?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Cronobacter sakazakii is a food-borne, conditionally pathogenic bacterium that mainly infects neonates, especially premature infants. Previous studies have indicated that an important route of infection for C. sakazakii is through infant formula, suggesting a high stress resistance of the bacterium. RpoS is a σ-factor that is closely related to the bacterial resistance mechanisms. In this study, a C. sakazakii BAA894 model strain was used. An rpoS-deficient mutant strain Δrpos was constructed using Red homologous recombination, and the differences between the mutant and the wild-type strains were compared. To investigate the functions of the rpoS gene, the membrane formation and cell wall properties of the strains were studied, and the tolerance of each strain to acid, osmotic pressure, desiccation, and drug resistance were compared. The results showed that the membrane formation ability in the mutant strain was increased, auto-aggregation was enhanced, motility, acid resistance and hyperosmotic resistance were alternated to different degrees, and desiccation resistance was stronger than observed in the wild type grown in LB medium but weaker than the wild type cultured in M9 medium. These results showed that rpoS is involved in environmental stress resistance in C. sakazakii BAA894. Finally, transcriptome analysis verified that the deletion of the rpoS gene caused differential expression of resistance-related genes and instigated changes in related metabolic pathways. These messenger RNA results were consistent with the functional experimental results and help explain the phenotypic changes observed in the mutant strain.
Introduction
Cronobacter sakazakii is a gram-negative bacterium, which is a food-borne opportunistic pathogen from the enterobacteriaceae family. Cronobacter was previously called Enterobacter sakazakii. In 2007, Iversen et al. [Citation1] proposed establishing a new genus including the original Enterobacter sakazakii, namely Cronobacter spp., and C. sakazakii as one of the seven species classified [Citation2,Citation3]. The genome of C. sakazakii BAA894 was the first sequenced in 2010 [Citation4]. C. sakazakii BAA894 can parasitic in the intestinal tract of human and warm-blooded animals, and can cause meningitis, enterocolitis and septicemia, and sepsis in infants, once infected [Citation5–9]. These infections were found to be related to infant formula milk (IFM), which is complex to process and has an extremely low water concentration [Citation10]. The method by which C. sakazakii infects organisms illustrates that its pathogenicity is closely related to its abilities relating to stress resistance.
RpoS is a sigma factor (σ38) of RNA polymerase, and is responsible for regulating the general stress response [Citation11]. It keeps bacteria alive in different external environments by inducing the expression of relevant genes and is believed to be extensively involved in cellular stress responses [Citation12]. As reported in E. coli, RpoS is not only an indispensable regulator of glucose-repressed acid resistance [Citation13], but it also plays regulatory roles in other resistance mechanisms, for example, RpoS can regulate the expression of the virulence genes [Citation14,Citation15]. At present, the gradual role of sigma factor RpoS in C. sakazakii has been focused upon, and RpoS has proved to be significantly important ensuring tolerance in C. sakazakii to acid, osmotic pressure, and oxidation [Citation16–18]. RpoS also plays a crucial role in C. sakazakii tolerance in relation to long-term extremely low water activity (aw) [Citation19]. It has also been reported that the expression of rpoS, hfq and ompA genes was up-regulated when Cronobacter enter the viable but nonculturable (VBNC) state after a 2 h period of desiccation tolerance [Citation20]. By entering the VBNC state, Cronobacter can maintain its virulence while escaping the typical colonies tests, and regulating rpoS affects bacterial abilities in relation to entering this state.
The rpoS gene has global regulatory significance, but its mechanism(s) of action in C. sakazakii BAA894 remains to be elucidated. In the present study, a new mutant strain ΔrpoS was constructed by knocking out gene rpoS in C. sakazakii BAA894, the differences in environmental stress resistance between wild-type C. sakazakii BAA894 and the ΔrpoS mutant were compared in terms of osmotic pressure, acid tolerance, desiccation tolerance, and drug resistance. Membrane formation and motility, which are supposedly related to bacterial adhesion and considered to be important for high stress enhance, were tested too. Transcriptomic analysis of ΔrpoS and wild-type strains were conducted, and the pathways showing significant changes were analyzed. This study aimed to investigate the role of sigma factor RpoS in C. sakazakii in relation to environmental stress tolerance by studying the changes of the ΔrpoS mutant in phenotypic experiments and transcriptomic analysis. The transcriptome data can also further clarify the mechanisms of RpoS in stress survival of this strain.
Materials and methods
Strains and media
Bacteria and plasmids used in this work are listed in . C. sakazakii cells were grown at 37°C in LB medium 5-g/L yeast extract, 10-g/L tryptone, and 10-g/L NaCl [Citation21] or M9 medium (17.1 g/L Na2HPO4 · 12H2O, 3 g/L KH2PO4, 4 g/L glucose, 1 g/L NH4Cl, 0.5 g/LNaCl, 0.24 g/L MgSO4, and 0.011 g/L CaCl2) [Citation22] with 200-rpm shaking.
Table 1. Strains and plasmids used in this study
Reagents and primers
Kits: Plasmid DNA Miniprep Kit (Bio Basic Canada Inc), TIANamp Bacteria DNA Kit and agarose gel DNA recovery kit (Tiangen Biochemical Technology (Beijing) Co., Ltd.), and SDS-PAGE Gel Quick Preparation Kit (Beyotime Institute of Biotechnology).
Molecular biology reagents: Ex Taq DNA polymerase and dNTPs (Bao Biological Engineering (Dalian) Co., Ltd), DNA restriction endonuclease, T4 DNA ligase and DNA Marker (Fermentas), GoldView II Nuclear Staining Dye (Beijing Solarbio Science & Technology Co., Ltd), agarose (Shanghai Generay Biotech Co., Ltd). PCR experiments were performed using Mastercycler from Eppendorf (Hamburg, Germany). The sequences of all primers used in this study are listed in .
