ABSTRACT
The clinicopathological value of microRNA-141-3p (miR-141-3p) and its prospective target genes in endometrial carcinoma (EC) remains unclear. The present study determined the expression level of miR-141-3p in EC via quantitative real-time PCR (RT-qPCR). RT-qPCR showed a markedly higher expression level of miR-141-3p in EC tissues than in non-EC endometrium tissues (P < 0.0001). The microarray and miRNA-seq data revealed upregulation of miR-141-3p. Integrated analysis based on 675 cases of EC and 63 controls gave a standardized mean difference of 1.737, confirmed the upregulation of miR-141-3p. The Kaplan-Meier survival curve showed that a higher expression of miR-141-3p positively corelated with a poorer prognosis. Combining the predicted targets and downregulated genes in EC, we obtained 271 target genes for miR-141-3p in EC. Two potential targets, PPP1R12A and PPP1R12B, were downregulated at both the mRNA and protein levels. This study indicates that the overexpression of miR-141-3p may play an important part in the carcinogenesis of EC. The overexpression of miR-141-3p may be a risk factor for the prognosis of patients with EC.
Introduction
Endometrial carcinoma (EC), also known as uterine corpus endometrial carcinoma (UCEC), is a malignant tumor that originates from the epithelium in the endometrium. As one of the most common gynecological malignant tumors, EC threatens women’s health and is also the third greatest cause of cancer-related death in women [Citation1–4]. Although the statistics are incomplete, an estimated 65,620 new cases and 12,590 deaths have occurred globally in 2020 [Citation5]. In developed countries, the incidence of endometrial cancer, at 5.9%, is higher than in developing countries (4.0%) [Citation6] and continues to show a rising trend. Despite the advances made in the treatment of EC, this cancer still has a poor prognosis, especially in patients with recurrence or metastasis after surgery or radiation [Citation7–10]. In addition, the molecular mechanism of EC has not been completely clarified.
The current absence of early diagnostic methods and the high incidence of tumor recurrence means that many patients with EC fail to receive the best treatment [Citation11,Citation12] and have poor survival. At the same time, the oppressive growth of the tumor directly affects female reproductive function and can have detrimental effects on the mental health of the patients [Citation13]. Amelioration of these adverse consequences, and particularly of the poor survival of patients with EC, therefore requires the identification of new diagnostic and prognostic indicators for EC.
One class of promising indicators are the microRNAs (miRNAs), which are unique endogenous small non-coding RNAs (around 18 to 25 nucleotides long) that regulate protein-coding genes at the post-transcriptional level. This regulation involves pairing with the base of the 3ʹ-untranslated region (UTR) of the target mRNA to inhibit mRNA translation or to promote mRNA degradation [Citation14–16]. Much research has shown that some miRNAs can cause dysregulation in EC and could therefore play a significant role in prognosis. For example, an upregulation of miR-486-5p has been detected in both tissues and serum samples from patients with EC [Citation17]. Similarly, a meta-analysis identified that a high level of miR-205 might lead to poor disease-specific survival in patients with EC [Citation18]. High expression of miR-21-5p or miR-940 has also been related to shorter overall survival [Citation19,Citation20]. Thus, the aberrant expression of miRNAs may modulate tumorigenesis and affect the prognosis of patients with EC.
One miRNA, miR-141, is abnormally expressed in many human malignancies. This miRNA normally takes part in several different cell processes, including the epithelial-to-mesenchymal transition (EMT), proliferation, migration, invasion and drug resistance [Citation21], but it also plays a crucial role in the incidence and progression of several cancers. One member of the miR-200 family, miR-141-3p, originates from the 3ʹ-end of the miR-141 hairpin structure [Citation22]. Three studies have reported upregulation of miR-141-3p in EC tissues and cells [Citation23–25]; however, all were small studies that utilized a single method to determine the expression of miR-141-3p. This upregulation therefore requires further validation, as no comprehensive study has yet been conducted. The potential mechanism underlying miR-141-3p involvement in EC has also not been established.
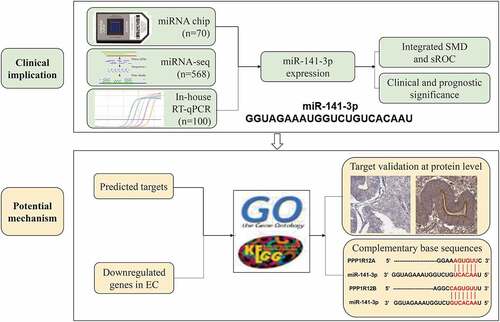
In present study, we examined the clinicopathological value of miR-141-3p in EC. The expression and prospective mechanism of miR-141-3p in EC tissues were analyzed to identify the prognostic significance of miR-141-3p and to elucidate its regulatory mechanism at molecular level.
Materials and methods
Expression level of miR-141-3p in clinical EC samples
Formalin-fixed, paraffin-embedded (FFPE) samples were obtained from patients with EC at the Department of Pathology, the First Affiliated Hospital of Guangxi Medical University from January 2015 to January 2020. Samples from patients who had been treated with radiotherapy or endocrine therapy were excluded. Ultimately, 70 EC tissues and 30 non-cancerous endometrial tissue were included in the study. Two pathologists selected and labeled typical tumor and non-cancerous tissue areas. The study was officially permitted by the ethics committee of the First Affiliated Hospital of Guangxi Medical University.
