ABSTRACT
Due to the important role of N6-methyladenosine (m6A) in breast cancer, single nucleotide polymorphisms (SNPs) in genes with m6A modification may also be involved in breast cancer pathogenesis. In this study, we used a public genome-wide association study dataset to identify m6A-SNPs associated with breast cancer and to further explore their potential functions. We found 113 m6A-SNPs associated with breast cancer that reached the genome-wide suggestive threshold (5.0E-05), and 86 m6A-SNPs had eQTL signals. Only six genes were differentially expressed between controls and breast cancer cases in GEO datasets (GSE15852, GSE115144, and GSE109169), and the SNPs rs4829 and rs9610915 were located next to the m6A modification sites in the 3ʹUTRs of TOM1L1 and MAFF, respectively. In addition, we found that polyadenylate-binding protein cytoplasmic 1 might have a potential interaction with rs4829 (TOM1L1) and rs9610915 (MAFF). In summary, these findings indicated that the SNPs rs4829 and rs9610915 are potentially associated with breast cancer because they had eQTL signals, altered gene expression, and were located next to the m6A modification sites in the 3ʹUTRs of their coding genes. However, further studies are still needed to clarify how genetic variation affects the epigenetic modification, m6A, and its subsequent functions in the pathogenesis of breast cancer.
1 Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer-related death among women worldwide [Citation1,Citation2]. Numerous potential risk loci and susceptible genes have been identified by genome-wide association studies (GWASs) in breast cancer [Citation3,Citation4]. Presently, high-penetrance genes (BRCA1, BRCA2, CDH1, STK11, PTEN, and TP53) and moderate-penetrance genes (ATM, CHEK2, PALB2, and BRIP1) are known to genetically predispose individuals to the development of breast cancer [Citation5]. Recently, a growing number of studies have suggested that genetic variations, especially single nucleotide polymorphisms (SNPs), contribute to breast cancer susceptibility [Citation6,Citation7]because SNPs located in protein-coding regions, as well as multiple SNPs located in non-coding regions, can alter gene expression and participate in epigenetic regulation.
N6-methyladenosine (m6A) modification is the most abundant internal modification of messenger RNA (mRNA) and non-coding RNA, such as microRNA, long non-coding RNA, and circular RNA in eukaryotes [Citation8,Citation9]. Emerging evidence has shown that m6A is involved in RNA metabolism and is dynamically regulated by methyltransferases, demethylases, and binding proteins [Citation10,Citation11]. Furthermore, it was confirmed that m6A plays an important role in breast cancer [Citation12,Citation13]. For example, the m6A demethylase FTO promoted breast cancer progression by inhibiting BNIP313, and the HBXIP-elevated methyltransferase METTL3 promoted breast cancer progression by inhibiting the tumor suppressor let-7 g [Citation12]. Recent studies have also suggested that SNPs may affect m6A by altering the RNA sequences of the target sites or crucial nucleotides [Citation14] and also contribute to the progression of breast cancer. Importantly, m6A-SNPs have the potential to regulate gene expression and mRNA stability, consequently leading to disease [Citation15,Citation16]. Yang et al. found that rs2416282 contributed to esophageal squamous cell carcinoma (ESCC) risk by regulating YTHDC2 expression [Citation16]. Recently, another study found that the SNP rs5746136 affected m6A modification and influenced the expression of SOD2 in bladder cancer [Citation15], and some METTL14 SNPs were similarly related to neuroblastoma [Citation17]. Zhuo also reported that WTAP SNPs may be genetic modifiers in hepatoblastoma [Citation18]. Ren et al. reported that the rs8400 AA genotype was correlated with increased expression of ALKBH5 and may be a risk factor for hepatoblastoma in the clinical stage III + IV subgroup [Citation19]. In glioma, studies found the m6A-SNPs, rs7766006 (WTAP), rs3738067 (YTHDF2), and rs9939609 (FTO), to increase cancer risk, whereas rs2293595 (YTHDC1), rs3813832 (YTHDC1), and rs8047395 (FTO) reduced the risk [Citation20].