Table 2. Primers for PCR amplification used in this study
Construction of gene knockout mutant of Cronobacter sakazakii
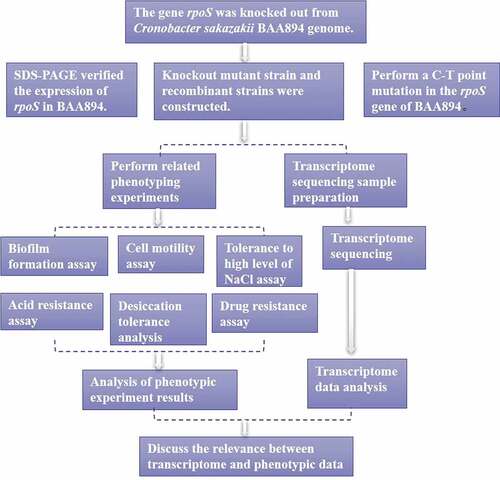
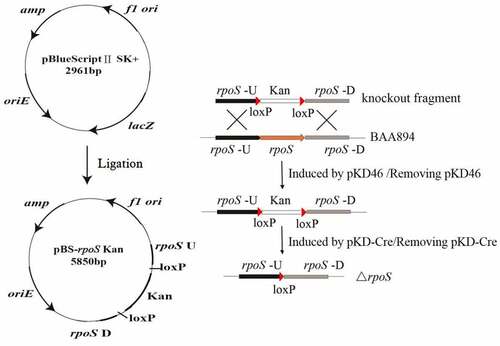
In this study, the target gene rpoS was knocked out using a C. sakazakii Red recombination system [Citation26,Citation27]. First, the upstream and downstream flanking sequences of the rpoS gene were amplified using the primer pairs rpoS-U-F/rpoS-U-R and rpoS-D-F/rpoS-D-R respectively retrieved from the C. sakazakii genome by PCR, and by introducing cleavage sites Kpn I (5ʹ) and BamH I (3ʹ) to the upstream homology arm, and Xba I (5ʹ) and Spe I (3ʹ) to the downstream homology arm. Next, the kanamycin resistance cassette kan fragment was amplified by PCR using primer pair kan-F and kan-R with pDTW202 as template, and enzyme digestion sites BamHI (5ʹ) and XbaI (3ʹ) were introduced to the product fragment. The three fragments obtained, rpoS-L, rpoS-R and kan, were digested with the corresponding DNA restriction endonucleases (Fermentas), and the recovered fragments were then ligated into the plasmid pBluescript II SK(+) digested by XhoI and PstI. The ligation product is transferred to E. coli JM109; then, the resulting transformants were screened on agar plates supplemented with kanamycin to obtain the knockout recombinant plasmid pBluescript-r-U-Fkan-r-D, with enzyme digestion verification containing the upstream and downstream homology arm of rpoS gene, and the kanamycin resistance cassette. Finally, the rpoS knockout fragment rpoS-U-Fkan-rpoS-D was amplified using the forward primer rpoS-U-F which targeted the upstream homology arm and the reverse primer rpoS-D-R targeting the downstream homology arm. The amplified product was recovered following verification with agarose gel electrophoresis. The knockout fragment was transferred into BAA894 competent cells containing pKD46, and the rpoS on the genome was replaced by loxP-kan-loxP. Following successful replacement, the temperature-sensitive plasmid pKD46 was removed via incubation at 42°C. Afterward, pKD-Cre was introduced into the cells, and the loxP site recombination was induced by Cre recombinase to remove the kan resistance gene. The pKD-Cre plasmid was removed via incubation at 42°C, and the ΔrpoS mutant was attained (). The primers are listed in .
The C-T mutation at position 601 of rpoS in C. sakazakii BAA894
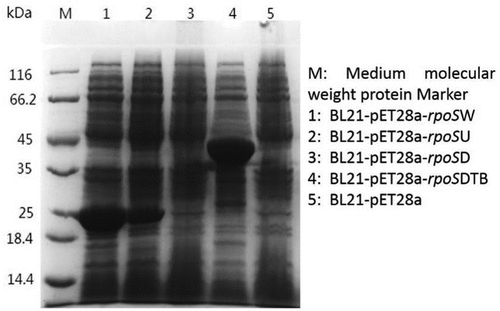
The rpoS sequence in C. sakazakii BAA894 had a CAG-TAG mutation at position 601 compared with other Cronobacter sakazakii, and this TAG (stop codon) at positions 601–603 mutated to CAG (glutamine), resulting in the strain rpoSDTB. Meanwhile, rpoSU (rpoS up part 1–603 bp), rpoSD (rpoS down part 633–993 bp), rpoSW (rpoS whole part 1–993 bp) and rpoSDTB were overexpressed in BL21, expression was identified using SDS-PAGE (Beyotime Institute of Biotechnology).
Overexpression of rpoSU, rpoSD, rpoSW and rpoSDTB
First, E. coli BL21 competent cells were prepared, transformed with the constructed pET28a-rpoSDTB, pET28a-rpoSU, pET28a-rpoSD and pET28a-rpoSW plasmids, and cultured onto LB agar plates supplemented with kan. After overnight incubation and colony PCR verification, the correct transformants were picked for plasmid extraction.
Total protein extractions from the five exponentially growing BL21 strains, BL21-pET28a-rpoSW, BL21-pET28a-rpoSU, BL21-pET28a-rpoSD, BL21-pET28a-rpoSDTB and BL21-pET28a, were collected via protein electrophoresis. 60 μL culture was added to 12 μL 5× Loading Buffer, boiled for 10 min, separated by SDS-PAGE (5% stacking gel and 12% separation gel) [Citation28,Citation29] and electrophoresis (Tricine-SDS-PAGE was used due to the small size of rposD), and the gel was stained with Coomassie Brilliant Blue (Beijing Solarbio Science & Technology Co., Ltd).
Stress resistance of mutant strains
Biofilm formation assay
In order to study the mutant strain change of membrane formation ability, the biofilm formation experiments were done according to some previous references [Citation30–32]. The overnight culture in LB or M9 medium was transferred into 5 mL of LB medium or M9 medium at an initial OD600 = 0.02; 1 mL of the culture was taken at OD600 = 1.0, and incubated without shaking at 37°C for 72 h. The culture was checked every 12 h, and fresh sterile LB medium was added when necessary to prevent reduction of the medium. After the biofilm had formed, all the medium liquid was pipetted out and the tube was washed three times with deionized water. To conclude, 1 mL of 0.3% Crystal violet solution was added, incubated without shaking for 3 min at room temperature, washed in deionized water and the biofilm was observed.
For the determination of biofilm formation ability, the tubes were washed with deionized water until the eluted water showed no purple coloration. Finally, 1 mL of 30% acetic acid was added into the tube, sonicated for 30 min; then, the absorbance of the solution at a wavelength of 597 nm was measured with 30% acetic acid as the control, this was recorded as the biofilm formation rate. Three replicates were undertaken for each sample.