The expression level of miR-141-3p in the FFPE tissue samples was evaluated by quantitative real-time polymerase chain reaction (RT-qPCR). The miR-141-3p primers were synthesized by Baobioengineering (Dalian) Co., Ltd. The specific primers were GCACACTGTCTGGTAAAGATGGAA. The formula 2−Δcq was employed to calculate the expression level of miR-141-3p [Citation26].
MiR-141-3p expression from miRNA-sequencing data
We searched The Cancer Genome Atlas (TCGA) database for level 3 miRNA sequencing data, including 546 EC tissues and 22 non-cancerous endometrial tissues. These were log2 transformed for subsequent analysis.
MiR-141-3p expression from miRNA-chip databases
We expanded our sample size by mining data from the ArrayExpress, Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) databases and from various literature databases. The search keywords were: endometrial cancer AND microRNA. All included study samples were EC and non-tumorous endometrial tissues or cell lines. Each dataset contained three or more pairs of cancer groups and non-tumorous endometrial control groups; no subjects had undergone any interventions. A total of 2 eligible miRNA microarrays were incorporated: GSE25405 and GSE35794. A flow diagram is shown in .
Integrated analysis of the clinical characteristics of miR-141-3p in EC
We conducted a comprehensive evaluation of the data from the three resources (in-house RT-qPCR, miRNA-seq and miRNA chips) by performing an integrated analysis using Stata software version 15.1 (TX, USA) to calculate the standard mean difference (SMD) and to draw a summary receiver operating characteristic (sROC) curve [Citation27–29]. Continuous variables were assessed by SMD with a 95% confidence interval (95%CI), and heterogeneity was evaluated by the chi-squared-based Q-test and the I2 statistics value. Fixed effect models could be utilized when heterogeneity was low (I2 ≤50% and P≥0.05); otherwise, we chose the random effects model. Publication bias was examined by Begg’s or Egger’s test. A two-tailed P ≥0.05 indicated no publication bias. Forest plots were drawn to show the sensitivity, specificity, positive likelihood ratio (LR+), negative likelihood ratio (LR-), diagnostic score and odds ratio.
Prognostic value of miR-141-3p in EC
Patients in the TCGA-UCEC cohort were included in the study, and samples with unknown survival time or status or with follow-up time less than 30 days were excluded. The median expression value of miR-141-3p was used to divide the samples into two groups with high and low expression. The survival rate of the two groups was compared by the Kaplan-Meier (K-M) method. Survival analysis and K-M curves were completed using the survival and survminer packages of R software.
Prediction of target genes
We examined the underlying mechanism of miR-141-3p in EC by performing a series of in silico investigations. First, the target genes of miR-141-3p were predicted using 11 online tools: DIANA microT-CDs, miRanda, miRDB, miRmap, miRNAMap, miRWalk, PicTar, PITA, RNA22, TargetMiner and TargetScan v7.2. Only genes that co-occurred in at least three platforms were deemed eligible.
An increase in miR-141-3p in EC was anticipated to reduce the expression of target genes. Therefore, we screened the downregulated genes from the TCGA-UCEC cohort with the limma package of R. The criteria were log2-fold changes (FC)<-1 and P <0.05. Following the theory of evidence-based medicine, we used the same method and standards to screen the downregulated genes in three EC-related microarrays from the GEO database: GSE17025, GSE63678 and GSE146889. Genes that appeared in at least two datasets were subjected to further analysis. The target genes of miR-141-3p were ultimately obtained by overlapping the predicted target genes from the online search with the identified downregulated genes [Citation30,Citation31].
Gene functional annotation and enrichment evaluation
We analyzed the function of potential target genes and related pathways of miR-141-3p in EC using the online Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 program [Citation32,Citation33] for the gene ontology (GO) term annotation, in addition to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The results were visualized with the ImageGP online platform and the GOplot package of R software. The online tool miRmap [Citation34,Citation35] was utilized to search for complementary base sequences between the two selected target genes and miR-141-3p. Their expression in EC was verified with the TCGA sequencing data, and the correlations between miR-141-3p and two targets were analyzed using starBase v3.0. Protein level expression was obtained from The Human Protein Atlas (THPA) database [Citation36–38].
Statistical analysis
We used SPSS version 25.0 software (IBM Corp., Armonk, NY, USA) and the Student’s t-test to estimate the differences in miR-141-3p expression between the EC samples and non-cancerous endometrium. The scatter plots and receiver operating characteristic (ROC) curves corresponding to each data set were drawn with GraphPad Prism 8. The area under the curve (AUC) of the ROC curve was used to evaluate the ability of miR-141-3p to distinguish EC. We considered a value of P < 0.05 (two-tailed) to be statistically significant.