Given that m6A-SNPs play a pivotal role in cancer [Citation18,Citation21–23] and the effect of m6A -SNPs in breast cancer is unclear, identifying m6A -SNPs associated with breast cancer is necessary and could provide a new annotation for the pathogenic mechanism of breast cancer risk loci identified by GWAS. Therefore, this study aimed to identify m6A-SNPs associated with breast cancer using a public GWAS database and the m6AVar database, and to demonstrate their potential functions.
2 Materials and methods
2.1 Screening for m6A -SNPs associated with breast cancer
The m6AVar database provides information for the annotation, visualization, and exploration of variants associated with m6A [Citation24,Citation25]. In the m6AVar database, variants were defined at three confidence levels, namely high confidence level when RNA modification sites were directly obtained from the m6A-Label-seq, DART-seq, m6A-REF-seq, or miCLIP experiment; medium confidence level when RNA modification sites were obtained from the MeRIP-Seq or m6A-Seal-seq experiment; and low confidence level when RNA modification sites were obtained by prediction of the whole transcriptome.
The intersection of SNPs, which reached the genome-wide suggestive threshold (5.0E-05), between the breast cancer GWAS and m6Avar databases was used to explore m6A-SNPs associated with breast cancer. In this study, breast cancer GWAS data were downloaded from the website (ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/MichailidouK_29059683_GCST004988/), and the m6A-SNP list was obtained from the m6Avar database (http://rmvar.renlab.org/download.html).
2.2 Expression quantitative trait loci (eQTL) analysis of m6A-SNPs
eQTL analysis can be used to evaluate the potential function of m6A-SNPs with eQTL signals in transcription regulation, such as altering protein binding, changing motifs, and affecting deoxyribonuclease. Hence, eQTL analysis was performed to investigate the breast cancer-associated m6A-SNPs that could affect RNA modification using the HaploReg browser (https://www.encodeproject.org/software/haploreg/) [Citation26,Citation27].
2.3 Gene Ontology (GO) enrichment analysis
GO enrichment analysis was performed using the Metascape tools (https://metascape.org/gp/index.html#/main/step1). GO biological process, GO molecular function, and GO cellular component were used to classify these genes according their functional annotation [Citation28].
2.4 Differential gene expression analysis
The GSE15852, GSE115144, and GSE109169 datasets were downloaded from NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) [Citation29,Citation30]. GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to identify the differentially expressed genes corresponding to m6A-SNPs with eQTL between controls and breast cancer cases. A significance level of 0.05 and |LogFC|≥2 were used for differential expression analysis.
2.5 Prediction of m6A-SNPs affecting RNA modification
To determine whether the m6A-SNPs affected RNA modification and predicted changes in m6A modification, we used the sequence-based RNA adenosine methylation site predictor (SRAMP), which is an online m6A modification prediction tool [Citation31]. Further, the USCS browser (GRCh37/hg19; https://genome.ucsc.edu/) includes abundant assemblies and annotations of the genomes of vertebrates and model organisms. Therefore, we used it to analyze the potential functions of m6A-SNPs [Citation32,Citation33].
3 Results
3.1 Identification of m6A-SNPs associated with breast cancer
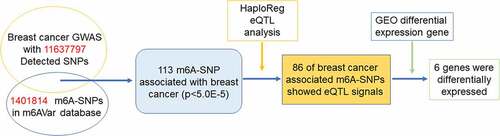
To identify m6A-SNPs associated with breast cancer, the intersection of the GWAS and m6AVar databases was analyzed. Of the 11,637,797 SNPs and 1,401,814 m6A-SNPs in the breast cancer GWAS and m6A Var databases (), we identified 113 m6A-SNPs, which reached the genome-wide suggestive threshold (5.0E-05), to be associated with breast cancer (Table S1).
Figure 2. Manhattan plot of identified m6A-SNPs associated with breast cancer. The Manhattan plot showed – log10(p.value) for each of 17,599 m6A-SNPs associated with breast cancer. There were 285 m6A-SNPs associated with breast cancer (p < 0.001), and 113 m6A-SNPs associated with breast cancer (p < 5.0E-5)
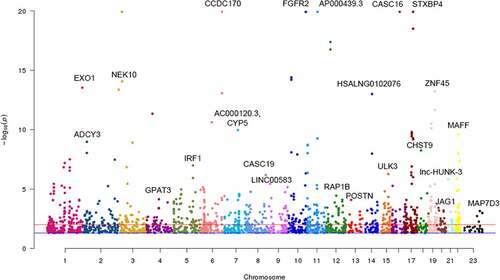
To check whether these m6A-SNPs have eQTL effects, the HaploReg browser was used, finding that 86 of these breast cancer-associated m6A-SNPs had eQTL signals. Subsequently, we found that of the 86 m6A-SNPs with eQTL signals, 31 belonged to the high confidence category, 20 to the medium confidence category, and 35 to the low confidence category. Additionally, among these 86 m6A-SNPs, 72 were loss-of-function SNPs and 14 were gain-of-function SNPs (Table S1).
