):2918–2927. .
- Saliba E, Debillon T, Auvin S, et al. [Neonatal arterial ischemic stroke: review of the current guidelines]. Arch Pediatr. 2017;24(2):180–188. .
- Kessner SS, Schlemm E, Cheng B, et al. Somatosensory deficits after ischemic stroke. Stroke. 2019;50(5):1116–1123. .
- Nave AH, Lange KS, Leonards CO, et al. Lipoprotein (a) as a risk factor for ischemic stroke: a meta-analysis. Atherosclerosis. 2015;242(2):496–503. .
- Favate AS, Younger DS. Epidemiology of ischemic stroke. Neurol Clin. 2016;34(4):967–980.
- Chauhan G, Debette S. Genetic risk factors for ischemic and hemorrhagic stroke. Curr Cardiol Rep. 2016;18(12):124.
- Castilla-Guerra L, Del Carmen Fernandez-Moreno M, Colmenero-Camacho MA. Statins in stroke prevention: present and future. Curr Pharm Des. 2016;22(30):4638–4644.
- Gelbenegger G, Postula M, Pecen L, et al. Aspirin for primary prevention of cardiovascular disease: a meta-analysis with a particular focus on subgroups. BMC Med. 2019;17(1):198. .
- Marklund M, Wu JHY, Imamura F, et al. Biomarkers of dietary omega-6 fatty acids and incident cardiovascular disease and mortality. Circulation. 2019;139(21):2422–2436. .
- Perez-Caballero L, Torres-Sanchez S, Bravo L, et al. Fluoxetine: a case history of its discovery and preclinical development. Expert Opin Drug Discov. 2014;9(5):567–578. .
- Benfield P, Heel RC, Lewis SP. Fluoxetine. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic efficacy in depressive illness. Drugs. 1986;32(6):481–508.
- Robert A, Schultz IR, Hucher N, et al. Toxicokinetics, disposition and metabolism of fluoxetine in crabs. Chemosphere. 2017;186:958–967.
- Sun Y, Sun X, Qu H, et al. Neuroplasticity and behavioral effects of fluoxetine after experimental stroke. Restor Neurol Neurosci. 2017;35(5):457–468. .
- Marwari S, Dawe GS. (R)-fluoxetine enhances cognitive flexibility and hippocampal cell proliferation in mice. J Psychopharmacol. 2018;32(4):441–457.
- Marcinkute M, Afshinjavid S, Fatokun AA, et al. Fluoxetine selectively induces p53-independent apoptosis in human colorectal cancer cells. Eur J Pharmacol. 2019;857:172441.
- Alboni S, van Dijk RM, Poggini S, et al. Fluoxetine effects on molecular, cellular and behavioral endophenotypes of depression are driven by the living environment. Mol Psychiatry. 2017;22(4):552–561. .
- He Y, Cai Z, Zeng S, et al. Effect of fluoxetine on three-year recurrence in acute ischemic stroke: a randomized controlled clinical study. Clin Neurol Neurosurg. 2018;168:1–6.
- He YT, Tang BS, Cai ZL, et al. Effects of fluoxetine on neural functional prognosis after ischemic stroke: a randomized controlled study in China. J Stroke Cerebrovasc Dis. 2016;25(4):761–770. .
- Asadollahi M, Ramezani M, Khanmoradi Z, et al. The efficacy comparison of citalopram, fluoxetine, and placebo on motor recovery after ischemic stroke: a double-blind placebo-controlled randomized controlled trial. Clin Rehabil. 2018;32(8):1069–1075. .
- Corbett AM, Sieber S, Wyatt N, et al. Increasing neurogenesis with fluoxetine, simvastatin and ascorbic acid leads to functional recovery in ischemic stroke. Recent Pat Drug Deliv Formul. 2015;9(2):158–166. .
- Das A, Gorospe M, Panda AC. The coding potential of circRNAs. Aging (Albany NY). 2018;10(9):2228–2229.
- Patop IL, Kadener S. CircRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127.
- Li LJ, Leng RX, Fan YG, et al. Translation of noncoding RNAs: focus on lncRNAs, pri-miRNAs, and circRNAs. Exp Cell Res. 2017;361(1):1–8. .
- Holdt LM, Kohlmaier A, Teupser D. Molecular functions and specific roles of circRNAs in the cardiovascular system. Noncoding RNA Res. 2018;3(2):75–98.
- Altesha MA, Ni T, Khan A, et al. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588–5600. .
- Floris G, Zhang L, Follesa P, et al. Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol. 2017;54(7):5156–5165. .
- Lu S, Yang X, Wang C, et al. Current status and potential role of circular RNAs in neurological disorders. J Neurochem. 2019;150(3):237–248. .
- Arnaiz E, Sole C, Manterola L, et al. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99.
- Xia X, Tang X, Wang S. Roles of circRNAs in autoimmune diseases. Front Immunol. 2019;10:639.