Results
In the current study, we explored the clinical significance of the overexpression of miR-141-3p in EC tissues and its prospective mechanism via in house RT-qPCR, miRNA chips and miRNA-seq. The K-M survival curve illustrated that patients with higher expression of miR-141-3p had a poorer prognosis. Through combining the predicted targets and downregulated genes in EC, we obtained 271 candidate target genes for miR-141-3p in EC. Moreover, we utilized the candidate targets to perform GO and KEGG enrichment analysis and found these targets enriched in some tumor-related pathways, such as ‘PI3K-Akt signaling pathway’ and ‘Ras signaling pathway’. Two potential targets enriched in ‘proteoglycans in cancer’ pathway, PPP1R12A and PPP1R12B, were downregulated at both the mRNA and protein levels.
Expression level of miR-141-3p in clinical EC samples based on qRT-PCR
The expression level of miR-141-3p was markedly higher in EC tissues than in non-EC endometrial specimens (P < 0.001, ). The AUC of the ROC curve was 0.7933 (95%CI: 0.6934–0.8932, P < 0.0001, ).
Figure 2. The expression of miR-141-3p in endometrial carcinoma (EC) and corresponding noncancerous tissues . (a, b) Scatter plot and receiver operating characteristic (ROC) curve of in-house quantitative real-time polymerase chain reaction (RT-qPCR). (c, d) Scatter plot and ROC curve of TCGA-UCEC cohort. (e, f) Scatter plot and ROC curve of GSE25405 dataset. (g, h) Scatter plot and ROC curve of GSE35794 dataset. (i) Forest plot of standard mean difference (SMD) of miR-141-3p expression in EC and non-EC groups. (j) Funnel plot of Egger’s test for pubilication bias

Expression level of miR-141-3p in the TCGA database in EC
Consistent with RT-qPCR results, the upregulation of miR-141-3p expression was greater in EC tissues than in non-EC endometrial tissues (P < 0.0001, ). The AUC of miR-141-3p was 0.8667 (95%CI: 0.7828–0.9507, P < 0.0001, ). By contrast, the expression level of miR-141-3p showed no clear differences in the low-grade versus the high-grade EC groups.
MiR-141-3p expression levels in microarray data of EC
Both microarrays revealed increasing trends in the expression of miR-141-3p in the EC tissue samples but not in the non-cancerous endometrial tissue samples ().
Integrated analysis of data from RT-qPCR, miRNA-seq and miRNA microarrays
We further confirmed the expression of miR-141-3p in EC by performing two types of integrated analysis using data from three sources: RT-qPCR, the TCGA-UCEC cohort and the GEO database ().
Table 1. The charateristics of the four cohorts included in the integrated analysis
Due to data heterogeneity (I2 = 90.4%, P < 0.001), we utilized the random effects model to combine the SMD. We observed a significantly increasing expression of miR-141-3p in EC tissue compared with non-cancerous endometrial tissue (SMD = 1.737, 95%CI: 0.692–2.783, P = 0.001, ). No publication bias was observed according to the Egger test (P = 0.786, ).
The combined sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, diagnostic score and odds ratio were 0.84, 0.76, 3.52, 0.21, 2.80 and 16.44, respectively (), and the area under the sROC curve was 0.87 (95%CI: 0.84–0.90, ). A Deeks’ funnel plot showed no publication bias (P = 0.055, ).
Figure 3. Pooled diagnostic indices for the integrated analysis based on four included studies. (a) The pooled sensitivity was 0.84 (0.81– 0.86). (b) The pooled specificity was 0.76 (0.64– 0.85). (c) The pooled positive likelihood ratio (LR) was 3.52 (2.26– 5.47). (d) The pooled negative LR was 0.21 (0.17– 0.27). (e) The pooled diagnostic score was 2.80 (2.18– 3.41). (f) The pooled diagnostic OR was 16.44 (8.89 −30.39)

Prognostic value of miR-141-3p in EC
The K-M curve indicated a prominent difference in the survival rate between patients with low expression of miR-141-3p and patients with high expression. Those with lower expression levels tended to have a more favorable survival outcome ().
Potential targets of miR-141-3p in EC
In this study, we obtained the predicted targets of miR-141-3p using 11 online tools. After overlapping, 1744 targets were selected. A further 2698 EC-related genes with low expression were identified from four datasets. The potential targets of miR-141-3p in EC were ultimately extracted by combining the predicted targets and those downregulated in EC. This finally identified 271 genes for subsequent GO annotation and KEGG pathway enrichment analysis ().
Signaling pathway enrichment analysis of the chosen targets
We further described the potential molecular biological functions of the 271 predicted target genes involved in EC by performing signaling enrichment analyses (). The targets of miR-141-3p were markedly enriched in the following biological process (BP) terms of GO: cell adhesion (n = 21) and positive regulation of glycogen biosynthetic process (n = 4). Enriched terms for the cellular component (CC) were focal adhesion (n = 19) and plasma membrane (n = 84). The target genes were also clustered with growth factor binding (n = 5) and ‘transcription factor activity, sequence-specific DNA binding’ (n = 28) for molecular function (MF). The visualization of the GO term enrichment is shown in the bubble plot in .
Table 2. KEGG pathway enrichment analysis of miR-141-3p related genes
The most striking KEGG pathway terms were focal adhesion (n = 12), the Rap1 signaling pathway (n = 11), proteoglycans in cancer (n = 10), microRNAs in cancer (n = 11), the PI3K-Akt signaling pathway (n = 12), the Ras signaling pathway (n = 9) and the ErbB signaling pathway (n = 7) ().