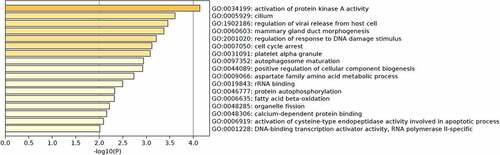
According to GO enrichment analysis, we found that the molecular functions of these m6A-SNPs were associated with rRNA binding, calcium-dependent protein binding, DNA-binding transcription activator activity, and RNA polymerase II-specific binding. In addition, the functions of the genes with these m6A-SNPs were enriched in various biological processes, such as the activation of protein kinase A activity, mammary gland duct morphogenesis, and autophagosome maturation ().
3.2 Six corresponding genes of eight m6A-SNPs were differentially expressed in breast cancer
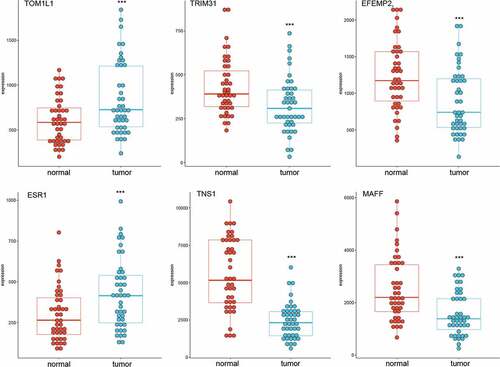
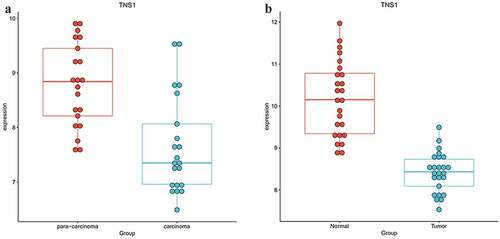
To study whether these m6A-SNPs may affect the expression level of their corresponding genes, differential gene expression analysis using GEO2R was performed, finding that only six genes, namely TOM1L1 (rs1802212; rs4829), ESR1 (rs2747655), MAFF (rs2267372; rs9610915), TNS1 (rs1424916), EFEMP2 (rs2234458), and TRIM31 (rs2023472), were differentially expressed in the GSE15852 dataset (p < 0.05) (). Specifically, TOM1L1 and ESR1 were highly expressed, but MAFF, TNS1, EFEMP2, and TRIM31 were significantly downregulated in breast cancer (). Moreover, we found that only TNS1 was differentially expressed in the GSE115144 and GSE109169 datasets (). Taken together, these results suggest that eight m6A-SNPs (rs1802212, rs4829, rs2747655, rs2267372, rs9610915, rs1424916, rs2234458, and rs2023472) may affect the expression level of their corresponding genes.
Table 1. Six genes of eight m6A-SNPs associated with breast cancer were differentially expressed between controls and breast cancer in the GSE15852
3.3 TOM1L1 (rs4829) and MAFF (rs9610915) may affect m6A modification in breast cancer
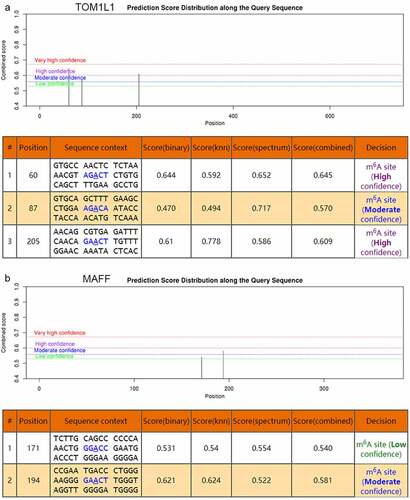
To investigate the possible m6A methylation sites of TOM1L1 (rs1802212; rs4829), MAFF (rs2267372; rs9610915), and TNS1 (rs1424916), their transcript sequences were predicted at the SRAMP website. The results showed that five m6A-SNPs belonged to the high or medium confidence category, including rs1802212 (TOM1L1), rs4829 (TOM1L1), rs2267372 (MAFF), rs9610915 (MAFF), and rs1424916 (TNS1) (). Based on the SRAMP website, we found moderately-highly convincible m6A-modified predicted peaks near TOM1L1 (rs4829) and a moderately convincible m6A-modified predicted peak near MAFF (rs9610915) ().