Cell motility assay
The motility experiment was performed following the methodology of Clemmer KM et al. [Citation33], with modifications. Bacterial motility was measured on modified semi-solid LB agar plates and M9 agar plates. The agar content of the plate was 0.25% (w/v). The overnight culture was diluted to OD600 = 0.2, and 1.5 µL of this solution was added to the semi-solid agar plate. After complete absorption, the plate was incubated at 25°C for 48 h and then photographed to record the movement of the bacteria.
Tolerance to different concentrations of NaCl
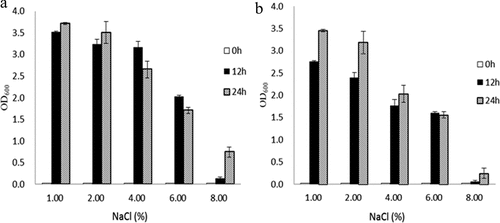
In line with the method published by Fakruddin [Citation15], a single colony was inoculated into 5 mL of LB liquid medium and incubated overnight. The overnight culture was then transferred into LB medium supplemented with different NaCl concentrations (1.0%, 2.0%, 4.0%, 6.0%, 8.0% and 10.0%) at an initial OD600 ≈ 0.02, and incubated at 37°C, 200 rpm. The OD600 value was measured at 12 h and 24 h, and three replicates were included for each group. These experiments were repeated twice.
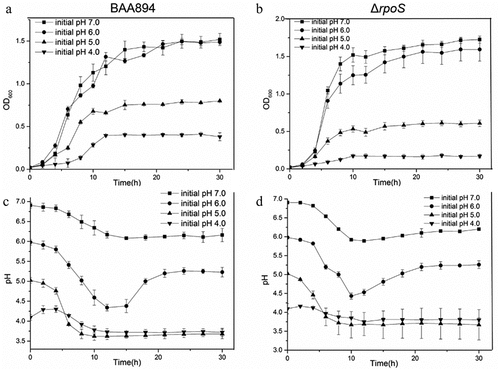
Acid resistance assay
The acid resistance assay was performed following the methodology of Fakruddin [Citation15] with modifications. The growth curves under different acidity and medium conditions, and the pH changes of the media during growth were determined. The strains were inoculated at 37°C, 200 rpm, and different media pH values of 7.0, 6.0, 5.0 and 4.0 were initiated. The cultures were then incubated in shaking flasks and sampled every 1–2 h. The OD600 of the samples were measured using UV spectrophotometry as the maximum absorption peak of C. sakazakii was 600 nm, and the valid range of the spectrophotometer was 0.3–0.9; samples with OD600 out of this range were adjusted and re-measured. The growth curve was plotted according to the mean value of OD600 at each sampling point, and the standard deviations were calculated and labeled. Meanwhile, the pH meter was used to measure acidity changes in each sample at all time points. The mean and standard deviation for each sample were calculated after obtaining stable readings, and the curves were plotted accordingly.
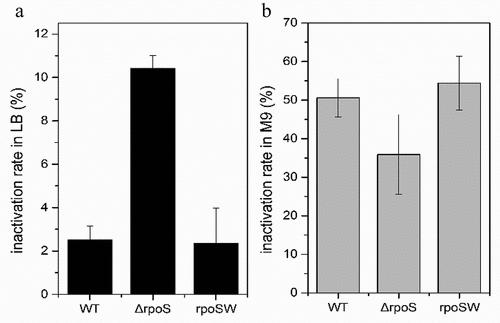
Desiccation tolerance analysis
This experiment was performed as previously published [Citation19], with some modifications. As the dryness resistance abilities of thallus can change in different environments, tests were conducted in both LB and M9. The strains were cultured on plates; then, a single colony was cultured in 5 mL liquid LB or M9 broth until the cells reached the logarithmic phase. After OD600 values had been measured in the different microbial cultures, a 100 mL portion of each culture was transferred into wells in a matched 96-well microtiter plate. Subsequently, the plate was transferred into a sterile dryer with dehydrated silica gel. The dryer was placed in a sterile incubator (DHP-2042BS, Huabei, Tianjin, China), which was constantly kept at 37°C. Six days later, the 96-well microtiter plates were placed onto a clean bench, and 100 mL/well of fresh LB or M9 medium was added; then, the plates were shaken at 200 rpm at 37°C for 3 h to re-culture. The liquid from each well was then transferred to a new 96-well microtiter plate, and the OD600 was detected. Each strain was tested in triplicate, and each test was repeated three times. The desiccation tolerance was performed with inactivation rate and calculated using the following formula:
OD0 represents the initial OD600 of each strain, and OD1 represents the OD600 of each strain after re-cultured for 3 h.
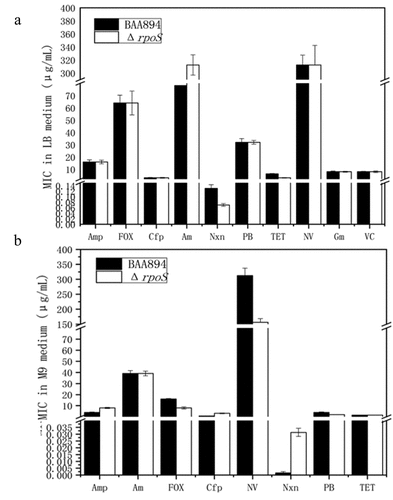
Drug resistance of BAA894-ΔrpoS
The minimum inhibitory concentrations (MICs) were determined for each strain, according to the method reported [Citation21,Citation34]. Antibiotic stock solutions were prepared at the following concentrations: ampicillin 2.56 mg/mL, cefoxitin 2.56 mg/mL, norfloxacin 0.16 mg/mL, polymyxin 0.64 mg/mL, tetracycline 2.00 mg/mL, amoxicillin 50.00 mg/mL, vancomycin 0.64 mg/mL, gentamycin 0.64 mg/mL, cefoperazone 1.00 mg/mL, and novobiocin 50.00 mg/mL. The antibiotics were filtered and then diluted accordingly. Firstly, 198 μL of LB medium was added to the first well of the plate, and 100 μL of LB medium was added to the remaining wells. Then, 2 μL of stock solution was added to the first well, mixed well, and 100 μL of this solution was transferred into the second well; then 100 μL of that medium was transferred into the following well for sequential dilutions until the seventh well was reached, from which 100 μL of the medium was discarded. The eighth well was supplemented with no drugs and served as the growth control. Overnight cultures were inoculated into 5 mL LB liquid medium at an initial OD600 = 0.02, incubated for around 2 h until OD600 = 0.5 was achieved; then, the solution was diluted with LB medium at 1:1000, and 100 μL of this diluted culture was added to each well. After the plate had been sealed, it was incubated for 16–20 h in a water-proof incubator at 37°C, and the results were observed thereafter. Four replicates were included for each sample.