We then focused on two target genes, PPP1R12A and PPP1R12B, for subsequent analysis, as both are members of the same gene family and enriched in the pathway of focal adhesion, proteoglycans in cancer and regulation of the actin cytoskeleton. and show the base-complementary pairing between miR-141-3p and these two genes. We also utilized the mRNA-seq data from the TCGA-UCEC cohort and validated the downregulation of both PPP1R12A and PPP1R12B in EC tissues at the mRNA level (, ). The AUC of ROC curves showed these two genes both had a favorable capacity to distinguish EC (, ). We then used starBase v3.0 to confirm the negative correlation between miR-141-3p and these two targets (, ). We also verified the lower expression of PPP1R12A () and PPP1R12B () in EC tissues than in non-cancerous endometrial tissues at the protein level using immunohistochemical data from the THPA database.
Figure 6. Relationship between miR-141-3p and PPP1R12A in endometrial carcinomam (EC). (a) Complementary base sequences of miR-141-3p and PPP1R12A. (b, c) Scatter plot and receiver operating characteristic (ROC) curve of PPP1R12A in EC and non-EC tissues from TCGA-UCEC. (d) Negative correlation between miR-141-3p and PPP1R12A in EC on the basis of starBase v3.0. (e) The expression of PPP1R12A protein in normal glandular cells of uterus endometrium (Female aged 39, patient ID: 4569, antibody HPA071956, high staining). (f) The expression of PPP1R12A protein in EC cells (Female aged 58, patient ID: 2621, antibody HPA071956, staining not dected). (g) The expression of PPP1R12A protein in EC cells (Female aged 79, patient ID: 7339, antibody HPA071956, staining not dected)

Figure 7. Relationship between miR-141-3p and PPP1R12B in endometrial carcinomam (EC). (a) Complementary base sequences of miR-141-3p and PPP1R12B. (b, c) Scatter plot and receiver operating characteristic (ROC) curve of PPP1R12B in EC and non-EC tissues from TCGA-UCEC. (d) Negative correlation between miR-141-3p and PPP1R12B in EC on the basis of starBase v3.0. (e) The expression of PPP1R12B protein in normal glandular cells of uterus endometrium (Female aged 51, patient ID: 3491, antibody HPA024640, low staining). (f) The expression of PPP1R12B protein in EC cells (Female aged 70, patient ID: 2118, antibody HPA024640, staining not dected). (g) The expression of PPP1R12B protein in EC cells (Female aged 51, patient ID: 3481, antibody HPA024640, staining not dected)

Discussion
The novelty of the present study is its combined use of multiple detection approaches (RT-qPCR, miRNA-seq and microarrays) to identify the clinical role of miR-141-3p in EC and its use of integrated analysis to evaluate the significance of miR-141-3p in EC. Moreover, our multicenter samples were collected from Polan (n = 22), Japan (n = 48), China (n = 100) and USA (n = 568). The large sample size (n = 738) also allowed us to draw more convincing conclusions regarding the involvement of miR-141-3p regulation in the tumorigenesis and progression of EC. Collectively, we comprehensively reported the overexpression of miR-141-3p from the aspects of clinical value and molecular regulatory mechanisms.
Altered expression of miR-141-3p has been found in hepatocellular carcinoma [Citation39,Citation40], pancreatic cancer [Citation41], T cell acute lymphoblastic leukemia [Citation42] and non-small cell lung cancer [Citation43,Citation44]. Some recent studies have also reported the dysregulation of miR-141-3p in EC [Citation23–25]. In the present study, we confirmed the upregulated expression of miR-141-3p with miRNA microarrays using FFPE tissue samples from 49 patients with EC. A similar upregulation of miR-141-3p was also observed previously in four EC cell lines (HEC1A, HEC1B, HEC50, and Ishikawa) [Citation23]. Another two studies [Citation24,Citation25] also reported RT-qPCR findings for 33 and 34 cases of EC and demonstrated overexpression of miR-141-3p in EC. However, all these previous studies were small and used only a single method for determining upregulation. By contrast, we used data from in-house RT-qPCR, a TCGA-UCEC cohort and other high throughput datasets and were able to demonstrate a consistent increasing trend for miR-141-3p expression in EC tissues.
Our integrated analysis, which included 675 cases of EC tissues and 63 non-EC tissues, also supported these findings, with an overall SMD value of 1.737 (95%CI: 0.692–2.783, P= 0.001). The sROC curve also indicated that miR-141-3p may have a moderate ability to distinguish between patients with EC and noncancerous individuals (AUC = 0.87, 95%CI: 0.84–0.90). The SMD and AUC may reflect a more comprehensive and objective expression level of miR-141-3p globally in EC. Therefore, we were able to confirm the increasing trend of miR-141-3p expression in EC and to provide evidence that the upregulation of miR-141-3p expression may trigger the progression of carcinogenesis in EC.