Figure 6. The genomic sequence of TOM1L1 and MAFF transcripts were used to predict the m6A modification on website (http://www.cuilab.cn/sramp). There were moderately-highly convincible m6A-modified predicted peaks near TOM1L1 (rs4829) and a moderately convincible m6A-modified predicted peak near MAFF (rs9610915)
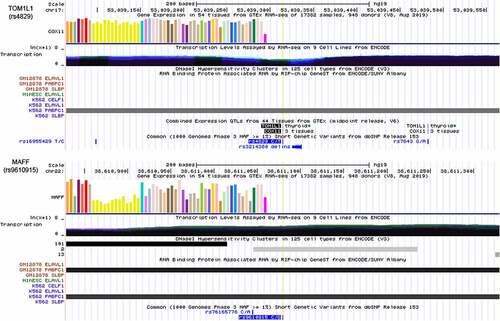
We next used the USCS browser to analyze the potential functions of the m6A-SNPs. An integrative analysis of the potential functions of rs4829 and rs9610915 is shown in . The SNP rs4829 was located in the 3ʹUTR of TOM1L1 on chromosome 17 and rs9610915 was located in the 3ʹUTR of MAFF on chromosome 22 (). This finding suggested that these two SNPs may affect the m6A modification site and regulate gene expression to participate in breast cancer pathogenesis.
Figure 7. Integrative analysis of the potential function of rs4829 and rs9610915 SNPs by querying USCS. The SNP rs4829 was located on the 3’UTR of the TOM1L1 gene on chromosome 17 and rs9610915 was located on the 3’UTR of the MAFF gene on chromosome 22. Further, RIP-chip GeneST from ENCODE/SUNY Albany data suggested that polyadenylate-binding protein cytoplasmic 1 (PABPC1) might have a potential interaction with rs4829 (TOM1L1) and rs9610915 (MAFF)
4 Discussion
m6A plays an important role in breast cancer, but the effect of m6A-SNPs in breast cancer is unclear. Identifying m6A-SNPs associated with breast cancer will provide a new annotation for the pathogenic mechanism of breast cancer risk loci identified by GWAS; therefore, we identified m6A-SNPs associated with breast cancer and explored their potential functions. We found 113 m6A-SNPs that were associated with breast cancer, and 86 m6A-SNPs that had eQTL signals.
Additionally, there were 31 m6A-SNPs belonging to the high confidence category; further, 75% of the m6A-SNPs were distributed in intron regions, 20% were distributed in the 3ʹUTR region, and 5% were distributed in the 5ʹUTR region. However, there were no SNPs of m6A regulators (METTL3, METTL14, METTL16, WTAP, VIRMA, RBM15, FTO, ALKBH5, YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, IGF2BP1, IGF2BP2, IGF2BP3, HNRNPA2B1, and EIF3A) in the m6A-SNPs associated with breast cancer (Table S1). This finding indicated that SNPs of these 18 m6A regulators may not affect breast cancer.