Transcriptome analysis
The main procedures for transcriptomic analysis included: total RNA extraction, DnaseI digestion, mRNA isolation, mRNA fragmentation, mRNA reverse transcription, cDNA synthesis and library quality checks. Qualified libraries were used for cDNA sequencing with Illumina HiSeqTM2000 technology. The RPKM (Reads per kilobase transcriptome per million mapped reads) method [Citation35] was used to calculate gene expression [Citation11], and the obtained gene expression levels were used directly for differential gene expression analysis. In this study, differentially expressed genes were defined as genes with both an FDR ≤ 0.05 by Benjamini’s test [Citation36,Citation37] and with a fold difference of higher than 2. After the differentially expressed genes were screened, KEGG [Citation38] Pathway clustering analysis was performed on this basis.
In microorganisms, coordination between genes is important for gene function, and thus the functional properties of genes in metabolic process can be studied based on pathway clustering analysis. KEGG is a public database for metabolic pathway analysis. For differentially expressed genes, significant enrichment based on KEGG pathways can be tested in the context of the entire genome to highlight significantly enriched pathways. This analysis was performed by calculating the p-value of each Pathway, corrected by Benjamini’s test, and by using a Q-value ≤0.05 as the threshold to determine significantly enriched pathways.
In this study, the biological functions associated with the differentially expressed genes were determined by clustering analysis of KEGG Pathways based on the significant differences in gene expression, and the involved metabolic pathways were used as references to study the mechanism of tolerance alteration in the mutant strain.
Results
In order to clarify the function of RpoS related to stress resistance within C. sakazakii BAA894, an rpoS gene related DNA fragment deletion mutant strain was constructed using Red homologous recombination technology. The expression and function of the internal mutation of rpoS by these mutant strains was then analyzed. Following this the mutant strains were analyzed based on biofilm formation ability, motility, resistance to high permeability, acid resistance, and desiccation tolerance. The findings showed that the high permeability resistance and acid resistance of the ΔrpoS mutant strain were weakened, dryness resistance increased in LB medium and decreased in M9 medium, and drug resistance changed significantly. Finally, transcriptome analysis confirmed that the rpoS gene deletion resulted in differential expression of genes related to stress resistance and changes in related metabolic pathways, these were consistent with the other experimental results.
Verification of rpoSU, rpoSD, rpoSW and rpoSDTB overexpression
The expression of rpoS in BAA894 cells and the effect of stop codon mutations on the expression levels were verified using SDS-PAGE (). The protein sizes were determined using the protein marker sizes (14.4, 18.4, 25, 35, 45, 66.2 and 116 kDa). Lanes 2 and 3 show proteins with similar sizes of around 25 kDa, and the respective genes were inferred to be around 600 bp accordingly, which corresponds to the first 603 bp of rpoS. Lane 3 showed no significantly overexpressed proteins. The size of the entire RpoS protein is between 35 kDa and 45 kDa, which is about 40 kDa, and an obvious protein band of this size could be observed in lane 4 of BL21-pET28a-rpoSDTB. Based on the protein expression analysis, it was verified that the partial rpoS before the stop codon in BAA894 was overexpressed. When expressed using a strong promoter, the remaining part of RpoS, after the mutation site, showed a band at a corresponding position on the Tricine-SDS-PAGE, but the expression levels seemed to be lower than other overexpressed proteins, which therefore requires further verification.
Resistance-related phenotyping experiments
Bacteria biofilm formation ability
Biofilms are formed when bacteria cells interact with each other under changing culture conditions, which ensures the viability of the colony. The process of biofilm formation includes five stages: reversible adhesion, irreversible adhesion, microcolony expansion, development and maturation of biofilm, and dissociation of the biofilm. Following formation of the biofilm, a complex metabolic network is formed between the microbial community and the glycocalyx within the biofilm, which transports oxygen and nutrients into the membrane and removes the metabolic waste from the membrane. Therefore, some researchers regard the biofilm as a primitive circulatory system. In this study, the membrane formation abilities of the BAA894 wild type, ΔrpoS mutant and rpoSW complementary strain were conducted in LB and M9 medium, respectively ().
Figure 3. Analysis of the biofilm formation among the BAA894, mutant ΔrpoS and recombinant strain rpoSW
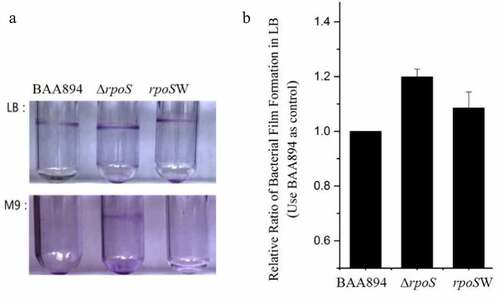
In the LB medium (in which IPTG and Amp were supplemented to the complementary strain), the membrane formation abilities of the ΔrpoS mutants were significantly enhanced comparing to that of BAA894, and the rpoSW complementary strain was able to complement the wild-type phenotype. In M9 medium (where both mutant strains were supplemented with IPTG and Amp), a blue band is shown on the wall of the test tube of ΔrpoS, whereas both the wild type and complementary strains showed no obvious bands.
As shown in , quantitative assays of biofilm formation ability were performed for the wild type, ΔrpoS mutant and complementary strains in LB medium. As observed in M9 medium, the BAA894 and rpoSW strains had almost no biofilm formation, similar to results from the qualitative experiments. The wild type was used as a control with a value of 1. The results showed that membrane formation in the mutant strain was increased and the complementation of the entire BAA894 rpoS gene (rpoSW) achieved good complementary effects.
Combining the results of both culture conditions, the absence of rpoS resulted in an increased membrane formation ability, indicating that the ΔrpoS mutant may adhere more readily to the material surface. This increased capacity may be a compensatory swarming response by the bacteria when the regulatory effects of rpoS are reduced.
Motility assay of the wild-type, mutant and complementary strains
Pili structure can directly affect the interaction and adherence of cells, both of which affect the stress resistance of bacteria. Motility is one of the direct means by which changes in pili structure and number can be analyzed. In this study, the motility of wild-type, ΔrpoS mutant and complementary strains were studied in different medium agar plates ().