Some studies have reported a relationship between miR-141-3p and the development of various malignant tumors. For example, miR-141-3p expression inhibited cell proliferation and affected the development of clear cell renal cell cancer [Citation45], breast cancer [Citation46] and prostate cancer [Citation47]. By contrast, the downregulation of miR-141-3p was also related to bone metastasis of prostate cancer. Another study identified miR-141-3p as a promoter of EC cell proliferation [Citation24]. Li et al. [Citation44] found that overall survival was poorer in non-small cell lung cancer (NSCLC) patients with low miR-141-3p expression than with high expression. Similarly, overall survival was longer in breast cancer patients with high expression of miR-141-3p than with low expression [Citation48]. Yang et al [Citation49]. reported that a high expression of miR-141-3p might promote the progression of bladder cancer and lead to poor prognosis. Overall, the dysregulation of miR-141-3p appears to play either a suppressing or a promoting role in the development of different tumors and in patient survival.
Our Kaplan-Meier survival analysis for the TCGA-UCEC cohort indicated that prognosis tended to be better in patients with EC with low miR-141-3p levels than with high levels, suggesting for the first time that miR-141-3p may have prognostic significance in EC. Our findings also indicate that miR-141-3p upregulation may be a prognostic risk factor for EC, in accordance a similar suggestion for bladder and breast cancer. However, unlike our results, miR-141-3p expression appears to be a protective factor in NSCLC; this might reflect a suppressing effect due to the targeting of different genes [Citation44]. Overall, however, the dysregulation of miR-141-3p appears to affect the prognosis for certain tumors, and it specifically seems to be a factor leading to the poor prognosis in EC. However, the current prognostic value of miR-141-3p expression is based on the level of tissue samples. If the prognostic value of miR-141-3p expression could be implemented under noninvasive conditions, such as detection from body fluid, its clinical value will be even greater. Such experiments are required to be carried out in the future.
We also evaluated the potential targets that were regulated by miR-141-3p and that might explain its tumor-promoting role. Previous studies have shown that miR-141-3p acts as an oncogene in cervical cancer by suppressing its FOXA2 target [Citation50]. Thus far, only one candidate, DAPK1, has been verified as a target gene of miR-141-3p in EC [Citation24]. We therefore extended the investigation of potential targets of miR-141-3p in EC by extracting predicted targets that showed low expression trends in EC tissues. We recognized that the expression of some targets would certainly be dysregulated only at the protein level and that this dysregulation would not appear as changes in the mRNA level. However, our analysis revealed that many target genes were concentrated in the classical cancer-related pathways, such as focal adhesion, the PI3K-Akt signaling pathway and cancer-related proteoglycans.
Ultimately, we focused on two target genes, PPP1R12A and PPP1R12B, as these are both members of the myosin phosphatase-targeting protein (MYPT) family. Previous studies have implicated the MYPT family in the progression of several diseases, such as pulmonary hypertension, Parkinson’s disease, cancer and vasospasm [Citation51–53]. PPP1R12A (protein phosphatase 1 regulatory subunit 12A) is also known as MYPT1 and is located on chromosome 12q15-q21.2. The human PPP1R12A gene is expressed in numerous tissues, but especially in tissues with abundant smooth muscles [Citation54]. PPP1R12A is mainly related to the RhoA/ROCK signaling pathway, and the activity of PPP1R12A can be suppressed by ROCK-induced phosphorylation [Citation55]. PPP1R12A expression is downregulated in ovarian cancer tissues, and this promotes cell proliferation [Citation56]. The copy number of PPP1R12A has also been reported as an independent predictor of overall survival and recurrence in stage III colorectal cancer patients [Citation57]. Two studies have reported a relationship between alteration of PPP1R12A expression and tumourigenesis [Citation58,Citation59]. Overall, PPP1R12A expression appears to be altered in certain tumors and affects tumorigenesis. However, the dysregulation of PPP1R12A in EC has not been reported until now.
PPP1R12B (protein phosphatase 1 regulatory subunit 12B, also known as MYPT2) is located on chromosome 1q32.1 [Citation60]. The protein encoded by PPP1R12B regulates the construction of muscle cells [Citation61]. Unfortunately, no previous study has reported a relationship between PPP1R12B and cancers, so a role for PPP1R12B in the development of tumors still needs to be identified. In the present study, we found that both PPP1R12A and PPP1R12B were downregulated target genes of miR-141-3p in EC. The downregulating trend was also verified at the protein level from the THPA database. Establishment of a clinical significance for PPP1R12A and PPP1R12B downregulation in EC still requires further studies with larger sample sizes, and the underlying mechanism awaits further exploration.
The present study has some limitations. One is that a relationship between the high expression level of miR-141-3p and several important clinical parameters was not confirmed. A larger sample size is needed to establish the clinicopathological implications of miR-141-3p expression. The targets PPP1R12A and PPP1R12B identified here also require further evaluation.
Conclusion
This study integrated the data from in-house RT-qPCR, GEO and TCGA cohorts to demonstrate that miR-141-3p expression is upregulated in EC tissue. This higher expression of miR-141-3p might serve as a biomarker of poor prognosis in patients with EC. By acting as an oncogenic miRNA, miR-141-3p appears to participate in the development of EC through its regulatory axes with target genes. However, more in vitro and in vivo experiments are needed to uncover the underlying molecular mechanisms of miR-141-3p.
Authors’ contributions
LJ Yang: project development and manuscript editing.
L Gao: data analysis and tables preparing.