We also found that six corresponding genes of eight m6A-SNPs were differentially expressed in breast cancer, and the SNPs rs4829 and rs9610915 were located next to the m6A modification site in the 3ʹUTR of TOM1L1 and MAFF, respectively, suggesting that these two SNPs may affect the m6A modification site and regulate gene expression to play a role in breast cancer. Studies have shown that TOM1L1 is associated with breast cancer. For example, Chevalier et al. found that TOM1L1 amplification may enhance the metastatic progression of ERBB2-positive breast cancers by regulating the ERBB2-driven proteolytic invasion [Citation34,Citation35]. There is also evidence that MAFF regulates cancer pathogenesis. For instance, Martínez-Hernández et al. found 17 variants of MAFF in chronic myeloid leukemia (CML), and the rs9610915 SNP of MAFF was significantly associated with CML [Citation36]. Further, RIP-chip GeneST from the ENCODE/SUNY Albany data suggested that polyadenylate-binding protein cytoplasmic 1 (PABPC1) might have a potential interaction with rs4829 (TOM1L1) and rs9610915 (MAFF) (). It is known that PABP1 participates in the initiation of translation and stabilization of mRNA and that PABPC1 can encode PABP1 protein and promote tumor progression of gliomas and hepatocellular carcinomas [Citation37,Citation38]. In breast cancer, studies have indicated that PABPC1 mediates SNHG14-induced oncogenic effects [Citation39].
5 Conclusion
In this study, we used a comprehensive analysis of GWAS raw data and summary statistics combined with eQTL and differential gene expression analyses to identify breast cancer-associated m6A-SNPs. We found that two SNPs (rs4829 and rs9610915) had eQTL signals, altered gene expression, and were located next to the m6A modification sites in the 3ʹUTRs of their coding genes and that PABPC1 might have a potential interaction with these two SNPs. These findings indicated that the SNPs rs4829 and rs9610915 may be potentially associated with breast cancer. However, the relationship between these m6A-SNPs and breast cancer risk and their potential roles in the pathogenesis of breast cancer still require clarification.
Highlights
113 m6A-SNPs associated with breast cancer (5.0E-05), and 86 m6A-SNPs had eQTL signals.
SNPs rs4829 and rs9610915 had eQTL signals, altered the expression, located next to the m6A 3’UTR modification site of their coding genes.
SNPs rs4829 and rs9610915 may be potentially associated with breast cancer.
PABPC1 might have a potential interaction with SNPs rs4829 and rs9610915.
Author contributions
GB.Z and P.H designed the research. ZX.X, YW.Z, JY.J, and XW.Z performed the experiments. XP.H, XL.Y and YF.S analysed the data. All authors drafted and reviewed the manuscript.
Supplemental Material
Download ()Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Mattiuzzi C, Lippi G. Current cancer epidemiology. J Epidemiol Glob Health. 2019;9:217–222.
- Fahad Ullah M. Breast cancer: current perspectives on the disease status. Adv Exp Med Biol. 2019;1152:51–64.
- Nones K, Johnson J, Newell F, et al. Whole-genome sequencing reveals clinically relevant insights into the aetiology of familial breast cancers. Ann Oncol. 2019;30:1071–1079.
- Ferreira MA, Gamazon ER, Al-Ejeh F, et al. Genome-wide association and transcriptome studies identify target genes and risk loci for breast cancer. Nat Commun. 2019;10:1741.
- Shiovitz S, Korde LA. Genetics of breast cancer: a topic in evolution. Ann Oncol. 2015;26:1291–1299.
- Barrdahl M, Canzian F, Lindström S, et al. Association of breast cancer risk loci with breast cancer survival. Int J Cancer. 2015;137:2837–2845.
- Hamdi Y, Ben Rekaya M, Jingxuan S, et al. A genome wide SNP genotyping study in the Tunisian population: specific reporting on a subset of common breast cancer risk loci. BMC Cancer. 2018;18:1295.
- Chen M, Wong CM. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer. 2020;19:44.
- Dai D, Wang H, Zhu L, et al. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124.
- Anderson SJ, Kramer MC, Gosai SJ, et al. N(6)-methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in arabidopsis. Cell Rep. 2018;25:1146–57.e3.
- Liu J, Sun G, Pan S, et al. The Cancer Genome Atlas (TCGA) based m(6)A methylation-related genes predict prognosis in hepatocellular carcinoma. Bioengineered. 2020;11:759–768.
- Cai X, Wang X, Cao C, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19.
- Niu Y, Lin Z, Wan A, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46.
- Chen K, Song B, Tang Y, et al. RMDisease: a database of genetic variants that affect RNA modifications, with implications for epitranscriptome pathogenesis. Nucleic Acids Res. 2021;49(D1):D1396–D1404
- Liu H, Gu J, Jin Y, et al. Genetic variants in N6-methyladenosine are associated with bladder cancer risk in the Chinese population. Arch Toxicol. 2021;95(1):299–309
- Yang N, Ying P, Tian J, et al. Genetic variants in m6A modification genes are associated with esophageal squamous-cell carcinoma in the Chinese population. Carcinogenesis. 2020;41:761–768.