Figure 4. Swimming motility of the wild-type, rpoS null mutant and expression recombinant strains on M9 or LB medium
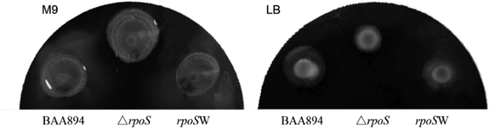
When the bacteria were in M9 medium (M9+ IPTG), it was observed that the concentric circles of the BAA-ΔrpoS colony were the largest and most obvious among the three strains. This size and shape indicated that motility was stronger. Additionally, the colony morphology was slightly flatter and more mucilaginous than that of the wild type, whereas the rpoSW complemented colony showed similar morphological and motility traits to those of the wild-type colonies. In comparison to the M9 medium, differing motilities between the strains growing on LB (LB+IPTG) agar plates were not obvious, and the motility of the mutant was weaker than that of the wild type.
Tolerance to high NaCl concentrations
As shown in , when the medium contained normal concentrations of NaCl (i.e. 1.0% NaCl), the bacteria reached higher concentrations than those other high NaCl media. The growth of the BAA894 wild-type bacteria showed a decreasing trend with increased concentrations of NaCl. The bacteria showed basically no growth when the medium contained 8.0% NaCl, the highest density reached only 2.03 when 6.0% NaCl was supplemented. The highest density was observed at 12 h when supplemented with more than 4% salt, which decreased again at 24 h. In general, the growth of the wild-type bacteria decreased with increased NaCl concentrations. ΔrpoS showed similar growth patterns to BAA894, but grew at a slower rate than BAA894. The growth rate of BAA894 was higher than that of ΔrpoS under normal culture conditions, which was mainly caused by the longer adaptation stage of ΔrpoS, resulting in a significantly lower OD value at 12 h than that of BAA894; however, the difference in OD value at 24 h had reduced. The ΔrpoS mutant was less tolerant to high osmotic environments, which is consistent with the stress-resistance-related functions of the rpoS gene. This may be related to the transport or synthesis of ion or compatible solutes.
Acid tolerance assay
M9 medium was used to study the changes in acid tolerance in the mutant strains, the strains grow slowly in this medium; therefore, the differences in growth curves are more obviously reflected.
The growth of C. sakazakii BAA-894-ΔrpoS in the M9 medium under different pHs (7.0, 6.0, 5.0, 4.0) showed that the growth of the mutant was significantly slower than that of the wild type at pH 5.0 and pH 4.0. Medium measurements found that the mutant strain adjusted to the medium pH earlier than the wild type, but the pH of the wild type under pH 6.0 conditions increased rapidly after entering stationary phase and reached a higher final value than that of the mutant. The highest adjusted pH value of the wild type at pH 4.0 was higher than that of the mutant, which might explain the better growth of the wild type under low pH conditions ().
Desiccation tolerance assay
Resistance to desiccation is one of the important characteristics of C. sakazakii. In order to investigate the role of RpoS in regulating the C. sakazakii desiccation resistance, the survival rates of the wild-type BAA894, ΔrpoS mutant, and rpoSW were tested under varying desiccation conditions.
The desiccation tolerance assay showed that, in LB medium, the desiccation resistance of the mutant strain decreased than the wild type and the inactivation rate reached 4 times that of the wild type. The mutant strain desiccation resistance in M9 medium was slightly higher than that of the wild type, whilst its inactivation rate decreased by about a third (). In addition, rpoSW rates complemented the desiccation resistance observed in the mutant strain.
Drug resistance assays on BAA894 and ΔrpoS
Minimum inhibitory concentrations (MICs) were determined for the wild-type and ΔrpoS mutant strains, respectively.
The mutant strain was tested for drug resistance in both LB and M9 medium conditions (). In the M9 medium, compared with the wild-type BAA894, the mutant strain showed reduced resistance to the cephalosporins cefoxitin and coumarins neomycin, and increased MIC to norfloxacin, which prevents bacterial DNA replication by blocking its DNA gyrase. When grown in LB media, the mutant strain showed increased sensitivity to norfloxacin and tetracycline, and decreased sensitivity to amoxicillin. The mutant strain showed higher susceptibility to polymyxin B in both LB and M9 media. The reduced MIC to β-lactam and peptide antibiotics in the mutant strain suggested that the rpoS deletion may have caused some structural changes in the cell membrane and wall, making it more fragile.
Transcriptome analysis under different conditions
Overall comparison of transcriptomes of BAA894 and ΔrpoS mutant strains
In LB medium, the total number of annotated genes was 3927 for ATCC BAA894 and 3900 for the ΔrpoS mutant strain. A total of 3973 annotated genes were obtained from the two strains, with 3854 crossed, of which 1778 genes were up-regulated and 2195 genes were down-regulated in the mutant. After accounting for the FDR correction, 123 genes were significantly up-regulated, and 260 genes were significantly down-regulated. When cultured in the M9 medium, 74 genes were significantly up-regulated and 19 genes were significantly down-regulated after accounting for the FDR correction.
Overall pathways enrichment of differentially expressed genes in BAA894 and ΔrpoS mutant strains
In LB medium, the three most significantly enriched pathways in the ΔrpoS mutant strains were flagellar assembly, microbial metabolism in diverse environments and nitrogen metabolism, and all three pathways were significantly down-regulated when compared to the wild type. Among them, the pili synthesis pathways were all down-regulated, and over 75% of the genes in metabolic pathways of different environments were downregulated. Few nitrogen-metabolism-related genes were identified, but the genes in this pathway were significantly down-regulated, and the number of down-regulated genes was more than 6 times that of up-regulated genes. The overall observations showed that the differentially expressed genes caused by rpoS deletion in LB medium were mainly down-regulated, and all of the pathways also showed a trend of down-regulation ().
Figure 9. KEGG pathway cluster analysis of differently expressed genes in ΔrpoS, using BAA894 as a control
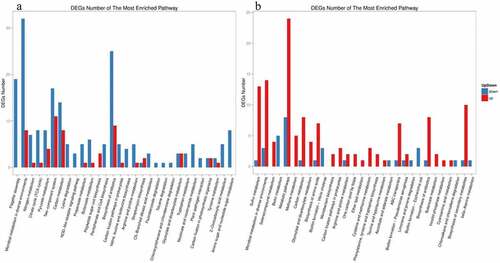
In M9 medium, sulfur metabolism, microbial metabolism in diverse environments and selenocompound metabolism were the most significantly altered pathways, they were all up-regulated. Among them, all the genes within the selenium-containing compound metabolism pathway were up-regulated. shows that the differentially expressed pathways caused by rpoS deletion in M9 medium were mostly up-regulated, but in general, the number of genes significantly differentially expressed in M9 media was less than that in LB media.