YN Guo: study designing, experiments performing, and data analysis.
ZQ Liang: data analysis, figures and tables preparing, and manuscript writing.
DM Li: data collection and manuscript writing.
YL Tang: data collection and data interpretation.
YH Liu: data analysis and manuscript writing.
WJ Gao: data collection and data interpretation.
JJ Zeng: experiments performing and data interpretation.
L Shi: experiments performing and paper revising.
KL Wei: project development and study design.
G Chen: analysis supervising, project development and manuscript editing.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the author(s).
Ethics approval
The study proposal was ratified by Committee on Ethics of the First Affiliated Hospital of Guangxi Medical University.
Highlights
1. miR-141-3p is upregulated in endometrial carcinoma (EC) tissues.
2. Overexpression of miR-141-3p positively corelates with a poorer prognosis.
3. PPP1R12A and PPP1R12B, target genes of miR-141-3p, are downregulated in EC.
4. Overexpression of miR-141-3p may play an important part in the carcinogenesis of EC.
Acknowledgements
The authors would like to thank The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), Stata software version 15.1 (TX, USA), Database for Annotation, Visualisation and Integrated Discovery (DAVID), SPSS version 25.0 software (IBM Corp., Armonk, NY, USA), The Human Protein Atlas (THPA), ImageGP online platform and R software.
Data availability statement:
Not applicable.
Additional information
Funding
References
- Tabuchi Y, Hirohashi Y, Hashimoto S, et al. Clonal analysis revealed functional heterogeneity in cancer stem-like cell phenotypes in uterine endometrioid adenocarcinoma. Exp Mol Pathol. 2019;106:78–88. . PubMed PMID: 30503404.
- Wang Y, Liu D, Jin X, et al. Genome-wide characterization of aberrant DNA methylation patterns and the potential clinical implications in patients with endometrial cancer. Pathol Res Pract. 2019;215(1):137–143. . PubMed PMID: 30449607.
- Wang F, Wang B, Long J, et al. Identification of candidate target genes for endometrial cancer, such as ANO1, using weighted gene co-expression network analysis. Exp Ther Med. 2019;17(1):298–306. . PubMed PMID: 30651795.
- Silverberg SG, Gilks CB. The most important discoveries of the past 50 years in gynaecological pathology. Histopathology. 2020;76(1):6–10. . PubMed PMID: 31846536.
- Siegel RL, Miller KD, Jemal A, et al. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. Epub 2020/01/09. PubMed PMID: 31912902.
- Amant F, Mirza MR, Koskas M, et al. Cancer of the corpus uteri. Int J Gynaecol Obstet. 2018;143(Suppl 2):37–50. . PubMed PMID: 30306580.
- Gao L, Xie Z-C, Pang J-S, et al. A novel alternative splicing-based prediction model for uteri corpus endometrial carcinoma. Aging (Albany NY). 2019;11(1):263–283. . PubMed PMID: 30640723.
- Lu H, Ju -D-D, Yang G-D, et al. Targeting cancer stem cell signature gene SMOC-2 overcomes chemoresistance and inhibits cell proliferation of endometrial carcinoma. EBioMedicine. 2019;40:276–289. . PubMed PMID: 30594556.
- Wang Y, Xu M, Yang Q. A six-microRNA signature predicts survival of patients with uterine corpus endometrial carcinoma. Curr Probl Cancer. 2019;43(2):167–176. . PubMed PMID: 29567372.
- Yang S, Wang H, Li D, et al. Role of endometrial autophagy in physiological and pathophysiological processes. J Cancer. 2019;10(15):3459–3471. . PubMed PMID: 31293650.
- Brooks RA, Fleming GF, Lastra RR, et al. Current recommendations and recent progress in endometrial cancer. CA Cancer J Clin. 2019;69(4):258–279. . PubMed PMID: 31074865.
- Nie D, Yang E, Li Z, et al. Pretreatment thrombocytosis predict poor prognosis in patients with endometrial carcinoma: a systematic review and meta-analysis. BMC Cancer. 2019;19(1):73. . PubMed PMID: 30646853.
- Williams AT, Ganesan R. Role of the pathologist in assessing response to treatment of ovarian and endometrial cancers. Histopathology. 2020;76(1). DOI:10.1111/his.13994. PubMed PMID: 31846531.
- Moran Y, Agron M, Praher D, et al. The evolutionary origin of plant and animal microRNAs. Nat Ecol Evol. 2017;1(3):27. . PubMed PMID: 28529980.
- Du J, Zhang F, Zhang L, et al. MicroRNA-103 regulates the progression in endometrial carcinoma through ZO-1. Int J Immunopathol Pharmacol. 2019;33:2058738419872621. . PubMed PMID: 31456452.
- Bao W, Zhang Y, Li S, et al. miR‑107‑5p promotes tumor proliferation and invasion by targeting estrogen receptor‑α in endometrial carcinoma. Oncol Rep. 2019;41(3):1575–1585. . PubMed PMID: 30569100.
- Zheng X, Xu K, Zhu L, et al. MiR-486-5p act as a biomarker in endometrial carcinoma: promotes cell proliferation, migration, invasion by targeting MARK1. Onco Targets Ther. 2020;13:4843–4853. . PubMed PMID: 32547110.