- Zhuo Z, Lu H, Zhu J, et al. METTL14 gene polymorphisms confer neuroblastoma susceptibility: an eight-center case-control study. Mol Ther Nucleic Acids. 2020;22:17–26.
- Zhuo ZJ, Hua RX, Chen Z, et al. WTAP gene variants confer hepatoblastoma susceptibility: a seven-center case-control study. Mol Ther Oncolytics. 2020;18:118–125.
- Ren H, Zhuo ZJ, Duan F, et al. ALKBH5 gene polymorphisms and hepatoblastoma susceptibility in chinese children. J Oncol. 2021;2021:6658480.
- He J, Yuan L, Lin H, et al. Genetic variants in m(6)A modification core genes are associated with glioma risk in Chinese children. Mol Ther Oncolytics. 2021;20:199–208.
- Hua RX, Liu J, Fu W, et al. ALKBH5 gene polymorphisms and Wilms tumor risk in Chinese children: a five-center case-control study. J Clin Lab Anal. 2020;34:e23251.
- Ma L, Hua RX, Lin H, et al. The contribution of WTAP gene variants to Wilms tumor susceptibility. Gene. 2020;754:144839.
- Bian J, Zhuo Z, Zhu J, et al. Association between METTL3 gene polymorphisms and neuroblastoma susceptibility: a nine-centre case-control study. J Cell Mol Med. 2020;24:9280–9286.
- Zheng Y, Nie P, Peng D, et al. m6AVar: a database of functional variants involved in m6A modification. Nucleic Acids Res. 2018;46:D139–d45.
- Luo X, Li H, Liang J, et al. RMVar: an updated database of functional variants involved in RNA modifications. Nucleic Acids Res. 2021; 49(D1):D1405–D1412
- Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–81.
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4.
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
- Zhu X, Tao L, Yao J, et al. Identification of collaboration patterns of dysfunctional pathways in breast cancer. Int J Clin Exp Pathol. 2014;7:3853–3864.
- Pau NIB, Zakaria Z, Muhammad R, et al. Gene expression patterns distinguish breast carcinomas from normal breast tissues: the Malaysian context. Pathol Res Pract. 2010;206:223–228.
- Zhou Y, Zeng P, Li YH, et al. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91.
- Lin W, Xu H, Wu Y, et al. In silico genome-wide identification of m6A-associated SNPs as potential functional variants for periodontitis. J Cell Physiol. 2020;235:900–908.
- Mo X, Lei S, Zhang Y, et al. Genome-wide enrichment of m(6)A-associated single-nucleotide polymorphisms in the lipid loci. Pharmacogenomics J. 2019;19:347–357.
- Chevalier C, Collin G, Descamps S, et al. TOM1L1 drives membrane delivery of MT1-MMP to promote ERBB2-induced breast cancer cell invasion. Nat Commun. 2016;7:10765.
- Chevalier C, Roche S, Bénistant C. Vesicular trafficking regulators are new players in breast cancer progression: role of TOM1L1 in ERBB2-dependent invasion. Mol Cell Oncol. 2016;3:e1182241.
- Martínez-Hernández A, Gutierrez-Malacatt H, Carrillo-Sánchez K, et al. Small MAF genes variants and chronic myeloid leukemia. Eur J Haematol. 2014;92:35–41.
- Su R, Ma J, Zheng J, et al. PABPC1-induced stabilization of BDNF-AS inhibits malignant progression of glioblastoma cells through STAU1-mediated decay. Cell Death Dis. 2020;11:81.
- Zhang H, Sheng C, Yin Y, et al. PABPC1 interacts with AGO2 and is responsible for the microRNA mediated gene silencing in high grade hepatocellular carcinoma. Cancer Lett. 2015;367:49–57.
- Dong H, Wang W, Mo S, et al. Long non-coding RNA SNHG14 induces trastuzumab resistance of breast cancer via regulating PABPC1 expression through H3K27 acetylation. J Cell Mol Med. 2018;22:4935–4947.