Analysis of significant differential pathways
Bacteria can improve population tolerance through cell-to-cell interactions in stressful environments. Cell surface hydrophobicity, pili, and transport proteins can directly affect tolerance, while chemotaxis and population effect signaling molecules are involved to reflect the changes in tolerance through intercellular communication within the cell population. In this study, we analyzed the transcription of genes involved in flagella assembly, cellular chemotaxis, quorum sensing and ABC transport in LB medium, as they were significantly regulated pathways. Meanwhile, the membrane transport and signal transduction pathways were analyzed in M9 medium. The comparison between ΔrpoS mutant and wild type, and the results are shown in .
Figure 10. (a) Transcriptional ratios of the genes relevant to KEGG pathway significantly enriched in LB medium. (1): Flagellar assembly, (2): Bacteria chemotaxis, (3): Quorum sensing, (4): ABC transport. (b) Transcriptional ratios of the genes relevant to KEGG pathway significantly enriched in M9 medium. (1): membrane transport, (2): Signal transduction. (c) Transcriptional ratios of the genes relevant to Microbial metabolism in diverse environments in LB (1) and M9 (2). Log2R represents the ratio of gene expression in the mutant strain ΔrpoS versus the same gene in the BAA894 wild strain at logarithmic scale to base 2
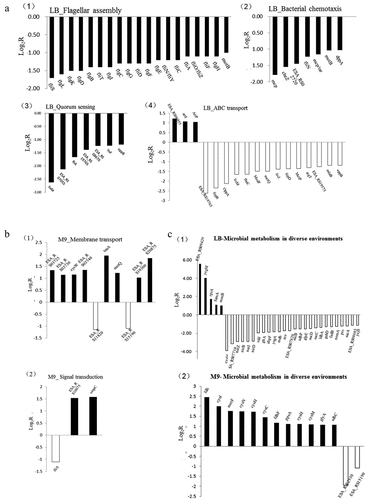
The mutant strain showed reduced motility in LB medium ((1))), and transcriptome analysis also showed down-regulation of all genes involved in the pili synthesis pathway in the mutant strain cultured in LB medium. The pili protein FliS presented as the most significantly down-regulated gene, followed by the hook proteins FlgL and FlgR, the pili basal-body rod protein FlgB, and the motility switch protein. These consistent down-regulations of flagellar assembly and function-related proteins were also consistent with the reduced motility observed in the rpoS mutant strain when in LB medium. The mutant strains were also downregulated in all bacterial chemotaxis and quorum sensing pathways, including MCP and protein phosphatase CheZ ((a(2). (3))), and some ABC transport-related proteins such as substrate binding protein OppA in the population sensing, which are actually related to cell perception, environmental response and motility. Most of the differential genes are also involved in the ABC transport system of membrane active transport, and as shown in (a(4)), the ABC transporter pathway was highlighted as one of the significantly down-regulated pathways.
In M9 medium, the pathways related to membrane transport and signal transduction were significantly up-regulated in the mutant (). Genes tauA and metQ encode taurine ABC transporter substrate binding protein and methionine ABC transporter substrate binding protein, respectively, and cysW was associated with sulfate permease to enhance active sulfide transport, in fact, the differential enrichment of the sulfur-metabolism-related pathway was the most significant seen in the M9 medium. In the signal transduction pathways, OmpC was associated with outer membrane permeability, and its upregulation indicated enhanced membrane permeability. In addition, the upregulation of motility-related ESA_RS10075 (trg) in bacteria grown in M9 medium indicated increased motility, which was consistent with the results of the motility assay. It was also of interest that the pathways related to the cell secretion system were also upregulated under M9 environments.
Cells can sense stress by recognizing the signal receptors in response to the external environment. These are then transmitted by messenger molecules such as ppGpp to stress proteins and σS regulators, and the transcriptional regulator σS regulates the transcription function of RNA polymerase (RNAP) by binding to the promoter or the nearby region to achieve the transduction of signal to final activation of gene expression in vivo. In this study, we compared and analyzed the differentially expressed genes in microbial metabolism in diverse environment pathways both in LB and M9 medium using transcriptomic analysis (). When compared to the wild type, the genes that were down-regulated the mutant strain in LB medium mainly included cysteine synthase cysM, 2-oxoglutarate dehydrogenase sucB, nitrite reductase catalytic subunits nirB, nirD, and nitrate reductase subunits narH, narZ, indicating a decrease in the metabolism of nitrogen and sulfur by the bacterium. Up-regulated genes were mainly related to formic acid metabolism. ESA_RS09420 formic acid dehydrogenase subunit and ybcF carbamate kinase were up-regulated in LB medium. Meanwhile in the M9 medium, sulfite reductase subunit cysHIJN and other sulfite synthesis and transformation-related genes were up-regulated, while those down-regulated included dimethylallyl transferase and membrane protein ESA_RS11100. The significant changes in this pathway demonstrate that rpoS removal can cause corresponding changes in cellular responses to the environment.
Discussion
RpoS is an important global regulator, and Alvarez et al. [Citation5] showed that σ-factor is the main signal switch in response to environmental stress, besides which, it is also an important gene which influences C. sakazaki to enter the VBNC state after extreme stress. During the process of our research, we also found that bacterial culturability decreased in the absence of rpoS gene expression. As the main σ-factor during the stationary phase of the cell, its effect and mechanism on environmental stress in C. sakazakii requires further investigation.
The biofilm formation abilities and cell motility are indirectly related to environmental tolerance. Microorganisms cannot stay in a specific environment during growth, but more often grow as a population within the environment; therefore, the study of bacterial tolerance should consider some properties related to cellular interactions. As a product of bacterial stress regulation in response to changing environmental conditions, the biofilm plays an important role in the survival and growth of bacteria under less favorable conditions [Citation39]. The adhesion of bacteria to materials depends more on pilis, and the bacterial adhesion not only affects the resistance of multiple external shocks, but also is related to the host invasion, which determines the virulence of bacteria to a certain extent.