- Yang J, Chen M, Cao RY, et al. The role of circular RNAs in cerebral ischemic diseases: ischemic stroke and cerebral ischemia/reperfusion injury. Adv Exp Med Biol. 2018;1087:309–325.
- Yang X, Ji H, Yao Y, et al. Downregulation of circ_008018 protects against cerebral ischemia–reperfusion injury by targeting miR-99a. Biochem Biophys Res Commun. 2018;499(4):758–764. .
- Cui J, Zhou B, Ross SA, et al. Nutrition, microRNAs, and human health. Adv Nutr. 2017;8(1):105–112. .
- Zhao S, Yan L, Zhao Z, et al. Up-regulation of miR-203 inhibits the growth of cervical cancer cells by inducing cell cycle arrest and apoptosis. Eur J Gynaecol Oncol. 2019;40(5):791–795.
- Liu W, Miao Y, Zhang L, et al. MiR-211 protects cerebral ischemia/reperfusion injury by inhibiting cell apoptosis. Bioengineered. 2020;11(1):189–200. .
- Jiao H, Chen R, Jiang Z, et al. MiR-22 protect PC12 from ischemia/reperfusion-induced injury by targeting p53 upregulated modulator of apoptosis (PUMA). Bioengineered. 2020;11(1):209–218. .
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. .
- Li Y, Huo C, Lin X, et al. Computational identification of cross-talking ceRNAs. Adv Exp Med Biol. 2018;1094:97–108.
- Qian L, Yu S, Chen Z, et al. The emerging role of circRNAs and their clinical significance in human cancers. Biochim Biophys Acta Rev Cancer. 2018;1870(2):247–260. .
- Li X, Yang L, Chen -L-L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442.
- Longa EZ, Weinstein PR, Carlson S, et al. Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke. 1989;20(1):84–91. .
- Li M, Zhao L, Li S, et al. Differentially expressed lncRNAs and mRNAs identified by NGS analysis in colorectal cancer patients. Cancer Med. 2018;7(9):4650–4664. .
- Wang Z, Shang P, Li Q, et al. ITRAQ-based proteomic analysis reveals key proteins affecting muscle growth and lipid deposition in pigs. Sci Rep. 2017;7:46717.
- Zhou C, He Y, Li X. Cerebral achromatopsia secondary to ischemic stroke. Neurol India. 2018;66(2):573–575.
- Heneghan C, Mahtani KR. Routine fluoxetine in stroke. BMJ Evid Based Med. 2019;24(5):195–196.
- van der Worp HB. Fluoxetine and recovery after stroke. Lancet. 2019;393(10168):206–207.
- Hancu G, Carcu-Dobrin M, Budau M, et al. Analytical methodologies for the stereoselective determination of fluoxetine: an overview. Biomed Chromatogr. 2018;32:1.
- Pham VT, Nguyen TQ, Dao UPN, et al. On the interaction between fluoxetine and lipid membranes: effect of the lipid composition. Spectrochim Acta A Mol Biomol Spectrosc. 2018;191:50–61.
- Wang G, Han B, Shen L, et al. Silencing of circular RNA HIPK2 in neural stem cells enhances functional recovery following ischaemic stroke. EBioMedicine. 2020;52:102660.
- Peng X, Jing P, Chen J, et al. The role of circular RNA HECTD1 expression in disease risk, disease severity, inflammation, and recurrence of acute ischemic stroke. J Clin Lab Anal. 2019;33(7):e22954. .
- Han B, Zhang Y, Zhang Y, et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14(7):1164–1184. .
- Bai Y, Zhang Y, Han B, et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. 2018;38(1):32–50. .
- Wu F, Han B, Wu S, et al. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335-3p/TIPARP. J Neurosci. 2019;39(37):7369–7393. .
- Chen Z, Gao Y, Gao S, et al. MiR-135b-5p promotes viability, proliferation, migration and invasion of gastric cancer cells by targeting Krüppel-like factor 4 (KLF4). Arch Med Sci. 2020;16(1):167–176. .
- Duan Q, Sun W, Yuan H, et al. MicroRNA-135b-5p prevents oxygen-glucose deprivation and reoxygenation-induced neuronal injury through regulation of the GSK-3β/Nrf2/ARE signaling pathway. Arch Med Sci. 2018;14(4):735–744. .
- Di Pietro V, Porto E, Ragusa M, et al. Salivary microRNAs: diagnostic markers of mild traumatic brain injury in contact-sport. Front Mol Neurosci. 2018;11:290.
- Yang N, Gao X, Qu X, et al. PIDD mediates radiation-induced microglia activation. Radiat Res. 2016;186(4):345–359. .
- Bock FJ, Krumschnabel G, Manzl C, et al. Loss of PIDD limits NF-κB activation and cytokine production but not cell survival or transformation after DNA damage. Cell Death Differ. 2013;20(4):546–557. .