- Wu Z, Tang H, Xiong Q, et al. Prognostic role of microRNA-205 in human gynecological cancer: a meta-analysis of fourteen studies. DNA Cell Biol. 2020;39(5):875–889. . PubMed PMID: 32354230.
- Wang C, Li Q, He Y, et al. MicroRNA‑21‑5p promotes epithelial to mesenchymal transition by targeting SRY‑box 17 in endometrial cancer. Oncol Rep. 2020;43(6):1897–1905. . PubMed PMID: 32236579.
- Zhou Z, Xu Y-P, Wang L-J, et al. miR-940 potentially promotes proliferation and metastasis of endometrial carcinoma through regulation of MRVI1. Biosci Rep. 2019;39(6). DOI:10.1042/BSR20190077. PubMed PMID: 31085718.
- Gao Y, Feng B, Han S, et al. The roles of MicroRNA-141 in human cancers:</L> from diagnosis to treatment. Cell Physiol Biochem. 2016;38(2):427–448. . PubMed PMID: 26828359.
- Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5(3):115–119. . PubMed PMID: 19182522.
- Jayaraman M, Radhakrishnan R, Mathews CA, et al. Identification of novel diagnostic and prognostic miRNA signatures in endometrial cancer. Genes Cancer. 2017;8(5–6):566–576. . PubMed PMID: 28740575.
- Cui Z, An X, Li J, et al. LncRNA MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression and inhibits endometrial carcinoma cells proliferation. Biomed Pharmacother. 2018;104:223–228. . PubMed PMID: 29775889.
- Snowdon J, Zhang X, Childs T, et al. The MicroRNA-200 family is upregulated in endometrial carcinoma. PLoS ONE. 2011;6(8):e22828.
- Huang H-Q, Chen G, Xiong -D-D, et al. Down-regulation of microRNA-125b-2-3p is a risk factor for a poor prognosis in hepatocellular carcinoma. Bioengineered. 2021;12(1):1627–1641. . PubMed PMID: 33949293.
- Chen S-W, Lu H-P, Chen G, et al. Downregulation of miRNA-126-3p is associated with progression of and poor prognosis for lung squamous cell carcinoma. FEBS Open Bio. 2020;10(8):1624–1641. . PubMed PMID: 32598517.
- Wang -S-S, Huang Z-G, Wu H-Y, et al. Downregulation of miR-193a-3p is involved in the pathogenesis of hepatocellular carcinoma by targeting CCND1. PeerJ. 2020;8:e8409. . PubMed PMID: 32095323.
- Liang C-Y, Li Z-Y, Gan T-Q, et al. Downregulation of hsa-microRNA-204-5p and identification of its potential regulatory network in non-small cell lung cancer: RT-qPCR, bioinformatic- and meta-analyses. Respir Res. 2020;21(1):60. . PubMed PMID: 32102656.
- Chen Y-J, Guo Y-N, Shi K, et al. Down-regulation of microRNA-144-3p and its clinical value in non-small cell lung cancer: a comprehensive analysis based on microarray, miRNA-sequencing, and quantitative real-time PCR data. Respir Res. 2019;20(1):48. . PubMed PMID: 30832674.
- Gao L, Yan S-B, Yang J, et al. MiR-182-5p and its target HOXA9 in non-small cell lung cancer: a clinical and in-silico exploration with the combination of RT-qPCR, miRNA-seq and miRNA-chip. BMC Med Genomics. 2020;13(1):3. . PubMed PMID: 31906958.
- Huang DW, Sherman BT, Lempicki RA, et al. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1). DOI:10.1093/nar/gkn923. PubMed PMID: 19033363.
- Huang DW, Sherman BT, Lempicki RA, et al. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. . PubMed PMID: 19131956.
- Vejnar CE, Blum M, Zdobnov EM, et al. miRmap web: comprehensive microRNA target prediction online. Nucleic Acids Res. 2013;41(W1):W165–W8. . PubMed PMID: 23716633.
- Vejnar CE, Zdobnov EM. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012;40(22):11673–11683. . PubMed PMID: 23034802.
- Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507. . PubMed PMID: 28818916.
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. . PubMed PMID: 25613900.
- Thul PJ, Åkesson L, Wiking M, et al. A subcellular map of the human proteome. Science. 2017;356(6340). DOI:10.1126/science.aal3321. PubMed PMID: 28495876.
- Liu C-Z, Ye Z-H, Ma J, et al. A qRT-PCR and gene functional enrichment study focused on downregulation of miR-141-3p in hepatocellular carcinoma and its clinicopathological significance. Technol Cancer Res Treat. 2017;16(6):835–849. . PubMed PMID: 28436261.
- Hou X, Yang L, Jiang X, et al. Role of microRNA-141-3p in the progression and metastasis of hepatocellular carcinoma cell. Int J Biol Macromol. 2019;128:331–339. . PubMed PMID: 30695725.
- Sun J, Zhang Y. LncRNA XIST enhanced TGF-β2 expression by targeting miR-141-3p to promote pancreatic cancer cells invasion. Biosci Rep. 2019;39(7). DOI:10.1042/BSR20190332. PubMed PMID: 31213574.