The increased ability of the mutant strains in this study to form biofilm indicates an increased adhesion property within the mutant strain [Citation40]. However, in the motility assay, mutant strain mobility was reduced in LB, suggesting that the increased auto-aggregation ability and membrane formation ability in this medium may not be caused by the pili synthesis, but may be related to other cell membrane alterations. rpoS gene modulates expression of 10% of the genome in Gram-negative bacteria, including flagella and fimbriae synthesis genes, membrane transport genes, and signal transmission genes, etc. Based on these regulations, a large part of genes involved in biofilm formation and motility are affected, especially in harsh environments. PmrA/PmrB (polymyxin resistance A/B) regulatory system is one of the two-component regulatory systems (TCS), and it is also known as an important stress tolerance and biofilm-related gene in C. sakazakii [Citation41–43]. PmrA/PmrB mainly affect the biofilm formation by regulating genes (lpxA, eptAB, cpxAR) related to lipopolysaccharide (LPS) modify [Citation44]. Unlike PmrA/PmrB regulatory system, rpoS not only affects virulence-related stress tolerance, but also affects many metabolic reactions and survival states of the strains.
In this study, the tolerance assay to different concentrations of NaCl indicated a reduced tolerance of the mutant strain ∆rpoS to high osmotic pressure. The acid tolerance assay showed that the acid tolerance of the ΔrpoS deletion mutant strain was reduced, and the growth assay in M9 medium indicated that the mutant strain grew worse than the wild type under acidic conditions. The desiccation tolerance of the mutant strain decreased in LB medium and increased in M9 medium. In LB medium, the desiccation tolerance of the cells might be mainly derived from the synthesis of compatible solutes, and the deletion of rpoS gene thus leads to reduced tolerance due to its positive regulation of a variety of intracellular synthesis and transport pathways. However, in M9 medium, the cells reduced the intensity of many cellular metabolism pathways and slowed growth down due to limited carbon and nitrogen sources. Meanwhile, the cells consumed a large amount of carbon sources to synthesize extracellular secretions to protect the cells from the stress environments, which also shifted the way of desiccation tolerance from intracellular regulation to the structural changes of membrane walls and appendages. Therefore, the regulatory effect of RpoS in the restricted medium was not as important as observed in normal medium, and the increased desiccation resistance of the mutant strain in M9 medium may be related to the survival status of the organism in different environments.
The drug resistance of the mutant strain was tested in both LB and M9 media. In the M9 medium, the mutant strain ∆rpoS showed reduced resistance to cefoxitin from the cephalosporins and neomycin from the coumarins, and increased MIC to norfloxacin, which prevents bacterial DNA replication by blocking the DNA gyrase. The mutant strain grew slower than the wild type, and its DNA replication rate may be lower than that of the wild type, thus showing lower resistance to these antibiotics. The reduction in MIC to β-lactam and peptide antibiotics suggests that the rpoS deletion may have caused some structural changes in the cell membrane and cell wall, making them more fragile.
In this study, we conducted biofilm formation ability, motility, acid resistance, desiccation tolerance, high permeability and antibiotic resistance phenotypic experiments. In the transcription analysis, we indicated that cell surface hydrophobicity, pili, and transport proteins directly affected tolerance, while chemotaxis and population effect signaling molecules were involved to reflect the changes in tolerance through intercellular communication within the cell population. Therefore, we analyzed the transcription activities of genes involved in flagella assembly, cellular chemotaxis, quorum sensing and ABC transport in LB medium, as they are significantly regulated pathways in LB medium (). Meanwhile, the membrane transport and signal transduction pathways were analyzed in M9 medium, as they can help explain the performance of the mutant strains in M9 medium (). We also compared and analyzed the differentially expressed genes in microbial metabolism in diverse environment pathways both in LB and M9 medium using transcriptomic analysis (). Cells can sense stress by recognizing the signal receptors in response to the external environment, and the transcriptional regulator σS regulates the transcription function of RNA polymerase (RNAP) by binding to the promoter or the nearby region to achieve the transduction of signal to final activation of gene expression in vivo. Analysis of the significantly differentially expressed genes of this pathway in the mutant strain can explain the results of the acid tolerance and high permeability experiments.
Conclusion
In this article, we investigated the overall regulatory σ factor RpoS for environmental stress resistance in C. sakazakii BAA894. First, the gene rpoS was knocked out of the genome by Red homologous recombination; then, the trials with acid, osmotic pressure, desiccation and drug tolerance were carried out. The results showed that mutant strain ΔrpoS membrane formation ability significantly enhanced, motility became enhanced, hyperosmotic resistance increased, acid tolerance decreased, whilst drying tolerance decreased in LB medium and increased in M9. Finally, we analyzed the transcriptome of the mutant to confirm the experiment results. These experiments concluded that RpoS played a critical regulatory role in C. sakazakii BAA894 in relation to withstanding environmental stresses. All these conclusions are meaningful toward understanding the infection mode of C. sakazaki in newborns and help to prevent these infections.
Highlights
1) Constructed rpoS deletion mutant strain using Red homologous recombination technology.
2) The ΔrpoS biofilm formation was enhanced, and motility changed oppositely in LB or M9 media.
3) ΔrpoS exhibited resistance changes in relation to hypertonicity, acid, dryness, and antibiotics.
4) Transcriptomic analysis results were mostly consistent with the other experiments.
Compliance with ethics requirements
This article does not contain any studies with human or animal subject.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Iversen C, Lehner A, Mullane N, et al. Identification of “Cronobacter” spp. (Enterobacter sakazakii). J Clin Microbiol. 2007;45(11):3814–3816.
- Stephan R, Grim CJ, Gopinath GR, et al. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov. Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int J Syst Evol Microbiol. 2014;64(Pt10):3402–3410.
- Álvarez-Ordóñez A, Begley M, Hill C. Polymorphisms in rpoS and stress tolerance heterogeneity in natural isolates of Cronobacter sakazakii. Appl Environ Microbiol. 2012;78(11):3975–3984.
- Kucerova E, Clifton SW, Xia XQ, et al. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One. 2010;5(3):e9556. Published 2010 Mar 8.
- Feeney A, Kropp KA, O’Connor R, et al. Cronobacter sakazakii: stress survival and virulence potential in an opportunistic foodborne pathogen. Gut Microbes. 2014;5:711–718.
- Forsythe SJ. Updates on the Cronobacter Genus. Annu Rev Food Sci Technol. 2017;9:2.1–2.22.
- Hunter CJ, Petrosyan M, Ford HR, et al. Enterobacter sakazakii: an emerging pathogen in infants and neonates. Surg Infect (Larchmt). 2008;9:533–539.
- Lai K. Enterobacter sakazakii infections among neonates, infants, children, and adults: case reports and a review of the literature. Medicine (Baltimore). 2001;80(2):113–122.
- Bowen A, Braden C. Invasive Enterobacter sakazakii disease in infants. Emerg Infect Dis. 2006;12:1185–1189.
- Shaker R, Osaili T, Al-Omary W, et al. Isolation of Enterobacter sakazakii and other Enterobacter spp. from food and food production environments. Food Cont. 2007;18:1241–1245.
- Cheville AM, Arnold KW, Buchrieser C, et al. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1996;62(5):1822–1824.
- Stephan L, Paolo L. SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J Bacteriol. 2004;186:7186–7195.
- Castanie-Cornet MP, Penfound TA, Smith D, et al. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535.
- Laaberki MH, Janabi N, Oswald E, et al. Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157: H7:interplay between Ler, GrlA, HNS and RpoS. Int J Med Microbiol. 2006;296(4–5):197–210.
- Fakruddin M, Rahaman M, Ahmed MM, et al. Stress tolerant virulent strains of Cronobacter sakazakii from food. Biol Res. 2014;47(63):0717–6287.
- Alvarez-Ordonez A, Cummins C, Deasy T, et al. Acid stress management by Cronobacter sakazakii. Int J Food Microbiol. 2014;178:21–28.
- Feeney A, Sleator RD. An in silico analysis of osmotolerance in the emerging gastrointestinal pathogen Cronobacter sakazakii. Bioeng Bugs. 2011;2(5):260–270.
- Burgess CM, Gianotti A, Gruzdev N, et al. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int J Food Microbiol. 2016;221:37–53.
- Du XJ, Wang XY, Dong X, et al. Characterization of the desiccation tolerance of Cronobacter sakazakii strains. Front Microbiol. 2018;9:2867.
- Jameelah M, Dewanti-Hariyadi R, Nurjanah S. Expression of rpoS, ompA and hfq genes of Cronobacter sakazakii strain Yrt2a during stress and viable but nonculturable state. Food Sci Biotechnol. 2018;27(3):915–920. Published 2018 Jan 18.
- Liu L, Li Y, Wang X, et al. A phosphoethanolamine transferase specific for the 4ʹ-phosphate residue of Cronobacter sakazakii lipid A. J Appl Microbiol. 2016;121(5):1444–1456.
- Chen S, Zhou Q, Tan X, et al. The global response of Cronobacter sakazakii cells to amino acid deficiency. Front Microbiol. 2018;9:1875. Published 2018 Aug 14.
- Jain C. Overexpression and purification of tagged Escherichia coli proteins using a chromosomal knock-in strategy. Protein Expr Purif. 2006;46(2):294–298.
- Heermann R, Zeppenfeld T, Jung K. Simple generation of site-directed point mutations in the Escherichia coli chromosome using Red(R)/ET(R) recombination. Microb Cell Fact. 2008;7:14.
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645.
- Albermann C, Trachtmann N, Sprenger GA. A simple and reliable method to conduct and monitor expression cassette integration into the Escherichia coli chromosome. Biotechnol J. 2010;5(1):32–38.
- De Mey M, Maertens J, Boogmans S, et al. Promoter knock-in: a novel rational method for the fine tuning of genes. BMC Biotechnol. 2010;10:26.
- Lelong C, Aguiluz K, Luche S, et al. The Crl-RpoS regulon of Escherichia coli. Mol Cell Proteomics. 2007;6(4):648–659.
- Spector MP, Aliabadi Z, Gonzalez T, et al. Global control in Salmonella typhimurium: two-dimensional electrophoretic analysis of starvation-, anaerobiosis-, and heat shock-inducible proteins. J Bacteriol. 1986;168(1):420–424.
- Kouidhi B, Zmantar T, Hentati H, et al. Cell surface hydrophobicity, biofilm formation, adhesives properties and molecular detection of adhesins genes in Staphylococcus aureus associated to dental caries. Microb Pathog. 2010;49(1–2):14–22.
- Kirov SM, Castrisios M, Shaw JG. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect Immun. 2004;72(4):1939–1945.
- Cerca N, Pier GB, Vilanova M, et al. Quantitative analysis of adhesion and biofilm formation on hydrophilic and hydrophobic surfaces of clinical isolates of Staphylococcus epidermidis. Res Microbiol. 2005;156(4):506–514.
- Clemmer KM, Bonomo RA, Rather PN. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology (Reading). 2011 Sept;157(Pt 9):2534–2544. Epub 2011 Jun 23. PMID: 21700662; PMCID: PMC3352170.
- Wang Z, Wang J, Ren G, et al. Influence of core oligosaccharide of lipopolysaccharide to outer membrane behavior of Escherichia coli. Mar Drugs. 2015;13(6):3325–3339. Published 2015 May 27.
- Li R, Yu C, Li Y, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967.
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188.
- Zhao H, Peddada SD, Cui X. Mixed directional false discovery rate control in multiple pairwise comparisons using weighted p-values. Biom J. 2015;57(1):144–158.
- Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 2001;25(4):409–418.
- Kolecka A, Chorvat D Jr, Bujdakova H. The impact of growth conditions on biofilm formation and the cell surface hydrophobicity in fluconazole susceptible and tolerant Candida albicans. Folia Microbiol (Praha). 2015;60(1):45–51.
- Silva-Dias A, Miranda IM, Branco J, et al. Adhesion, biofilm formation, cell surface hydrophobicity, and antifungal planktonic susceptibility: relationship among Candida spp. Front Microbiol. 2015;6:205.
- Bao X, Yang L, Chen L, et al. Virulent and pathogenic features on the Cronobacter sakazakii polymyxin resistant pmr mutant strain s-3. Microb Pathog. 2017;110:359–364.
- Bao X, Jia X, Chen L, et al. Effect of polymyxin resistance (pmr) on biofilm formation of Cronobacter sakazakii. Microb Pathog. 2017;106:16–19.
- Xu Z, Liu Z, Soteyome T, et al. Impact of pmrA on Cronobacter sakazakii planktonic and biofilm cells: a comprehensive transcriptomic study. Food Microbiol. 2021;98:103785.
- Hua J, Jia X, Zhang L, et al. The characterization of two-component system PmrA/PmrB in Cronobacter sakazakii. Front Microbiol. 2020;11:903. Published 2020 June 17.