- Zhou R, Mo W, Wang S, et al. miR-141-3p and TRAF5 network contributes to the progression of T-Cell acute lymphoblastic leukemia. Cell Transplant. 2019;28(1_suppl):59S–65S.
- Liu X, Wang M, Cui Y. LncRNA TP73-AS1 interacted with miR-141-3p to promote the proliferation of non-small cell lung cancer. Arch Med Sci. 2019;15(6):1547–1554. . PubMed PMID: 31749884.
- Li W, Cui Y, Wang D, et al. MiR-141-3p functions as a tumor suppressor through directly targeting ZFR in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;509(3):647–656. . PubMed PMID: 30611568.
- Liep J, Kilic E, Meyer HA, et al. Cooperative effect of miR-141-3p and miR-145-5p in the regulation of targets in clear cell renal cell carcinoma. PloS One. 2016;11(6):e0157801. . PubMed PMID: 27336447.
- Zhang Y, Li J, Jia S, et al. Down-regulation of lncRNA-ATB inhibits epithelial-mesenchymal transition of breast cancer cells by increasing miR-141-3p expression. Biochem Cell Biol. 2019;97(2):193–200. . PubMed PMID: 30352165.
- Li J-Z, Li J, Wang H-Q, et al. MiR-141-3p promotes prostate cancer cell proliferation through inhibiting kruppel-like factor-9 expression. Biochem Biophys Res Commun. 2017;482(4):1381–1386. . PubMed PMID: 27956179.
- Sun S, Ma J, Xie P, et al. Hypoxia-responsive miR-141-3p is involved in the progression of breast cancer via mediating the HMGB1/HIF-1α signaling pathway. J Gene Med. 2020;(10):e3230. doi:10.1002/jgm.3230. PubMed PMID: 32436353.
- Yang X, Wang P. MiR-188-5p and MiR-141-3p influence prognosis of bladder cancer and promote bladder cancer synergistically. Pathol Res Pract. 2019;215(11):152598. . PubMed PMID: 31562019.
- Li J-H, Zhang Z, Du M-Z, et al. microRNA-141-3p fosters the growth, invasion, and tumorigenesis of cervical cancer cells by targeting FOXA2. Arch Biochem Biophys. 2018;657:23–30. . PubMed PMID: 30222949.
- Guan R, Xu X, Chen M, et al. Advances in the studies of roles of rho/rho-kinase in diseases and the development of its inhibitors. Eur J Med Chem. 2013;70:613–622. . PubMed PMID: 24211637.
- Scotto-Lavino E, Garcia-Diaz M, Du G, et al. Basis for the isoform-specific interaction of myosin phosphatase subunits protein phosphatase 1c beta and myosin phosphatase targeting subunit 1. J Biol Chem. 2010;285(9):6419–6424. . PubMed PMID: 20042605.
- Grassie ME, Moffat LD, Walsh MP, et al. The myosin phosphatase targeting protein (MYPT) family: a regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1δ. Arch Biochem Biophys. 2011;510(2):147–159. . PubMed PMID: 21291858.
- Takahashi N, Ito M, Tanaka J, et al. Localization of the gene coding for myosin phosphatase, target subunit 1 (MYPT1) to human chromosome 12q15-q21. Genomics. 1997;44(1):150–152. . PubMed PMID: 9286714.
- Khasnis M, Nakatomi A, Gumpper K, et al. Reconstituted human myosin light chain phosphatase reveals distinct roles of two inhibitory phosphorylation sites of the regulatory subunit, MYPT1. Biochemistry. 2014;53(16):2701–2709. . PubMed PMID: 24712327.
- Munoz-Galvan S, Felipe-Abrio B, Verdugo-Sivianes EM, et al. Downregulation of MYPT1 increases tumor resistance in ovarian cancer by targeting the hippo pathway and increasing the stemness. Mol Cancer. 2020;19(1):7. . Epub 2020/01/14. PubMed PMID: 31926547; PubMed Central PMCID: PMCPMC6954568.
- Zhang C, Li A, Li H, et al. PPP1R12A copy number is associated with clinical outcomes of stage III CRC receiving oxaliplatin-based chemotherapy. Mediators Inflamm. 2015;2015:417184. . PubMed PMID: 26113782.
- Chirino YI, García-Cuellar CM, García-García C, et al. Airborne particulate matter in vitro exposure induces cytoskeleton remodeling through activation of the ROCK-MYPT1-MLC pathway in A549 epithelial lung cells. Toxicol Lett. 2017;272:29–37. . PubMed PMID: 28279687.
- Lin Z-Y, Chen G, Zhang Y-Q, et al. MicroRNA-30d promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer. Mol Cancer. 2017;16(1):48. . PubMed PMID: 28241827.
- Fujioka M, Takahashi N, Odai H, et al. A new isoform of human myosin phosphatase targeting/regulatory subunit (MYPT2): cDNA cloning, tissue expression, and chromosomal mapping. Genomics. 1998;49(1):59–68. . PubMed PMID: 9570949.
- Ito M, Nakano T, Erdodi F, et al. Myosin phosphatase: structure, regulation and function. Mol Cell Biochem. 2004;259(1/2):197–209. . PubMed PMID: 15124925.